Abstract
Mass spectrometry-based metabolomics relies on MS2 data for structural characterization of metabolites. To obtain the high-quality MS2 data necessary to support metabolite identifications, ions of interest must be purely isolated for fragmentation. Here we show that metabolomic MS2 data are frequently characterized by contaminating ions that prevent structural identification. Although using narrow-isolation windows can minimize contaminating MS2 fragments, even narrow windows are not always selective enough and they can complicate data analysis by removing isotopic patterns from MS2 spectra. Moreover, narrow windows can significantly reduce sensitivity. In this work we introduce a novel, two-part approach for performing metabolomic identifications that addresses these issues. First, we collect MS2 scans with less stringent isolation settings to obtain improved sensitivity at the expense of specificity. Then, by evaluating MS2 fragment intensities as a function of retention time and precursor mass targeted for MS2 analysis, we obtain deconvolved MS2 spectra that are consistent with pure standards and can therefore be used for metabolite identification. The value of our approach is highlighted with metabolic extracts from brain, liver, astrocytes, as well as nerve tissue and performance is evaluated by using pure metabolite standards in combination with simulations based on raw MS2 data from the METLIN metabolite database. An R package implementing the algorithms used in our workflow is available on our laboratory website (http://pattilab.wustl.edu/decoms2.php).
Untargeted metabolomics aims to simultaneously screen thousands of small molecules for alterations in concentration between biological samples.1 To maximize the breadth of ions covered with high-mass accuracy, typical untargeted metabolomic workflows use quadrupole time-of-flight (QTOF) or Orbitrap mass spectrometers.2-4 These workflows involve multiple steps (Supplementary Figure 1). First, the abundance of intact metabolites is quantified by collecting measurements in MS1 mode only. Next, the accurate mass of the peaks of interest, usually those that are dysregulated between sample groups, are searched in metabolite databases such as HMDB and METLIN to obtain putative identifications.5-7 Finally, all putative identifications must be structurally confirmed by performing a targeted MS2 analysis and matching these fragmentation data to those of a research standard.1,8,9
With advances in profiling software and metabolite databases, the application of untargeted metabolomics to generate putative lists of compounds whose concentrations are altered between biological samples has become relatively routine.10 Confirming putative assignments with structural data, however, remains a challenge for many laboratories and presents a critical barrier that has limited the widespread use of the metabolomic platform. Unlike MS2 data for peptides, MS2 data for metabolites are largely unpredictable.11 Consequently, structural assignments in metabolomics involve empirically matching both the intensities and the mass-to-charge values of each fragment in research MS2 spectra to the intensities and mass-to-charge values of each fragment in the MS2 spectra of standard compounds. It is important to emphasize that, to confirm a structural assignment, the intensity and mass-to-charge of all fragments detected must match the standard. Isobaric metabolites with very distinct structures can generate MS2 spectra that differ only by a single fragment, or in some cases, only by the intensities of their fragments (Supplementary Figure 2).12 As a further complication, the total number of naturally occurring metabolites is currently unknown and metabolite databases are largely incomplete.13,14 This means that a metabolomic investigator cannot structurally identify an endogenous small molecule in a biological system by simply finding the MS2 pattern in metabolite databases that provides the closest match to the research MS2 pattern. Rather, variations in the intensities of MS2 fragments or the presence/absence of a fragment are important for structural characterization and may be indicative of a novel metabolite that has not been previously characterized.
The major challenge in matching experimental MS2 spectra for metabolite identification is acquiring high-quality data. While high-quality MS2 data for research standards can generally be acquired easily, the molecular complexity of biological samples in addition to the low concentration of many interesting compounds frequently limits the data quality of MS2 spectra from research specimens. Low-quality MS2 spectra have unreliable fragment intensities and missing/additional fragments due to: (i) instrument noise (when the compound of interest is of low concentration), and/or (ii) from other compounds in the biological sample being isolated in the collision cell (when the sample has high molecular complexity). These MS2 artifacts are a major obstacle in metabolomic data interpretation, commonly prevent compounds from being identified, and limit publication of metabolomic results. We present a workflow to overcome these limitations and increase the number of ionized metabolites that can be identified by using MS2.
Experimental
Material
Cell culture media and reagents, including Dulbecco's Modified Eagle's Medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin, and trypsin-EDTA, as well as HPLC grade solvents (acetone, acetonitrile, and water) were purchased from Sigma-Aldrich (St. Louis, MO). Formic acid was purchased from Fluka (Sigma-Aldrich, St. Louis, MO).
Tissues
Liver, brain, and peripheral nerve tissues were dissected from black 6 mice. The tissues were washed with phosphate-buffered saline (PBS), frozen in liquid nitrogen (LN2), and then stored at −80 °C before extraction. Brain tissues were cut into 68 mg sections and liver tissues cut into 83 mg sections for extraction. The tissues were extracted according to the procedure described previously and as described below.15
Cell culture
Immortalized astrocytes were obtained from American Type Culture Collection (CRL-2005) and plated in DMEM containing 10% FBS, penicillin, and streptomycin. Cells were incubated at 37 °C with 5% CO2 and 95% humidity. Astrocytes were grown to confluence, and then detached by incubating in trypsin-EDTA (0.25%) for 5 min. The suspended astrocytes were spun down at 1000 g for 10 min. The cell pellet was collected after removing the supernatant and washing with PBS. The cell pellet was immediately extracted using the procedure below or frozen in LN2, and then stored at −80 °C.
Sample extraction
Tissues and cells were incubated in 600 μL of cold (−20 °C) acetone and vortexted prior to being incubated in LN2. The samples were thawed at room temperature and incubated in LN2 two more times prior to a 10 min sonication. After 1 hr at −20 °C, the samples were centrifuged at 13,000 rpm for 15 min and the resulting supernatant was stored at −20 °C. The precipitate was then mixed with 400 μL of methanol/water/formic acid in the ratio of 86.5/12.5/1.0 and sonicated prior to a 1-h incubation at −20 °C. After centrifugation, the supernatant was collected and transferred to that which was collected after acetone extraction. The solution was then dried with a vacuum concentrator at room temperature and redissolved in 100 μL of 95% acetonitrile/ 5% water for liquid chromatography/mass spectrometry (LC/MS) analysis.
Collection of LC/MS scans from Brain, Liver, and Astrocyte Metabolic Extracts
Analyses were carried out with an Agilent 1100 series HPLC system coupled to an Agilent 6520 QTOF mass spectrometer equipped with an electrospray ionization source. A reversed-phase (RP) column (Agilent ZORBAX C18-column, 5 μm, 150×0.5 mm) was used to separate the mixture. The solvent system for liquid chromatography was water with 0.1% formic acid (mobile phase A) and acetonitrile with 0.1% formic acid (mobile phase B). The solvent gradient was 2% B from 0-10 min, 10% B from 10-15 min, 20% B from 15-30 min, 95% B from 30-45, min and ended at 98% B. The flow rate was 20 μL/min. Sample was injected in 5-μL aliquots and ran in positive-ion mode. The spectra were collected with an acquisition rate of 1.01 spectra/sec with a mass range of 25-1500 mass units. Other scan rates were also tested and provided comparable results.
Estimating the number of contaminated scans
To estimate the portion of contaminated scans, we first detected features in LC/MS data using the centWave algorithm implemented in the XCMS R package.16 A feature is defined as an ion with a unique mass-to-charge ratio and a unique retention time. The peakwidth parameter was set to 10-120s and the ppm was set to 30. We then estimated contamination by considering all possible scans for which multiple features were within half of the MS2 isolation window. Isotopes were excluded from the analysis. The analysis was performed at MS2 isolation window widths of 1 and 9 m/z.
Estimating transmission efficiencies for different isolation windows
The transmission percentages of the quadrupole ion filter were assessed across the m/z range of 118 to 1222. Agilent ESI-L low concentration tuning mix (G1969-85000) was diluted 1/10 with acetonitrile and 5 μL injected with a flow of 10 μL/min and 0.1% v/v formic acid in 1:1 acetonitrile:water. Product ion scans with accelerating voltages of 0 were performed for each of the ions in the tune mix (m/z 118.086, 322.048, 622.028, 922.009, 1221.991). Following each set of product ion scans, an MS1 scan was performed to complete the cycle. This experiment was repeated with MS2 isolation window widths of 1 and 9 m/z. For each MS2 isolation window and mass, the intensity of the ion of interest from MS1 scans was compared to the intensity of the ion of interest from 0 V MS2 spectra to determine the transmission percentage.
MS2 isolation windows
Data related to this study were acquired from an Agilent 6520 QTOF, which is limited with respect to choice in MS2 isolation window width. Default settings on this instrument enable the use of 1.3, 4, and 9 Da MS2 isolation window widths only. It is important to note that the shifting-window deconvolution approach we describe herein could be used with any MS2 isolation window width, including broader isolation windows as discussed below. Notably, the shifting-window deconvolution strategy is generally not compatible with MSE data given that there is no isolation window to shift. It may still be possible to deconvolve MSE data with our approach, however, on the basis of retention time or sample-class correlation. Deconvolution of mass spectra on the basis of retention time was originally described for gas chromatography/mass spectrometry, but in our experiences it is much less effective compared to the shifting-window strategy for deconvolving LC/MS data.17,18
Estimating the number of features with sufficient intensity for MS2 analysis
QTOF product ion scans have decreased sensitivity compared to full MS1 scans due to: (i) loss of ions during quadrupole isolation (q) and (ii) the distribution of charge over multiple MS2 fragments (f). To estimate the lower limit of precursor ion intensities for which reliable MS2 data can be acquired with each isolation window, we estimated how both kinds of losses of intensity rescale the precursor ion abundance in the MS1 profiling scans. q is the transmission efficiency, estimated as the average of the transmission efficiencies of tune mix described above. f was estimated based on the distribution of fragment intensities in 20 V collision energy MS2 scans relative to precursor intensities in 0 V collision energy MS2 scans in raw METLIN data. We assumed that clean data necessary to identify a metabolite should contain all fragments with relative intensities above 10% in 20 V collision energy in the METLIN database. Of these ions, we defined f as the 5th percentile of the distribution given by relative fragment intensities in 20 V scans scaled by the precursor intensities in 0 V scans. Then, to evaluate the portion of detected features that could produce reliable MS2, we counted the amount of MS1 precursor ions intensities, i, that satisfied i * f * q ≥ m, where m is minimal signal threshold based on a 10:1 signal to noise ratio. We calculated the number of features above this threshold for brain, liver, and astrocyte samples based on the maximal feature intensities reported by the centWave peak finding described above. Of these ions, we defined f as the 5th percentile of the distribution given by relative fragment intensities in 20 V scans scaled by the precursor intensities at 0 V scans. It should be noted that the additional data processing associated with using wide MS2 isolation window widths may not be necessary for ions that are relatively intense, unless the researcher wants to retain the isotopic pattern in the MS2 spectrum.
Collection of MS2 scans for amino acids
All spectra were collected on the same instrument as LC/MS scans with an ESI source set to 120 eV ionization energy. MS2 scans of amino acid mixtures were collected using 0 and 20 V collision energy on a C18 RP column (Agilent ZORBAX C18-column, 5 μm, 150×0.5 mm). For each targeted amino acid, the isolation window was shifted about the mass of the precursor ion. On the Agilent 6520 QTOF used to generate this data, the offsets were obtained by centering the 9 Da isolation window at precursor mass −1 and −8 m/z offsets.
Collection of MS2 scans for astrocyte features
We targeted 50 randomly selected features for MS2 with 9 m/z shifting windows and 1 m/z fixed isolation windows in the astrocyte extracts described above. The shifting windows were positioned with the same offsets as for amino acids, at precursor mass-to-charge of interest −1 and −8 m/z. Both 0 and 20 V collision energy scans were collected since the quadrupole behaves differently in different mass ranges.
Generative model for the observed spectra
Because we collect MS2 spectra such that the contributions of precursor ions to the observed spectra are independent, we can represent the observed mass intensities as a weighted sum of all precursors with fragments in the spectrum. Letting oij represent the observed intensity of mass i in scan j, ski represents the relative intensity of mass i in the pure spectrum for precursor k, and mjk represents the total contribution of source k to scan j. The observations for mass j in scan i are written oij = ∑k mjk ski. Expressing this simultaneously for all observations in matrix notation, the expression is O=MS, where the rows of O are the observed MS2 spectra, the rows of M are the relative contributions of each precursor to each scan, and the rows of S are the unknown source spectra.
Estimating S
We obtain the entries in S by using two models. We begin by using cubic splines to estimate the entries of M with spectra collected at 0 V collision energy, positioned at the same precursor locations as the spectra collected at higher collision energies. This produces a set of third degree polynomials between consecutive precursor observations, such that at each observation the adjacent polynomials have the same first and second derivatives.19 The entries in M are made up of the appropriate polynomial evaluated at the time of each non-0 V scan in O. To then estimate the entries in S, we minimize the non-negative least squares objective: min (O – MS)T (O – MS), such that ∀ si,j ≥ 0. The cubic spline is fit using R package stats, and the non-negative least squares solution is obtained using the R package nnls.
Results and Discussion
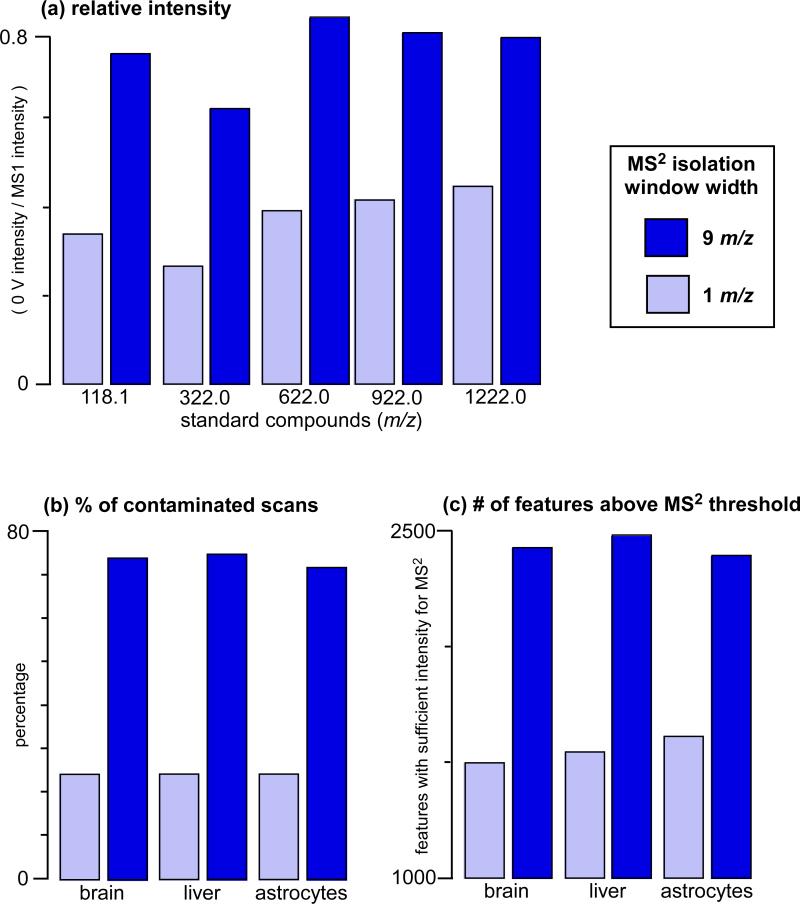
Untargeted metabolomic platforms often rely on quadrupoles to isolate ions of interest for fragmentation. When using a quadrupole, there is a tradeoff between sensitivity and specificity of ion isolation. For example, with the Agilent QTOF instrumentation, it is possible to improve sensitivity by a factor of ~2 if a broad 9 Da MS2 window is used for ion isolation instead of the narrow 1 Da isolation window (Figure 1a). Using broader isolation windows, however, introduces contaminating fragments from coeluting metabolites of similar mass. Based on metabolites extracted from astrocytes, brain, and liver tissue, we estimate that using a 9 Da MS2 isolation window results in more than 75% of MS2 scans having fragments from more than one precursor ion (Figure 1b). Although MS2 spectra characterized by fragments from more than one precursor ion is not a problem specific to metabolites, such MS2 patterns are particularly challenging in untargeted metabolomics because they cannot be interpreted de novo and therefore prevent structural identification of the fragmented compounds. The loss in specificity of 9 Da MS2 isolation windows is necessary, however, because only ~1500 metabolomic features are present at intensities sufficient to obtain high-quality MS2 data in these research specimens when using an isolation window of 1 Da.20 In contrast, increasing the isolation window to 9 Da increases the number of features with sufficient intensity to obtain high-quality MS2 data to ~2400 (Figure 1c, also see Experimental). Thus, the decreased specificity increases the number of features that can be targeted for identification by nearly 75% in these tissues based on signal intensity, but the caveat is that a major portion of the MS2 data from these ions are contaminated by coeluting species.
Figure 1. Comparison of MS2 isolation window widths.
(a) Effect of MS2 isolation window width on signal intensity. Ions measured are small molecule standards from Agilent's ESI-TOF tuning mix solution. (b) With an MS2 isolation window of 9 m/z, more than 75% of acquired scans have peaks that cannot be purely isolated in the collision cell for MS2 analysis and therefore their MS2 spectra may contain artifacts that hinder metabolite identification. With an MS2 isolation window of 1 m/z, the proportion of acquired scans with peaks that cannot be purely isolated in the collision cell is reduced to ~20%. (c) By using an MS2 isolation window of 9 m/z instead of 1 m/z, sensitivity is improved such that ~900 more metabolite peaks (i.e., features) can be structurally characterized by MS2 analysis.
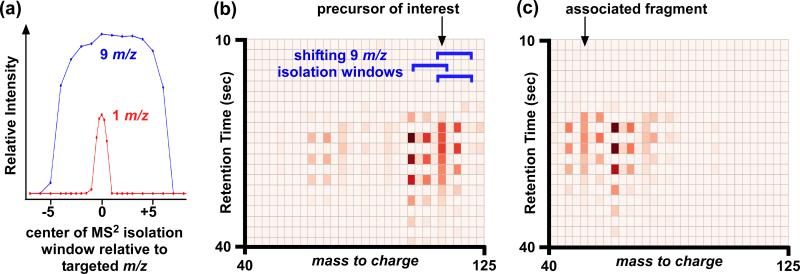
Here we present a strategy that enables the sensitivity gain of using 9 Da MS2 isolation windows without losing the specificity that is needed for compound identification. Additionally our strategy prevents isotopic patterns from being lost in MS2 spectra, another disadvantage of using a 1 Da isolation window width. Our approach relies on the experimental deconvolution of MS2 data collected by using broad isolation windows. To accomplish the deconvolution, we rely on two sources of variability in the contributions of precursor ions to individual scans. The first source is the natural variability in the chromatographic retention of metabolites with similar mass (Figure 2). The second source of variability is introduced experimentally by shifting the mass-to-charge values targeted for fragmentation (Figure 2b-c). The latter ensures that sufficient variation between the ion of interest and the contaminants is present. Without the shifting isolation window step, fragments from precursor ions with highly correlated retention profiles would appear to originate from the same precursor and could not be deconvolved.
Figure 2. Basis of deconvolution approach.
(a) MS2 isolation window shapes for Agilent 6520 QTOF. Using a larger MS2 isolation window increases the likelihood of isolating more than 1 precursor in the collision cell for MS2 analysis, but also improves the relative intensity of ions transmitted. Representative (b) MS2 at 0 V and (c) MS2 at 20 V data used for deconvolution over a 30 second chromatographic window. Each box in the plots represents a single scan. Peak intensity is denoted by color, with bright red representing the most intense peaks. The blue brackets indicate the position of the 9 Da MS2 isolation window used. Ion intensities that are high and low in alternating scans (i.e., bright red followed by light red) are ions that are included and excluded respectively with shifts in the position of the MS2 isolation window. Our deconvolution approach is based on identifying matching patterns in the MS2 plots at 0 and 20 V. An example of a matched precursor and associated fragment is shown with arrows.
After collecting data with different precursor ions contributing in different proportions to each MS2 scan, we infer the source fragmentation spectra by fitting two models. The first model estimates the amount of each precursor ion entering the collision cell, while the second model uses those estimates to determine the portion of each fragment's intensity coming from each precursor (see Experimental). We call this method decoMS2 and have made an R package implementing these algorithms available on our website (pattilab.wustl.edu/deconMS2.php).
Application of decoMS2 to standards and biological samples
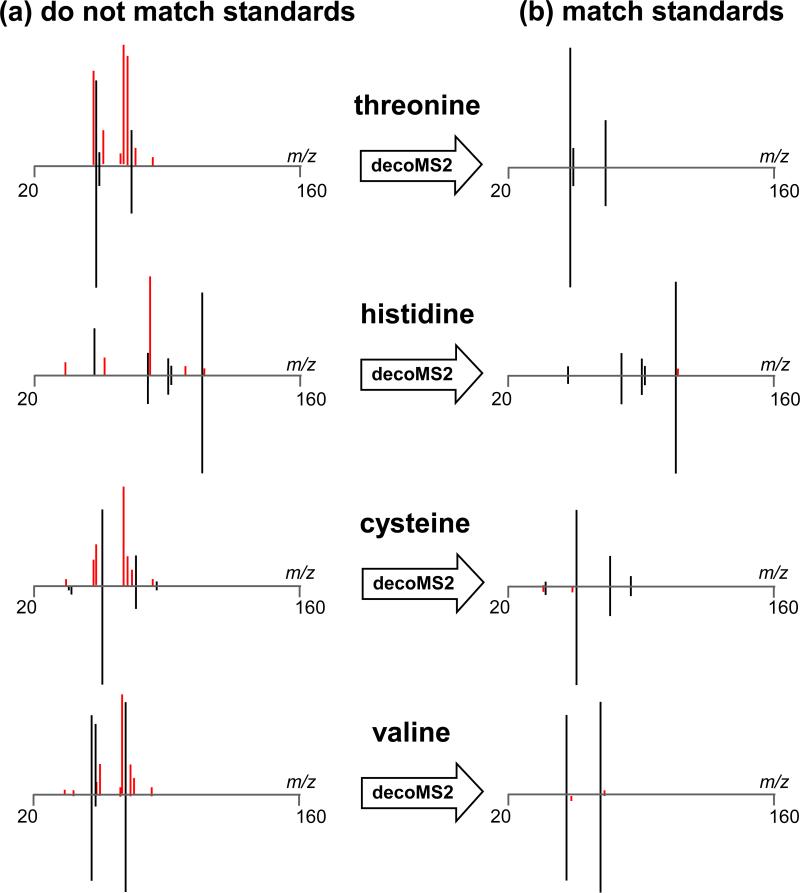
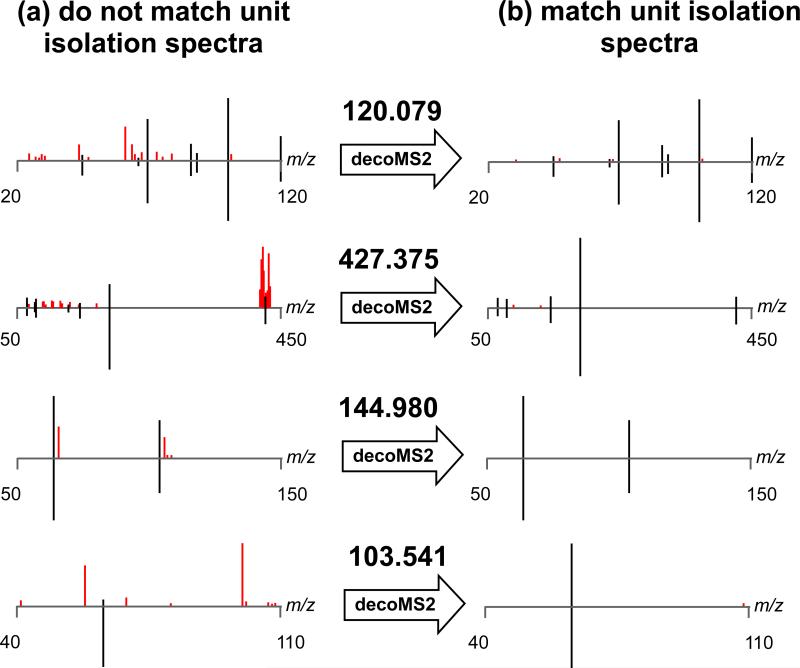
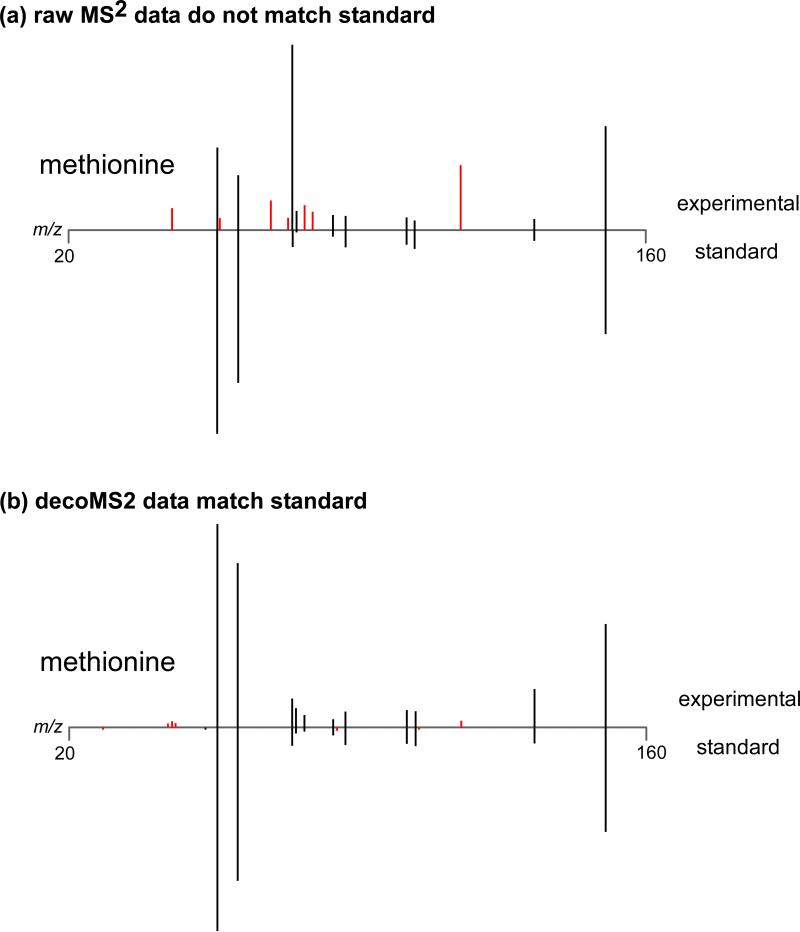
To illustrate the performance of our metabolomic deconvolution workflow in a controlled way, we mixed 10 amino acids in three sets of mixtures that produced cross-contaminated MS2 spectra (Figure 3a). We emulated a worst-case scenario of overlapping retention time profiles by collecting MS2 fragmentation spectra of the hydrophilic metabolites on a hydrophobic C18 column. Then, using variability from shifting isolation windows and applying our deconvolution algorithm, we were able to infer the true fragmentation profiles of the mixed amino acids (Figure 3b). These results confirm that our workflow increases sensitivity without the limitation of MS2 artifacts that hinder structural identification of metabolites. To validate that our approach is effective for analyzing the metabolic extracts derived from biological samples, we compared pure MS2 spectra acquired experimentally from astrocyte cell cultures by using a 1 Da isolation window to the MS2 spectra that we obtained from astrocyte cell cultures by deconvolving fragmentation data acquired from the same ions by using a 9 Da isolation window (Figure 4). As shown for the identified example of methionine (Figure 5), the fragmentation patterns obtained from each approach were in concordance.
Figure 3. decoMS2 applied to amino acid standards.
Amino acid standards were mixed and analyzed by LC/MS2, using a 9 Da MS2 isolation window. Because the amino acid standards were inadequately separated, each of their MS2 spectra were contaminated by additional precursors in the collision cell. (a) The experimental MS2 spectra for 4 representative amino acids are shown on the top of each plot. The standard MS2 spectra for each of these amino acids as obtained from pure model compounds is shown on the bottom. Fragments that match in the 2 spectra are colored black, while fragments that do not match are colored red. (b) After the application of decoMS2, the top experimental MS2 spectra of each amino acid are highly consistent with the MS2 spectra from their respective amino acid standards.
Figure 4. decoMS2 applied to a biological sample.
To evaluate the performance of decoMS2 on a large set of metabolites in a biological sample, we identified ions in the metabolic extract of astrocytes that were contaminated when isolated with a 9 Da MS2 isolation window but that were not contaminated when isolated using a 1 Da MS2 isolation window. (a) The experimental MS2 spectra for 4 representative accurate masses as obtained using a 9 Da MS2 isolation window are shown on the top of each plot. The experimental MS2 spectra for the same 4 compounds as obtained using a 1 Da MS2 isolation window is shown on the bottom of each plot. Fragments that match in the 2 spectra are colored black, while fragments that do not match are colored red. (b) After applying decoMS2 to the top spectra using the shifting window approach, the MS2 spectra obtained with a 9 Da MS2 isolation window are in concordance with those MS2 spectra obtained with a 1 Da MS2 isolation window.
Figure 5. decoMS2 applied to methionine from astrocytes.
(a) The metabolic extract of astrocytes was analyzed by using the standard metabolomic workflow and MS2 data for methionine were obtained. The raw experimental MS2 spectrum for methionine is shown on top and the MS2 spectrum from a pure standard is shown on bottom. Fragments that match in the 2 spectra are colored black, while fragments that do not match are colored red. The additional fragments in the experimental MS2 spectrum preclude identification of this compound as methionine. (b) After applying decoMS2, the experimental MS2 data matches the MS2 data of the methionine standard and thereby supports the structural assignment.
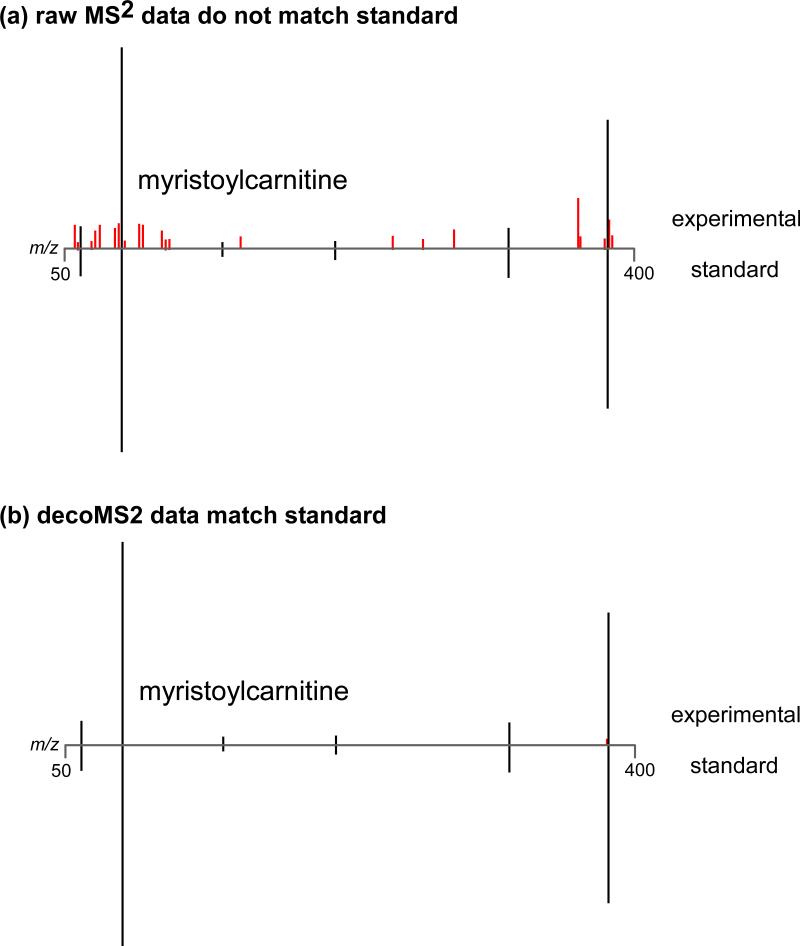
As a demonstration of the effectiveness of our method applied to a real experimental example, we highlight the structural characterization of myristoylcarnitine in peripheral nerve tissue. Peripheral nerve tissue was collected from two groups of mice and analyzed by a widely used metabolomic workflow that is referred to as “traditional” by some laboratories (Supplementary Figure 1).6 A dysregulated feature of interest was identified to have an m/z of 372.311. MS2 data were acquired for the feature and compared to the MS2 data of model compounds. Based on accurate mass and MS2 data, we hypothesized that this feature was myristoylcarnitine. However, the MS2 data from the research sample contained several additional peaks that prevented us from making a definitive assignment (Figure 6). By examining MS2 data acquired at 0 V, we determined that the MS2 spectra of the feature of interest were being averaged together with another feature of similar mass that eluted at the same retention time. With decoMS2, we were able to deconvolve the MS2 data and increase our confidence in the assignment of the peak as myristoylcarnitine.
Figure 6. decoMS2 applied to myristoylcarnitine from peripheral nerve tissue.
Peripheral nerve tissue was analyzed from 2 groups of mice using the standard metabolomic workflow. A feature of interest was identified to be dysregulated with statistical significance and was targeted for structural characterization. MS2 data were acquired for the feature and searched in metabolite databases. (a) The raw MS2 spectrum acquired for this feature is shown on top. The accurate mass of the feature was consistent with that of myristoylcarnitine and the MS2 data had similarities to the MS2 data of the myristoylcarnitine standard, which is shown on the bottom. Fragments that match in the 2 spectra are colored black. Fragments that do not match are colored red. Although we hypothesized that this feature of interest in the peripheral nerve tissue may be myristoylcarnitine, the additional fragments in the experimental MS2 spectrum precluded identification and publication of the compound as myristoylcarnitine. (b) After applying decoMS2, the experimental MS2 data matched the MS2 data of the myristoylcarnitine standard and thereby supported the structural assignment.
Applying the decoMS2-based workflow to untargeted metabolomics
Our approach for deconvolving metabolite MS2 spectra provides a valuable resource for improving the success rate of structural identification by using the traditional metabolomic workflow. In the traditional workflow (Supplementary Figure 1), only a small subset of the thousands of features detected in the samples are selected for structural characterization by MS2 analysis. Compound selection may be based on context, biological assays, meta-analysis, or simply statistical thresholds.21-23 For example, after analyzing hundreds to thousands of clinical specimens, an investigator may identify a single peak (i.e., a metabolomic feature) as a potential biomarker of disease.24 Under these circumstances, structural identification of this particular compound may largely define the overall success of the metabolomic analysis. A substantial proportion of the peaks detected, however, are challenging to identify because their MS2 data do not match the MS2 data of model compounds in standard libraries such as METLIN. As we have demonstrated above, an underlying problem is that the model compounds used to construct metabolomic libraries are pure, highly concentrated, and provide extremely clean data. Research samples, in contrast, are exceedingly complex and the metabolites of interest are often in very low concentration. As a result, the MS2 spectra of research samples are noisy and characterized by contaminating artifacts that limit metabolite identifications. By applying our deconvolution approach, investigators will improve their success rate of identifying compounds targeted for MS2 analysis. Our approach applied to the traditional workflow will be particularly valuable when investigators determine that the identification of a specific peak is of high priority to the success of the study but the peak cannot be identified by using conventional MS2 strategies because of limitations in instrument sensitivity combined with sample complexity.
In a broader context, the deconvolution approach developed here will enable innovative metabolomic workflows in which larger quantities of MS2 data are obtained at faster acquisition rates, characterized by increased noise/contamination. While a related workflow has been applied in proteomics (sometimes called SWATH MS), similar strategies have not been applied to metabolite analysis to date.25 The application of SWATH MS in proteomics relies on the ability to interpret chimeric MS2 data from multiple peptides de novo. This is possible, in part, because for some organisms the MS2 pattern for every occurring peptide is known.26 In contrast, our knowledge of MS2 patterns for metabolites is largely incomplete and thereby precludes the identification of unique precursor-product ion transitions. Instead, metabolite identification necessarily involves matching of complete MS2 spectra, and obtaining deconvolved spectra is essential for correctly evaluating metabolite matches.
The metabolite deconvolution strategy described here is thus an important prerequisite for the development of SWATH-like methods in metabolomics. Specifically, the proposed metabolomic workflow will provide 3 key advantages. First, it will introduce a standardized procedure for publishing metabolite identifications. Instead of laboratories applying different spectral editing strategies, decoMS2 will provide an unbiased mechanism for producing publication-quality MS2 spectra. Second, the workflow will reduce the number of metabolite assignments that are ruled out due to artificial differences between the MS2 data of the research sample and the MS2 data of standards. And, thirdly, the workflow will improve the sensitivity with which metabolites can be structurally characterized by MS2 analysis.
Conclusion
Untargeted metabolomic profiling relies on MS2 data to support metabolite assignments. To obtain MS2 data, compounds of interest are typically isolated for fragmentation by both chromatographic separation as well as quadrupole-based mass selection. We have shown, however, that a substantial portion of compounds are not purely isolated in the collision cell for MS2 fragmentation with traditional metabolomic workflows due to the molecular complexity of most biological samples. This creates contaminating fragments in the MS2 spectra of research samples and thereby prevents metabolite identifications from being made. These results suggest that metabolomic investigators should use as narrow an MS2 isolation window as possible to acquire fragmentation data, yet narrow isolation windows can severely limit sensitivity and still are sometimes not selective enough. To address this challenge, here we introduce a novel metabolomic workflow that simultaneously offers the specificity necessary for metabolite identifications without compromising analytical sensitivity. Our approach involves the experimental deconvolution of metabolomic MS2 data acquired by using wide MS2 isolation windows shifted over the precursor mass-to-charge region of interest. To accomplish the data analysis, an R package called decoMS2 implementing the algorithms used in our workflow is available on our website. With real examples from biological specimens, we have shown that decoMS2 enables the identification of metabolites that would not otherwise be possible. Our approach introduced here is a necessary prerequisite for the development of SWATH-like metabolomic methods in which large amounts of MS2 data accompanied by increased noise/contamination are acquired.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants R01 ES022181 and L30 AG0 038036.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Milne SB, Mathews TP, Myers DS, Ivanova PT, Brown HA. Biochemistry. 2013;52:3829–40. doi: 10.1021/bi400060e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinayavekhin N, Saghatelian A. Curr Protoc Mol Biol. 2010;1:1–24. doi: 10.1002/0471142727.mb3001s90. Chapter 30, Unit 30. [DOI] [PubMed] [Google Scholar]
- 3.Johnson CH, Gonzalez FJ. J Cell Physiol. 2012;227:2975–81. doi: 10.1002/jcp.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Anal Chem. 2010;82:3212–21. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 6.Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. Nat Biotechnol. 2012;30:826–8. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Nucleic Acids Res. 2009;37:D603–10. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patti GJ, Yanes O, Siuzdak G. Nat Rev Mol Cell Biol. 2012;13:263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopp F, Komatsu T, Nomura DK, Trauger SA, Thomas JR, Siuzdak G, Simon GM, Cravatt BF. Chem Biol. 2010;17:831–40. doi: 10.1016/j.chembiol.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. Anal Chem. 2012;84:5035–9. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kangas LJ, Metz TO, Isaac G, Schrom BT, Ginovska-Pangovska B, Wang L, Tan L, Lewis RR, Miller JH. Bioinformatics. 2012;28:1705–13. doi: 10.1093/bioinformatics/bts194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalisiak J, Trauger SA, Kalisiak E, Morita H, Fokin VV, Adams MW, Sharpless KB, Siuzdak G. J Am Chem Soc. 2009;131:378–86. doi: 10.1021/ja808172n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker M. Nat Meth. 2011;8:117–121. [Google Scholar]
- 14.Kind T, Scholz M, Fiehn O. PLoS One. 2009;4:e5440. doi: 10.1371/journal.pone.0005440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patti GJ, Yanes O, Shriver LP, Courade JP, Tautenhahn R, Manchester M, Siuzdak G. Nat Chem Biol. 2012;8:232–4. doi: 10.1038/nchembio.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tautenhahn R, Bottcher C, Neumann S. BMC Bioinformatics. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colby BN. J Am Soc Mass Spectrom. 1992;5:558–562. doi: 10.1016/1044-0305(92)85033-G. [DOI] [PubMed] [Google Scholar]
- 18.Norli HR, Christiansen A, Holen B. J Chromatogr A. 2010;1217:2056–64. doi: 10.1016/j.chroma.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed. Springer; New York City: 2008. p. 745. [Google Scholar]
- 20.Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Analytical chemistry. 2011;83:2152–61. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Carvalho LP, Zhao H, Dickinson CE, Arango NM, Lima CD, Fischer SM, Ouerfelli O, Nathan C, Rhee KY. Chem Biol. 2010;17:323–32. doi: 10.1016/j.chembiol.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manna SK, Patterson AD, Yang Q, Krausz KW, Idle JR, Fornace AJ, Gonzalez FJ. J Proteome Res. 2011;10:4120–33. doi: 10.1021/pr200310s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Nature. 2009;457:910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 25.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Mol Cell Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picotti P, Clement-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Rost H, Sun Z, Rinner O, Reiter L, Shen Q, Michaelson JJ, Frei A, Alberti S, Kusebauch U, Wollscheid B, Moritz RL, Beyer A, Aebersold R. Nature. 2013;494:266–70. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.