Abstract
Protein O-mannosylation is a special type of glycosylation that plays prominent roles in metazoans, affecting development and physiology of the nervous system and muscles. A major biological effect of O-mannosylation concentrates on the regulation of α-Dystroglycan, a membrane glycoprotein mediating cell – extracellular matrix interactions. Genetic defects of O-mannosylation result in the loss of ligand binding activity of α-Dystroglycan and causes congenital muscular dystrophies termed dystroglycanopathies. Recent progress in mass spectrometry and in vitro analyses has shed new light the mechanism of α-Dystroglycan glycosylation. However, this mechanism is underlain by complex genetic and molecular regulation that remain poorly understood. Protein O-mannosylation is evolutionarily conserved in metazoans, yet this pathway is simplified and more amenable to genetic analyses in invertebrate organisms, indicating that genetically tractable in vivo models could facilitate research in this area. This review will describe recent methodological strategies for studying protein O-mannosylation using in vitro and in vivo approaches.
Introduction
Protein O-mannosylation is an evolutionarily conserved type of glycosylation found in a wide range of metazoans, from insects to humans. Despite the fact that O-mannosyl glycans appear to be present on a number of glycoproteins, until now α-Dystroglycan (α-Dg) represents the only well-studied functional target of this modification (Chiba et al., 1997; Stalnaker et al., 2011b). More recent research in vertebrate and invertebrate species has significantly expanded our knowledge of O-mannosyl glycan structures and glycoproteins bearing this unique type of glycosylation (Fig. 1). The presence of O-mannosylation has been demonstrated for Drosophila Dystroglycan and its glycosylation has been analyzed by in vitro and in vivo approaches (Nakamura et al., 2010b). O-mannose-linked structures have been reported on receptor tyrosine phosphatase β (RPTPβ) that mediates cell signaling and regulates neural cell adhesion and migration (Abbott et al., 2008). Other confirmed targets of this modification include CD24, a mouse GPI-linked cell adhesion molecule that functions in the nervous and immune systems (Bleckmann et al., 2009), neurofascin, a neuronal cell adhesion molecule involved in the nervous system development and neural transmission (Pacharra et al., 2012), and IgG2 light chain antibody recombinantly expressed in Chinese hamster ovary cell culture (Martinez et al., 2007). Previous studies suggested that other O-mannosylated glycoproteins likely exist (Finne et al., 1979; Krusius et al., 1986; Wing et al., 1992; Yuen et al., 1997). This conclusion is consistent with recent glycomic analyses that indicated that α-Dg represents just a minor component of the O-mannosylated glycoproteome of mouse brain (Stalnaker et al., 2011a). However, the structure-function relationship of O-mannosylation of proteins other than α-Dg remains unknown. On the other hand, O-mannosyl glycans of α-Dg have been analyzed in a number of studies (Chiba et al., 1997; Harrison et al., 2012; Inamori et al., 2012a; Nakamura et al., 2010b; Stalnaker et al., 2010; Yoshida-Moriguchi et al., 2010), and these glycans have been demonstrated to play crucial roles in the development and physiology of the nervous system and muscles (reviewed in (Muntoni et al., 2011; Nakamura et al., 2010a; Stalnaker et al., 2011b)). Recent research has also implicated O-mannosylation of α-Dg in pathobiological processes such as cancer progression and viral infection (de Bernabe et al., 2009; Kunz et al., 2005; Yoshida-Moriguchi et al., 2010).
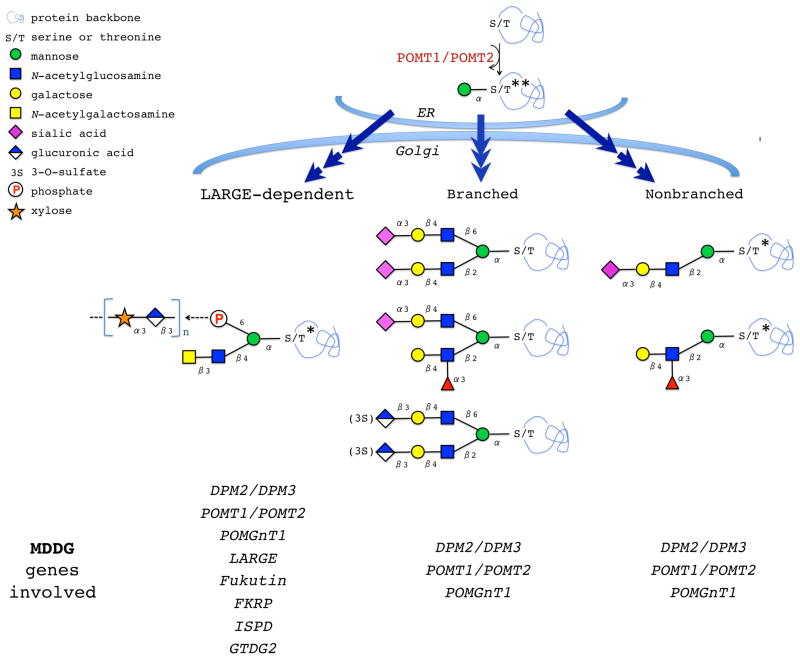
Figure 1. O-mannosylated glycans.
Biosynthesis of O-mannosyl glycans is initiated in the ER by a complex of two protein O-mannosyltransferases, POMT1/POMT2. The O-mannose-linked structures undergo further maturation in the Golgi. LARGE-dependent structures are thought to be responsible for Laminin-binding activity of α–Dg. MDDG-associated genes affecting (or predicted to affect) different structures are indicated. *, structures identified on mammalian α-Dystroglycan. **, the structure found on Drosophila Dystroglycan. Only mature structures are shown, however, O-mannosyl glycans with incomplete maturation have been also detected on glycoproteins (Stalnaker et al., 2011b).
Defects in the biosynthesis of O-mannosyl glycans are associated with a group of congenital muscular dystrophies (CMDs), such as Walker Warburg syndrome (WWS), Muscle-Eye-Brain disease (MEB), and Fukuyama congenital muscular dystrophy (FCMD) (Beltran-Valero de Bernabe et al., 2002; Kobayashi et al., 1998; van Reeuwijk et al., 2005; Yoshida et al., 2001). They all show various degrees of hypoglycosylation of α-Dg with concomitant loss of its ligand-binding activity (Fig. 2), and thus these syndromes are termed dystroglycanopathies (see more information about Muscular Dystrophy-Dystroglycanopathy, or MDDG, in OMIM 236670 (OMIM, 2012)). So far, eight genes have been shown to specifically affect the biosynthesis of O-mannosyl glycans of α-Dg and, when mutated, result in some form of MDDG. Two of these genes encode protein O-mannosyltransferases that initiate biosynthesis of O-mannosyl glycans on a protein backbone (POMT1 and POMT2) (Beltran-Valero de Bernabe et al., 2002; van Reeuwijk et al., 2005). Other genes encode glycosyltransferases, or glycosyltransferase-like proteins, that facilitate further elaboration of O-mannosyl glycans. They include β1,2-N-acetylglucosaminyltransferase 1 that elongates O-mannose with N-acetylglucosamine (POMGnT1) (Yoshida et al., 2001) and LARGE (aka LARGE1), a bifunctional glycosyltransferase with xylosyltransferase and glucuronyltransferase activities (Inamori et al., 2012a; Longman et al., 2003). LARGE synthesizes a carbohydrate chain of disaccharide repeats attached to O-mannose presumably via an unusual phosphate modification at the mannose 6-position, and this structure is essential for binding between α-Dg and its ligand Laminin (Barresi et al., 2004; Inamori et al., 2012a) (Fig. 2). Similar enzymatic activity has been demonstrated for LARGE2, a paralog of LARGE (Brockington et al., 2005; Fujimura et al., 2005; Grewal et al., 2005; Inamori et al., 2012b). Interestingly, LARGE2 has different pH optima of enzymatic activity and a distinct tissue-specific expression, suggesting that LARGE and LARGE2 may have complementary in vivo functions (Inamori et al., 2012b). Fukutin and Fukutin-related protein represent two glycosyltransferase-like proteins with LicD-domains with unknown activities that also affect the biosynthesis of highly glycosylated active form of α-Dg (Brockington et al., 2001b; Kobayashi et al., 1998). These proteins presumably function in the same pathway as LARGE, and the recent mouse Fukutin knockout revealed that the phosphate-linked carbohydrate modification of O-mannose required for Laminin binding is compromised in Fukutin mutants, while the phosphate modification of O-mannose itself remains intact (Beedle et al., 2012). Finally, ISPD (isoprenoid synthase domain containing) and GTDC2 (glycosyltransferase-like domain-containing protein 2) represent the two most recent additions to the group of genes associated with dystroglycanopathies, however the mechanism of their involvement in α-Dg glycosylation is still unknown (Manzini et al., 2012; Willer et al., 2012). In addition to the eight genes discussed above, mutations in dolichol-phosphate mannose synthase genes DPM2 and DPM3 have been implicated in MDDG (Barone et al., 2012; Lefeber et al., 2009). These enzymes are involved in the biosynthesis of dolichol-phosphate mannose (Dol-P-Man), a sugar donor substrate used in O-mannosylation and several other glycosylation pathways. Thus, DPM2 and DPM3 are expected to have pleiotropic effects on glycosylation that are not limited to protein O-mannosylation pathway.
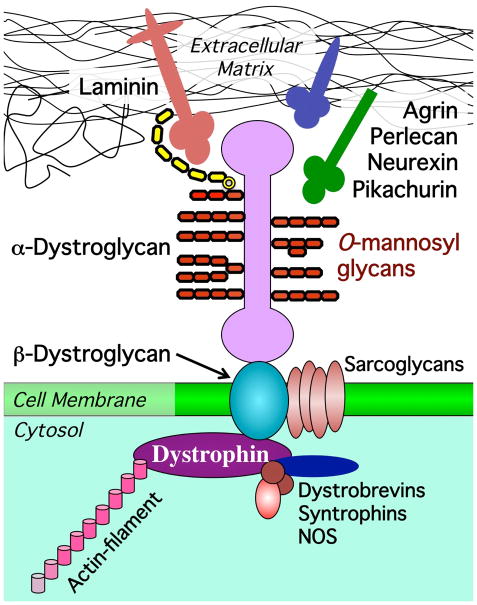
Figure 2. O-mannosyl glycans mediate interactions between α–Dg and extracellular matrix.
Dystroglycan interacts with basal lamina by binding ECM ligands (Laminin, Neurexin, Agrin, Perlecan, and Pikachurin). β-Dystroglycan is a transmembrane protein that connects extracellular (α-Dystroglycan) and intracellular (Dystrophin) components of the Dystrophin-associated glycoprotein complex (DGC). Dystrophin links the DGC to actin cytoskeleton. Other DGC-associated proteins include sarcoglycans, dystrobrevins, syntrophin, and NOS (nitric oxide synthase). Note that in addition to O-mannosylation, α–Dg has also O-linked mucin-type and N-linked glycans (not shown here). LARGE-dependent carbohydrate chain responsible for Laminin binding is shown in yellow.
Metazoan protein O-mannosyltransferases (POMTs) show evolutionary relationship to the multigene family of yeast O-mannosyltransferases (PMTs) that modify numerous secreted and cell wall proteins in fungi (Lommel and Strahl, 2009). Unlike fungi, animal organisms have only two O-mannosyltransferases that evolved to modify a relatively small number of glycoproteins, including Dystroglycan which is also conserved in metazoans. Invertebrate species have two protein O-mannosyltransferases highly homologous to their mammalian counterparts (Ichimiya et al., 2004; Lyalin et al., 2006; Nakamura et al., 2010b). However, invertebrates appear to be missing POMGnT1 and therefore their O-mannosyl glycans have simplified structures (Nakamura et al., 2010a). This was confirmed by mass spectrometry analysis of glycans of Drosophila Dg that revealed the presence of non-elongated O-mannose within the mucin-like domain (Nakamura et al., 2010b). Whether invertebrates have a glycan structure analogous to the one generated by LARGE is still unclear, while genetic evidence suggests this possibility since homologues of LARGE and/or FKRP are present in insect genomes (Brockington et al., 2001a; Grewal et al., 2005). Importantly, mutations in Drosophila POMTs and Dg result in muscle developmental defects, abnormal synaptic transmission and age-dependent muscle degeneration (Haines et al., 2007; Lyalin et al., 2006; Shcherbata et al., 2007; Wairkar et al., 2008), the phenotypes related to some important clinical findings associated with dystroglycanopathies (discussed in (Nakamura et al., 2010a)). Thus, Drosophila represents an attractive model organism for studying evolutionarily conserved mechanisms underlying the function of O-mannosylation in the nervous system and muscles of mammals.
In Drosophila, rotated abdomen (rt) and twisted (tw) genes encode protein O-mannosyltransferase enzymes POMT1 (aka RT) and POMT2 (aka TW), respectively (Ichimiya et al., 2004; Lyalin et al., 2006). It has been demonstrated that POMT1-POMT2 activity towards Dystroglycan is conserved between invertebrates and mammals, since Drosophila RT and TW can modify Drosophila DG in vitro and in vivo, and they can also use mammalian α-DG as an in vitro substrate (Ichimiya et al., 2004; Nakamura et al., 2010b). POMT1 and POMT2 function as an enzymatic heterocomplex, they co-localize within the ER, can be co-immunoprecipitated, and their co-expression is a prerequisite for protein O-mannosyltransferase activity in vitro and in vivo (Akasaka-Manya et al., 2006; Ichimiya et al., 2004; Lyalin et al., 2006; Manya et al., 2004; Nakamura et al., 2010b). The substrate specificity of POMT enzymes has been analyzed in vitro with polypeptide fragments of mammalian and Drosophila Dg recombinantly expressed in bacteria, or using short synthetic peptides corresponding to the mucin-type domain of α-DG (Ichimiya et al., 2004; Manya et al., 2004; Manya et al., 2007; Nakamura et al., 2010b). As a source of O-mannosyltransferase activity, these assays used membrane microsomal fraction prepared from mammalian or Drosophila cells. However, the substrate specificity revealed by these in vitro experiments is not consistent with the results of in cellulo assays that employed transgenic co-expression of POMTs together with their peptide substrates in cultured cells, followed by purification and analyses of O-mannosylated substrates (Breloy et al., 2008). The later approach indicated a potential requirement for some more distant structural determinants upstream of O-mannose attachment sites (Breloy et al., 2008). Mass spectrometry analysis of the whole extracellular domain of Drosophila Dg transgenically expressed in flies and modified in vivo by RT and TW unveiled some O-mannose-modified sequences that do not conform to the substrate preferences of mammalian POMTs. These experiments also discovered the presence of O-mannose outside of the mucin-type domain and revealed a surprising abundance of O-mannosyl glycans within the mucin-type domain of Dg (Nakamura et al., 2009). Thus, the acceptor specificity of O-mannosyltransferases appears to be complex and regulated by some yet to be identified factors. This specificity is not determined solely by a consensus sequence at modification sites but also appears to depend on some properties of the acceptor on a larger structural scale (Nakamura et al., 2010a). A similar feature of substrate recognition was shown for yeast PMT enzymes (Hutzler et al., 2007), which suggests that the mechanisms of O-mannosylation may be conserved within a broader family of eukaryotic O-mannosyltransferases. Interestingly, the analysis of Drosophila and mammalian substrates revealed that O-mannose can modify the same sites that can be used by mucin-type O-GalNAc modification (Nakamura et al., 2010b; Stalnaker et al., 2010). Since protein O-mannosylation occurs in the ER, while O-GalNAc is attached later, in the Golgi compartment of secretory pathway, this suggests that O-mannosylation could compete with O-GalNAc and therefore regulate the mucin-type glycosylation of protein substrates. This scenario indicates an intriguing possibility that some phenotypes of POMT1/2 mutations may be due to ectopic O-GalNAc modifications of Dg (Tran et al., 2012).
Below we will review the protocols developed for the analysis of O-mannosylation in animal cells using in vitro and in vivo approaches. We will discuss an in vitro assay for protein O-mannosyltransferase activity using purified fragments of Dystroglycan mucin-type domain as an acceptor and microsomal membrane fraction from cells co-expressing POMT1 and POMT2 as a source of enzymatic activity (Basic Protocol 1 with Support Protocols 1.1–3). We will also describe how to express in vivo and purify Drosophila Dystroglycan, and how to analyze its O-mannosylation by treatment with glycosidases, and using lectin and western blots (Basic and Alternate Protocols 2 with Support Protocols 2.1). In addition, we will review O-linked glycomics strategies that are based on mass spectrometry approaches and used to decipher the structure of O-mannosyl glycans (glycan composition analysis) and their location within polypeptide chains (site-mapping) (Protocols 3–6).
Basic Protocol 1: In vitro assay for O-mannosyltransferase activity
The assay can be used to study the enzymatic activity of protein O-mannosyltransferases in vitro (Ichimiya et al., 2004; Manya et al., 2008; Manya et al., 2004; Nakamura et al., 2010b). It is useful for testing different proteins acceptors as targets of O-mannosylation (Nakamura et al., 2010b), which can also reveal structural determinants in acceptor recognition (Manya et al., 2007). The in vitro O-mannosylation reaction will generate polypeptides with O-linked mannose that can be useful in further experiments, e.g. in the analysis of other enzymes mediating downstream steps in the biosynthesis of O-mannosyl glycan structures (Fig. 1). In the assay reaction, POMT1-POMT2 complex mediates the transfer of Man from sugar donor Dol-P-Man to a peptide acceptor (Endo and Manya, 2006). As an acceptor substrate, non-glycosylated polypeptides corresponding to mucin-type domain of Dg can be used, for example, a Dg fragment recombinantly produced in bacterial cells, eukaryotic cells lacking O-mannosyltransferase activity, or synthesized in vitro. As a source of O-mannosyltransferase activity, the assay uses microsomal membrane fraction prepared from cells co-expressing POMT1 and POMT2 (Support Protocol 1.1 and 1.2). Dol-P-Man with radioactively labeled mannose (e.g., [3H]Man) is used as a sugar donor to facilitate the detection of mannose transferred onto the acceptor. Below we include the protocol of protein O-mannosyltransferase assay that can be used with microsomal membrane fraction containing RT and TW (see Support Protocols 1.2–1.3 for expression of RT/TW and microsomal fraction purification), or other POMT1-POMT2.
Materials
Recombinant Dg-GST protein expressed and purified from E. coli (see Support Protocol 1.1)
Microsomal membrane fraction with POMT1 & POMT2 expression (see Support Protocols 1.2 and 1.3)
Dolichol phosphate-activated [3H]mannose (Dol-P-[3H]Man, 100 μCi/ml, 60 Ci/mmol, ARC)
Assay Buffer (see Reagents and Solutions)
PBS
Triton X-100
Scintillation counter, scintillation liquid and vials
Protocol Steps
Take an aliquot of [3H] Dol-P-Man sufficient for assays to be performed, e.g. 4 pmol per reaction, or 2.5 μl of the manufacturer’s stock solution of 100μCi/ml, 60 Ci/mmol Dol-P-[3H]Man in methanol:chlorophorm. Dry under a gentle stream of N2 at room temperature. Dissolve in the assay buffer (6 μl of assay buffer per 4 pmol of [3H] Dol-P-Man) by brief sonication using an inverted tip filled with ice-cold water.
Prepare the assay reactions on ice. For each reaction, mix the following ingredients dissolved in Assay Buffer: 10 μg of protein acceptor (Dg-GST fragment or BSA as a negative control) in 8 μl, 4 pmol [3H] Dol-P-Man dissolved in 6 μl, 80 μg of microsomal membrane fraction in 6 μl.
Incubate at 25°C for 1 hour.
Stop the assay reaction by adding 200 μl of PBS with 1% Triton X-100. Mix well by flicking the tube.
Remove insoluble material from the reaction mixtures by centrifugation at 10,000 g for 10 min at 4°C.
During the previous step, prepare GST-affinity beads by rinsing four times in PBS with 0.5% Triton X-100. Prepare 20 μl of beads per each assay reaction.
Load the pre-cleared supernatants from step 5 to pre-washed GST-beads, incubate with gentle nutation at 4°C for 1 hour.
Wash beads three times with 1 ml of PBS containing 0.5% Triton X-100.
Measure the incorporation of [3H] Man in the substrate trapped on GST-affinity beads using a scintillation counter.
Support Protocol 1.1: Expression in E. coli and purification of O-mannosyltransferase substrates
E. coli cells do not have protein O-mannosyltransferase activity, hence they can be conveniently used for the recombinant expression of protein substrates without O-mannose modifications. Several protein acceptors have been used in O-mannosyltransferase assays with Drosophila RT/TW enzymes; they all represent polypeptide fragments of mucin-type domains of mammalian or Drosophila Dg (Ichimiya et al., 2004; Nakamura et al., 2010b). Although currently there is no reliable algorithm to predict the location of O-mannosylation sites within a protein sequence, there is an apparent correlation between the presence of O-mannosylation and increased probability of multiple O-GalNAc sites within the mucin-type domain of Drosophila Dg, as predicted by NetOGlyc program (www.cbs.dtu.dk/services/netoglyc (Julenius et al., 2005)). Thus, this prediction can be potentially useful when designing expression constructs for the production of O-mannosyltransferase substrates based on Dg protein sequence (Nakamura et al., 2010b). So far, the in vitro O-mannosylation of other substrates has not been reported. The Dg fragments are expressed for as GST-tagged polypeptides, which facilitates their expression in a soluble form and purification using GST-affinity beads. The GST tag of a protein substrate is also exploited during in vitro O-mannosylation assay for the separation of [3H]Man-modified protein acceptor from non-incorporated radioactive sugar.
Materials
E.coli cells trasformed with pET-41-Dg-GST construct (e.g. using electroporation protocol (Sharma and Schimke, 1996)).
GST-affinity beads, e.g. glutathione agarose resin from Pierce (Thermo 15160)
Spectrophotometer for measuring bacterial cell density
LB medium for bacterial culture
IPTG
Wash Buffer, Elution Buffer, Dialysis Buffer (see Reagents and Solutions for composition)
Sonicator
0.45 micron low protein-binding syringe filter (Millipore)
Spectra/Por dialysis membrane, MWCO 15,000
Millipore centrifugal concentrators, MWCO 15,000
SDS-PAGE and Coomassie staining reagents
Protocol Steps
Incubate E. coli BL21(D3) cells transformed with pET-41-Dg-GST expression vector in 200 ml of LB on a shaker with vigorous agitation at 37°C overnight.
Induce the expression of Dg-GST with 0.4 mM IPTG when cells reach the density of OD600=0.6.
Continue incubation on a shaker at 28°C for 18 hours.
Harvest cells by centrifugation at 10,000 g, 4°C for 5 min. Decant the supernatant and resuspend the cell pellet in 25 ml ice-cold Wash Buffer (PBS, 0.1% NP-40, pH 7.5) with freshly added 0.1 % PMSF.
Lyse cells by sonication in 50 ml Falcon tubes on ice, applying 6 pulses of 20 sec sonication.
Complete cell lysis by incubating cells with gentle nutation for 20 min at +4°C.
Pre-clear the cell lysate by centrifugation at 14,000 g, 4°C for 15 min, followed by filtering the supernatant using 0.45 micron low protein-binding filter.
During the previous centrifugation step, prepare 200 μl GST-affinity beads by rinsing them four times in Wash Buffer.
Add GST-affinity beads to pre-cleared supernatant from step 7 and incubated overnight on a nutator at 4°C.
Assemble a mini-column using a pipette tip plugged with glass fiber. Apply the bead slurry from step 8 to the column and let it run through.
Wash the beads trapped in the column with 2 ml of Wash Buffer.
Wash once with 2 ml of PBS + 0.1% PMSF
Elute Dg-GST polypeptides with 0.5 ml of Elution Buffer (100 mM glutathione in 0.5M Tris pH 8.0): load the buffer onto the column, stop the flow, incubate column for 10 min, and collect the flow through. Repeat this elution step 2 times. Add collected fractions together to obtain 1.5 ml total volume of eluted protein in Elution Buffer.
Dialyze using Spectra/Por dialysis membrane (Spectrum, MWCO 15,000) in 2 L of Dialysis Buffer (20 mM Tris, 2 mM EDTA, 0.1% PMSF, pH 8) at 4°C for 4 hours. Repeat the dialysis 3 times.
Concentrate samples using Millipore centrifugal filters, e.g. with 15 kDa membrane cut-off filters, centrifuge at 12,000 g, 4°C for 20 min.
Analyze sample concentration and purity by PAGE and Coomassie staining. Normally, a 1/10 – 1/20 aliquot of the sample is sufficient for the analysis.
Store samples 4°C. For a long-term storage, the samples can be quickly frozen using liquid nitrogen or an ethanol-dry ice bath. Store samples at −80°C and avoid repeated freezing and thawing.
Support Protocol 1.2: Expression of Drosophila O-mannosyltransferases and Dystroglycan in vivo
The purpose of this procedure is to ectopically express proteins of interest (Drosophila O-mannosyltransferases RT and TW, or Dg) in vivo in flies. Drosophila tissue with expressed proteins can be used for the isolation of microsomal membrane fraction with O-mannosyltransferase activity (in case of co-expression of RT and TW), or for the purification of Dg protein for further analysis of its glycosylation (in case of Dg expression). Drosophila DG protein and protein O-mannosyltransferase enzymes RT (POMT1) and TW (POMT2) can be expressed in vivo using UAS-GAL4 expression system (Brand et al., 1994). To this end, the pUAST vector is used to make an expression construct that includes a protein coding sequence of interest, followed by an SV40 termination signal and placed under the control a minimal heat-shock promoter and five upstream-located copies of UAS sequence for binding of GAL4 transcriptional activator (Brand et al., 1994). This pUAST-based expression construct is assembled in vitro using molecular cloning techniques, verified by sequencing, and then inserted into Drosophila genome using embryo injection and P element-mediated transformation (Spradling and Rubin, 1982). Once the stable transgenic Drosophila lines carrying the constructs of interest are created using simple genetic manipulations, they are used for in vivo expression of corresponding proteins by crossing to a line carrying GAL4 driver with a particular temporal and cell-specific pattern of expression. Large collections of strains with different GAL4 drivers are available from several public Drosophila Stock collections (e.g., Bloomington Stock Center <http://flystocks.bio.indiana.edu/> or Kyoto Drosophila Genetic Resource Center <http://www.dgrc.kit.ac.jp/en/index.html>).
Materials
Transgenic Drosophila strains carrying UAS-constructs of interest, e.g. UAS-RT, UAS-TW, and UAS-ExDg-FLAG strains (available upon request from V.M.P.) (Lyalin et al., 2006; Nakamura et al., 2010b).
Materials and reagents for fly rearing, such as fly vials, food, environmental incubator for maintaining fly stocks at 25°C, 40% humidity (e.g., Percival DR36NL) (Roberts, 1998).
Microscope station (e.g., using Nikon SMZ645 microscope) with CO2 diffusion pads for fly sorting
PBS and 70% ethanol-water solutions
Glass dissection trays (e.g. Corning 7223-34)
Ethanol-dry ice or liquid N2 bath
Protocol Steps
-
Amplify stable transgenic Drosophila strains carrying UAS-based constructs of interest. Depending on the scale of the experiment, flies can be reared in population bottles having 25–30 ml of food and accommodating about 40–60 flies, or in fly vials with 7–10 ml of food and accommodating 10–20 flies.
Note: When selecting lines for in vivo expression, preferences should be given to the strains that can be propagated as homozygous genotypes. If homozygous-viable strains are not available, a balancer chromosome with a genetic marker suitable for the selection of desired genotypes among F1 generation can be used (e.g., TM6, Tb balancer can be conveniently used to select non-Tb larvae without TM6 chromosome). Using the amplified strains, collect males and virgin females to set up crosses in fresh vials or bottles using equal number of males and females. To ensure that all collected females are virgins, all adult flies are removed from the bottles or vials with parental populations, and then newly hatched flies are collected every 8 hours at 25°C. Reciprocal crosses can be set up for transgenic lines carrying autosomal transgenes and GAL4 drivers. For UAS transgenes or GAL4 drivers located on the X chromosome, it is preferable to use females for the crosses in order to ensure that all progeny will inherit the chromosome of interest.
Transfer parents to new vials or bottles with fresh food every 3–5 days to maintain the crossed flies in a healthy environment, maximize the yield and prevent overcrowding of progeny population.
Collect the F1 progeny of desired genotype and developmental stage using genetic and morphological markers, e.g. use Tb marker to select for flies without a TM6, Tb balancer chromosome, or use the size of the larvae and spiracle morphology to discriminate between instar stages.
Rinse collected Drosophila larvae two times in ice-cold 25% sucrose in dissecting trays to remove residual food, then they rinsed twice in ice-cold PBS. Aspirate PBS from dissecting trays with a Pasteur pipette. Residual PBS is removed by gently blotting with Kimwipe paper, and then larvae are transferred to 1.5 ml eppendorf tubes on ice. When collecting adults, flies are immobilized using CO2, transferred to dissection trays on ice and rinsed with cold PBS, flowed by a brief rinse with ice-cold 70% ethanol. The ethanol solution is quickly aspirated from the flies using Pasteur pipette, and flies are rinsed one more time with ice-cold PBS. After removing residual PBS with Kimwipe paper, flies are transferred to eppendorf tubes on ice.
Immediately proceed with homogenization of collected Drosophila to isolate microsomal membrane fractions with protein O-mannosyltransferase activity for in vitro assays (Support Protocol 1.3). An aliquot of tissue lysate can be saved for a western blot to analyze the amount of expressed proteins.
For the analysis of in vivo-expressed DG proteins, collected Drosophila can be immediately homogenized for protein purification (Support protocol 1.4). Alternatively, the samples can be quickly frozen using liquid nitrogen or an ethanol-dry ice bath and stored at −80°C for later experiments.
Support Protocol 1.3: Purification of microsomal fractions with O-mannosyltransferase activity
Microsomal membrane fraction can be purified as a source of protein O-mannosyltransferase activity for in vitro assays. Since POMT1 and POMT2 proteins are required to be simultaneously present within the cell in order to form enzymatically active complexes, their co-expression is a prerequisite for isolation of O-mannosyltransferase activity. The microsomal membrane fraction with protein O-mannosyltransferase activity has been purified from cultured mammalian cells with transgenically co-expressed or endogenous POMTs, from wildtype Drosophila tissues with endogenous RT and TW proteins, and from Drosophila transgenic strains with ectopic co-expression of RT and TW (see Support Protocol 1.2) (Ichimiya et al., 2004; Manya et al., 2008; Manya et al., 2004; Nakamura et al., 2010b). Ectopic co-expression of POMTs provides a way to increase the enzymatic activity of purified microsomal fractions. We include below a protocol for purification of the microsomal membrane fraction with O-mannosyltransferase activity from Drosophila larvae.
Materials
Homogenization Buffer, Assay Buffer (see Reagents and Solutions for composition)
Live Drosophila 3rd instar larvae (see Support Protocol 1.2)
Sonicator (e.g., Branson Sonifier 150) with microtip
Centrifuges: Eppendorf 5417R for low-speed centrifugation, and Beckman TL100
Ultracentrifuge with TLA 100.3 rotor for high-speed centrifugation
Bradford assay reagent (e.g., Sigma B6916)
Spectrophotometer with cuvettes for Bradford assay
Protocol Steps
Note: Protein O-mannosyltransferase activity of microsomal membrane fraction is not stable in vitro at room temperature, and precautions should be taken to maintain samples at 4°C during all purification steps.
Homogenize 3rd instar larvae in Homogenization Buffer using a glass Dounce homogenizer on ice. Use 10–15 μl of Homogenization Buffer per one larva. Approximately 40 larvae are sufficient for one experiment.
Sonicate on ice for 20 sec in an eppendorf tube at power level 2 using Branson Sonifier 150 with microtip. Repeat four times with 40 sec intermittent incubation on ice to prevent overheating of the sample.
Incubate with gentle nutation at 4°C for 20 min to complete tissue lysis.
Pre-clear the lysate by removing tissue debris with centrifugation at 800 g, 4°C for 10 min. Discard pellet, transfer supernatant to centrifuge tubes for high-speed centrifugation (13×15 mm tubes for Beckman TLA100.3 rotor)
Centrifuge at 100,000 g, 4°C in Beckman TL100 Ultracentrifuge using TLA 100.3 rotor for 1 hour.
Discard supernatant, resuspend the pellet representing membrane microsomal fraction in 20–40 μl of ice-cold Assay Buffer.
Estimate total protein concentration in the membrane microsomal fraction using Bradford assay (Sigma B6916). Use the microsomal fraction in in vitro assays as a source of protein O-mannosyltransferase activity within few hours.
Basic Protocol 2: Analysis of Drosophila Dystroglycan O-mannosylation using lectin blots
Mammalian α-Dg is a highly glycosylated protein bearing a variety of complex carbohydrate structures, including of N- and O-linked modifications (Chiba et al., 1997; Inamori et al., 2012a; Nakamura et al., 2010a; Yoshida-Moriguchi et al., 2010). Thus, a direct analysis of α-Dg glycosylation by lectins is usually not very informative. On the other hand, Drosophila Dg has fewer glycans, and their structures are less complicated (Nakamura et al., 2010a). Drosophila Dg or its extracellular domain, ExDg, can be expressed in vivo as a FLAG-tagged protein, purified, and its glycans can be analyzed by lectins (Nakamura et al., 2010b). In order to analyze O-linked structures attached to a protein, N-glycans can be removed prior to this analysis by PNGase F treatment to avoid their potential interactions with lectins. The specificity of lectin staining should be confirmed by a separate control experiment including incubation with the lectin in the presence of inhibiting sugar (e.g., using 0.2 M of Methyl α-D-Mannopyranoside, GalNAc or Gal for ConA, VVA or PNA, respectively). Ideally, all steps of the control and the main experiments should be carried out in parallel. The analysis of Drosophila Dg glycosylation can be performed for different mutant backgrounds, which can shed light on the involvement of other genes in the regulation of Dg glycosylation and function.
Materials
Purified ExDg-FLAG protein (see Support Protocol 2.1)
PNGase F glycosidase supplied with 10x denaturing, 10x G7 buffers (NEB P0704S)
Reagents and instrumentation for SDS-PAGE and western blot (e.g., from Bio-Rad)
BSA Fraction V (Roche)
TBST buffer (see Reagents and Solutions for composition)
Biotinylated lectins (e.g., ConA, VVA and PNA, Vector Laboratories)
Inhibiting sugars for sugar competition controls (e.g., α-methyl mannoside, GalNAc or Gal for ConA, VVA, PNA, respectively)
Instrumentation for chemiluminescent western blot analyses (e.g., ChemiDoc XRS system with Quantity One software, Bio-Rad).
Protocol steps
Remove N-linked glycans by incubating ExDg-FLAG protein purified on FLAG-affinity beads (see Support Protocol 2.1) with the PNGase F glycosidase. Use 500 units of PNGase F in a volume of 30μl of 1x G7 buffer per 2–3 μl of FLAG beads (corresponding to ExDg purified from 3–4 larvae). Incubate at 37°C with gentle agitation for 1 hour. As a control, set up in parallel a mock incubation reaction without adding the enzyme.
Release purified ExDg-FLAG from FLAG-affinity beads by incubating beads in SDS-PAGE lading buffer at 95°C for 5 min.
Cool briefly on ice and separate samples on 5% SDS-PAGE gel. Use the amount of sample corresponding to approximately 1–2 larvae per one lane.
Transfer gel-separated samples to nitrocellulose membrane.
Block the membrane with 2% BSA (Fraction V, Roche) in TBST (10mM Tris-HCl pH8.0, 150mM NaCl, 0.05% Tween 20) with gentle agitation at room temperature for 30 min.
Incubate the membrane with 2.5μg/ml of a biotinylated lectin at room temperature for 1 hour. For instance, use Concanavalin A for α-mannose-containing structures, VVA for α-GalNAc (the Tn antigen), and PNA for Galβ1,3GalNAc (the T-antigen) (Vector Laboratories).
Carry out the detection of lectin binding using Vectastain ABC kit with chemiluminescent detection (Vector).
Quantify blots using an instrument for western blot analysis (e.g., ChemiDoc XRS system, Bio-Rad).
Alternate Protocol 2: Analysis of Drosophila Dystroglycan O-mannosylation using glycosidase treatment and western blots
In addition to lectin blots (Basic Protocol 2), treatments by glycosidases with known specificities can also reveal the structure of glycans attached to ExDg. Glycosidase incubation time varies significantly for different glycosidases. Some glycosidase are highly active towards their substrates, and treatments with them require only ~1-hour incubation (e.g., PNGase F treatment, step 1 of this Protocol and Basic Protocol 2). The removal of O-linked mannose with Jack bean α-mannosidase can take up to 5–8 hours because the activity of this enzyme towards O-mannose is relatively low (step 3a of this Protocol). The removal of a specific sugar structure by glycosidase treatment leads to a change in molecular mass of the glycoprotein and thus can be detected by SDS-PAGE and western blot (e.g., (Nakamura et al., 2010b)). The analysis is facilitated by the presence of FLAG tag that provides a sensitive way of ExDg detection using western blot.
Materials
Purified ExDg-FLAG protein (see Support Protocol 2.1)
Glycosidase with a specific activity (e.g., α-mannosidase from Jack beans, Sigma)
Reagents and instrumentation for SDS-PAGE and western blot (e.g., from Bio-Rad)
BSA Fraction V (Roche)
TBST Buffer (see Reagents and Solutions for composition)
Mouse anti-FLAG M2 antibody (Sigma)
Anti-mouse HRP-conjugated secondary antibody (e.g., from Jackson ImmunoResearch Labs, catalog #11-035-003)
Pierce SuperSignal WestPico Chemiluminescent Substrate kit (Pierce).
Instrumentation for chemiluminescent western blot analyses (e.g., ChemiDoc XRS system with Quantity One software, Bio-Rad).
Protocol Steps
-
1
Perform PNGase F –mediated release of N-linked glycans as described in step 1 of Basic Protocol 2.
-
2
Rinse beads with 50μl of 1x G2 buffer (50mM sodium citrate pH4.5, NEB): add G2 buffer, mix beads by flicking the tube several times, separate beads by a brief centrifugation (800 g, 20 sec). Repeat twice.
-
3a
Treat beads with 0.4 units of α-mannosidase from Jack beans (Sigma) in 30μl of 1x G2 buffer for 3 hr at 37°C.
-
3b
As a control, set up a mock incubation without α-mannosidase.
-
4
Stop the reactions by adding 40μl of 2x SDS loading buffer.
-
5
Analyze the samples by western blot using standard protocol. Use anti-FLAG M2 as primary antibody at 1:4,000 dilution. Carry out detection using HRP-conjugated anti-mouse secondary antibody and chemiluminescent detection with Super Signal West Pico Chemiluminescent Substrate kit (Thermo Scientific).
-
6
Quantify blots using an instrument for western blot analysis (e.g., ChemiDoc XRS system with Quantity One software, Bio-Rad).
Support Protocol 2.1: Purification of in vivo-expressed Drosophila Dystroglycan using affinity beads
Drosophila model system offers a convenient possibility of in vivo expression of transgenic constructs in various cells and at different development stages using UAS-GAL4 ectopic expression system (Brand et al., 1994). Extracellular part of Dg can be expressed as a FLAG-tagged protein (ExDg-FLAG) while retaining its activity in vivo (Nakamura et al., 2010b). This secreted version of Dg can be purified using FLAG-affinity beads for the analysis of glycosylation by lectin or western blots (Protocol 2 and Alternate Protocol 2). While approximately 20 Drosophila larvae or pupae can be sufficient for a small-scale purification, the purification protocol can be scaled up for larger experiments.
Materials
Collected Drosophila with transgenically expressed ExDg-FLAG (see Support Protocol 1.2)
Glass Dounce homogenizer
Anti-FLAG M2 Affinity Gel (Sigma A2220)
Lysis Buffer (see Reagents and Solutions for composition)
Centrifuge Eppendorf 5417R
Protocol Steps
Homogenize collected Drosophila with transgenically expressed ExDg-FLAG (see Support Protocol 1.2) using a glass Dounce homogenizer on ice. Use 10–15 μl of Lysis Buffer per one larva or pupa.
Incubate with nutation at 4°C for 20 min to complete tissue lysis.
Remove insoluble material by centrifugation at 18,000 g, 4°C for 20 min.
During the previous centrifugation step, prepare FLAG affinity beads by rinsing them four times in Lysis Buffer.
-
Add the supernatant from the centrifugation step 3 to the FLAG beads. Use 10 μl of beads per volume of cell lysate corresponding to 20 larvae. Incubate on a nutator at 4°C for 2–4 hours.
Separate the beads from supernatant by low-speed brief centrifugation (800 g, 20 sec). Remove supernatant, resuspend beads in fresh ice-cold Lysis Buffer. Use 20–50 volumes of Lysis Buffer per one volume of beads. Incubate on a nutator at 4°C for 5 min. Repeat this centrifugation-washing step 3 more times. Purified ExDg bound to beads can be directly used in later assays. The purified sample can be stored at 4°C in the Lysis Buffer with Triton X-100 concentration decreased to 0.05% (150 mM NaCl, 50 mM Tris pH 7.4, 0.05% Triton X-100) for several weeks.
Basic Protocol 3: O-linked Glycomics (release, permethylation)
Mass spectrometry-based analyses of released and permethylated glycans have been previously described and allow for an in-depth characterization of the O-glycome from a variety of sources (Aoki et al., 2008; Haslam et al., 2006; Jang-Lee et al., 2007; Wada et al., 2010). The method relies on organic extraction and precipitation of all proteins from a biological source followed by release of the O-linked glycans via reductive beta-elimination. The resulting free glycans are then permethylated and analyzed via mass spectrometry. Analysis via direct infusion of permethylated glycans into a tandem mass spectrometer for MSn analysis is a powerful approach for detailed structural analysis but other approaches including MALDI-TOF and LC-MS/MS techniques can also be performed (Anumula and Taylor, 1992; Haslam et al., 2006; Jiao et al., 2011).
Materials
Intact isolated proteins (or proteolytic peptides).
Sodium Hydroxide
Sodium borohydride
Glacial acetic acid (AcOH)
Anhydrous dimethyl sulfoxide (dry DMSO)
Anhydrous methanol (dry MeOH)
Prepared base (see Support Protocol 3.2)
50% w/w sodium hydroxide solution
Iodomethane (MeI)
Nanopure water
Methylene chloride (DCM)
Screwtop tubes with, teflon lined caps, glass syringes, pasteur pipettes, and pipette bulbs
Centrifuge
Concentrator – attached to N2 gas to dry down sample
Nitrogen tank and regulator
Protocol Steps
Elimination
-
1
Weight out 38 mg (per one sample) of sodium borohydride (NaBH4) in a glass tube, add 500 μL of 50 mM NaOH resulted in ~ 2 M of NaBH4, and vortex.
-
2
Add 500 μL of 50 mM NaOH to intact isolated proteins (or proteolytic peptides).
-
3
Add 500 μL of 2 M of NaBH4 in the sample tube resulted in 1 mL of 1 M NaBH4.
-
4
Vortex and sonicate quickly the sample tube.
-
5
Incubate this for 16 – 18 hrs at 45 °C.
-
6
Remove the sample from the heat and allow it to cool to room temperature.
-
7
Neutralize the sample by adding 10 % AcOH dropwise with vortexing until bubbling stops (be careful to minimize sample loss by bubbling).
-
8
Clean up the sample using cation exchange column (see Support Protocol 3.1).
-
9
Move the column onto a screwtop tube and load the sample in the column.
-
10
Elute the sample into a glass screwtop tube using 5 – 6 mL of 5 % AcOH.
-
11
Dry sample in SpeedVac overnight.
-
12
Remove borate by methanol:glacial acetic acid (9:1) mixture.
-
13
Add 1.5 mL of MeOH/AcOH mixture in the sample tube with a glass pipette and transfer the sample into the screwtop tube.
-
14
Dry the sample down with dry N2 at 45 °C.
-
15
Repeat steps 13 and 14 twice.
When the sample is dry, sample can be permethylated.
Permethylation
-
16
Add 200 μL dry DMSO to the sample using a glass pipette or syringe.
-
17
Vortex to dissolve sample.
-
18
Add 300 μL of the prepared base (see Support Protocol 3.2) to the sample using a glass pipette or syringe. Comment: Prepare the base immediately prior to permethylation
-
19
Add 150 μL of iodomethane using a glass syringe (rinsed with dry DMSO) to the sample and then immediately cap the sample.
-
20
Vortex the sample vigorously for 5 min at maximum speed.
-
21
(Optional) Sonicate the sample for maximum 10 minutes or vortex the sample vigorously for another 5 min at maximum speed.
-
22
Add approximately 2 mL (1 pipette full) of nanopure water and mix well. The sample should turn cloudy as an indicator of a successful permethylation.
Note: If the sample does not turn cloudy, iodomethane may have evaporated before permethylation is completely done. May repeat permethylation.
-
23
Add approx. 2 mL of DCM (1 pipette full) using a glass pipette and vortex vigorously to extract the permethylated glycans.
-
24
Centrifuge and remove the aqueous (top) layer.
-
25
Rinse the DCM layer with approx. 2 mL of water (1 pipette full), vortex vigorously then centrifuge briefly, and remove the aqueous top layer.
-
26
Repeat step 10 two - four times (total rinsing would be 3–5 times).
-
27
After the final rinsing, transfer the DCM layer into another tube being careful not include any water.
-
28
Dry off the DCM using N2. Sample is ready for mass spectrometry analysis.
Support Protocol 3.1: Preparing cation exchange resin stock and packing chromatography column
In order to clean up the glycans for further analysis after release, cation exchange chromatography is performed as described.
Materials
Methanol
Chromatography column
Cation exchange resin
1M HCl
5% Acetic Acid
Add ~15 mL of resin to glass tube, add sufficient amount of 100% methanol to wash resin. Allow resin to incubate at room temperature. Remove methanol. Repeat this step 3 times.
Incubate resin overnight at room temperature in methanol.
Pour resin and methanol into 20mL chromatography column.
Rinse resin 3x with methanol, with 1M HCl until bubbles are gone, and 5% AcOH pushing through with air.
Keep the resin in glass tube with methanol at 4 °C and use this as a stock.
Pack the resin (1 mL bed volume) from the stock in the column.
Wash resin with methanol (x2), 1M HCl, and 5%AcOH forcing through with air until dry. Repeat this step twice.
Wash resin with 5%AcOH, start with air, and allow to pass through by gravity.
Support Protocol 3.2: Base preparation for permethylation
Preparation of fresh anhydrous base is crucial for complete permethylation and should be used the same day as prepared.
Materials
Glass screw cap tubes
50% w/w NaOH solution
Anhydrous methanol
Anhydrous DMSO
Glass Pipettes
Centrifuge for 10 cm × 13 cm tubes capable of 2,000 rpm
In a glass screw-cap tube (10 cm × 13 mm), combine 100 μL of 50% w/w sodium hydroxide solution (NaOH) using plastic pipits and 200 μL of anhydrous methanol (dry MeOH) using a glass pipette or syringe.
Vortex the mixture.
Add approx. 4 mL (2 pipits full) of anhydrous dimethyl sulfoxide (dry DMSO) using a glass pipette and vortex.
Centrifuge the tube (quick spin at 2,000 rpm at room temperature) and then remove DMSO, salts, and white residue from pellet.
Perform steps 3 and 4, total 3 through 5 five times to remove all water and white residues from the pellet.
Finally, add 1mL of DMSO and break down gel gently.
Basic Protocol 4: Preparing O-glycosylated peptides for MS analyses
Given the high degree of glycosylation than can occur on many O-glycoproteins, proteolytic digestion for shotgun proteomic analysis can yield poor results (Kobata, 1979; Nakamura et al., 2010b; Stalnaker et al., 2010). There is not an equivalent enzyme to N-glycanase (PNGaseF or PNGaseA) for O-glycoproteins. Thus, pretreatment of samples with a cocktail of glycosidases is often required to maximize proteolytic digestion. Commercial kits are available from multiple vendors and commonly include the enzymes PNGase F, galactosidase, N-acetylglucosaminidase, and sialidase. Note that many of these kits also contain O-glycanase that can remove core 1 structures. For site-mapping of core 1 and related structures, this enzyme should be omitted from the treatment, whereas, if optimal protein coverage is sought, the enzyme may be included. Mapping sites of O-glycoslyation on glycopeptides can be challenging given the lability of the glycosidic linkage to collision-induced dissociation frequently used for tandem mass spectrometry. However, beta-elimination can be used to remove the O-glycan resulting in a dehydro-amino acid (Greis et al., 1996). This resulting alpha,beta-unsaturated carbonyl is subject to conjugate (Michael) addition with a nucleophile that allows for a unique mass tag to be placed on the previously modified site of glycosylation. Nucleophiles that have been used for these methods include ammonia, dithiothreitol, and biotinylated primary amines (Rademaker et al., 1998; Vosseller et al., 2005; Wells et al., 2002).
Materials
Enriched glycoprotein sample
Cocktail of glycosidase enzymes (e.g. Enzymatic CarboRelease Kit, QA-Bio KE-DG01)
Trypsin (sequencing grade)
8M Urea in 40 mM ammonium bicarbonate, pH 8.1
27.5 mM iodoacetamide
Triethylamine
Sodium hydroxide
Ethanol
Trifluoroacetic acid
Nucleophile (dithiolthreitol or biotin pentylamine)
C18 Spin Columns (The Nest Group)
Protocol Steps
Treat glycoproteins with a cocktail of glycosidase enzymes as described by manufacturer (e.g., QA-Bio KE-DG01 or)
Add equal volume of 8M urea in 40 mM ammonium bicarbonate, pH 8.1, to sample.
Add dithiolthreitol to 5 mM final concentration and incubate at 37°C for 1 hour.
Add equal volume of 27.5 mM iodoacetamide to sample and incubate in dark for 45 minutes with quick vortexing every 15 minutes.
Dilute sample to final urea concentration of 0.8 M with 40 mM ammonium bicarbonate.
Digest with sequencing grade trypsin (typically 1:25 w/w of starting protein) overnight at 37°C.
Add Trifluoroacetic acid to 0.5% final concentration.
Purify with C18 reverse phase spin columns (Nest Group) using manufacturer’s protocol.
Dry desalted, eluted tryptic peptides in a SpeedVac system to complete dryness.
Resuspend desalted proteolytic glycopeptides in 1.5% triethylamine/0.15% NaOH/10% ethanol in an eppendorf tube.
Add nucleophile in excess (typically to 10 mM final concentration) and incubate sample at 50°C for 2 hours.
Add an equal volume of 2% trifluoracetic acid to quench the reaction.
Desalt sample using C18 spin columns according to manufacturer’s protocol (e.g. Nest Group, http://www.nestgrp.com/).
Basic Protocol 5: Glycopeptide Analysis via Neutral-loss triggered MSn approaches
Given that the glycosidic linkages are labile, the main peak often observed for a glycopeptide in a tandem mass spectrometer using collision-induced dissociation is the intact peptide with the neutral loss of the peptide. However, this can be used to one’s advantage in an ion-trap-based tandem mass spectrometer that is capable of trapping MS2 peaks and generating further fragmentation events (MSn). An instrument can be programmed to identify a major fragment in MS2 that corresponds to a particular loss (for examples 162 or 204 Da for Hexose or HexNAc, respectively). If such a neutral loss peak is observed, the instrument can isolate and fragment this species further to generate sequence information. Such approaches have been used routinely for phosphorylation (resulting in a dehydroamino acid scar where the phosphate once was) and more recently for O-glycopeptides (producing Ser/Thr residues without changes in mass where the glycan once was) (Jiang et al., 2008; Stalnaker et al., 2010). Given that O-glycopeptides result in “true” neutral loss peaks and do not leave behind a scar, the neutral loss approach is very sensitive at identifying glycopeptides but often times cannot reliably identify the exact residue modified when the peptide contains multiple hydroxyl-containing amino acids. This method benefits from no special care beyond the typical proteolytic digestion and desalting performed before analysis by LC-coupled tandem mass spectrometry. Only the method settings for the particular instrument used need be modified and analysis is restricted to those instruments capable of generating MS3 spectra.
Materials
No special materials required
Instrumentation
LTQ or an LTQ-Orbitrap hybrid instrument (ThermoFisher) is routinely used for these analyses, however multiple other mass spectrometers can be used as well.
Protocol
Tryptic peptides are analyzed by typical proteomic analysis with changes in the instrument method files. Pseudo-neutral loss-MS3 is chosen for analysis looking for a neutral loss peak as one of the 3 (5, for higher coverage but more false-positives) major fragments in MS2 that then triggers a MS3 event. A neutral loss table is generated that includes the m/z for +1, +2, +3, and +4 for Hexose, HexNAc, Deoxyhexose, and Sialic Acid (Neu5Ac, may also include Neu5Gc if looking at non-human samples). For example for hexose, the neutral loss table would include 162.1, 81.05, 54.03, and 40.53.
Basic Protocol 6: Glycopeptide Analysis via HCD-triggered ETD approaches
The current generation of hybrid mass spectrometers allows creative combinations of fragmentation energy to yield valuable information about an analyte. One such application is the use of higher-energy C-trap dissociation (HCD) of glycopeptides to yield abundant sugar oxonium ions (204.1 for example for HexNAc). HCD-analysis is rapid and sensitive and can be used to trigger another fragmentation event upon the presence of sugar oxonium ions in the mass spectrometer. In this case, electron transfer dissociation (ETD) of the precursor mass is used if HCD analysis detects a sugar oxonium ion (Zhao et al., 2011). Many post-translational modifications, including glycosylation, are quite stable in ETD allowing good backbone fragmentation of the peptide for sequence assignment and maintenance of the glycan on the modified amino acid for accurate site-mapping and glycan compositional analysis (Mikesh et al., 2006). In the particular format described above, analysis is restricted to a linear-ion trap/orbitrap mass spectrometer equipped with ETD capability. However, the method theoretically would work on any instrument capable of generating and scanning for oxonium ions generated by collisions that was also equipped to generate electron dissociations.
Materials
No special materials required
Instrumentation
Reverse-phase nano-high-performance liquid chromatography (HPLC) system (LC Packings, Dionex, etc.) connected to an LTQ-OrbitrapXL with ETD capability (ThermoFisher, or similar instrument).
Protocol
Tryptic peptides are analyzed by typical proteomic analysis with changes in the instrument method files. MS2 is acquired by HCD fragmentation of the top 5 most abundant ions from the full MS spectra. Importantly, the m/z range needs to be set to include the range from 125–300 m/z to capture single charged sugar oxonium ions. The instrument uses an oxonium ion list for Hexose, HexNAc, Deoxyhexose, and sialic acid (Neu5Ac, Neu5Gc may also be considered if looking at non-human samples). If an ion of sufficient intensity (differs depending on instrument) is present in the MS2 spectra within the tolerance of the instrument for a sugar oxonium ion (204.1 m/z for HexNAc for example), then the precursor ion is selected for ETD fragmentation.
Reagents and Solutions
Homogenization Buffer: 20 mM Tris (pH 8), 10 mM EDTA, 0.5% n-octyl-β-thioglucoside, 1mM PMSF (100 mM stock), 3 μg/ml pepstatin A, 1 μg/ml leupeptin, 1mM benzamidine HCl, 250 mM sucrose, 1 mM mercaptoethanol.
Assay Buffer: 20 mM Tris (pH 8), 10 mM EDTA, 0.5% n-octyl-β-thioglucoside, 2 mM mercaptoethanol.
Wash Buffer: PBS (phosphate-buffered saline), 0.1% NP-40, pH 7.5
Elution Buffer: 0.5M Tris, 100 mM glutathione, pH 8.0
Dialysis Buffer: 20 mM Tris, 2 mM EDTA, 0.1% PMSF, pH 8
TBST Buffer: 150 mM NaCl, 10 mM Tris pH 8.0, 0.05% Tween 20
G7 Buffer: 50 mM Sodium Phosphate pH 7.5, 1%NP-40
G2 Buffer: 50 mM Sodium Citrate pH 4.5
Lysis Buffer: 150mM NaCl, 50 mM Tris pH 7.4, 1% Triton X-100
Commentary
Background Information
O-linked mannosyl glycans were first found on a mammalian brain glycoprotein about thirty years ago, yet it was not until twenty five years later when the protein O-mannosyltransferase activity was reconstituted in vitro (Finne et al., 1979; Krusius et al., 1986; Manya et al., 2004). The first unambiguously identified glycoprotein bearing O-mannosyl glycans in vivo was α-dystroglycan, however, it was challenging to demonstrate in vitro that mammalian POMT1 and POMT2 can modify α-Dg with O-linked mannose (Chiba et al., 1997; Manya et al., 2004). The initial evidence of the enzymatic activity of these enzymes came from the work of Endo and coworkers who demonstrated that the microsomal fraction of HEK293T cells transfected with pcDNA3.1-based constructs expressing human POMT1 and POMT2 could modify a peptide acceptor with O-linked mannose in vitro (Manya et al., 2004). HEK293T cells showed only minimal endogenous O-mannosyltransferase activity, and the reconstitution of high level of this activity required co-transfection of both POMT constructs. In these experiments, O-mannosyltransferase activity was purified in vitro in the form of a microsomal membrane fraction. The isolated activity was found to be unstable above 25°C, and also required a special detergent, octyl-thioglucoside, for optimal function (Manya et al., 2004). As a sugar donor, protein O-mannosyltransferases use dolichol phosphate-activated mannose (Dol-P-Man) to modify Ser/Thr residues of protein acceptors. To demonstrate the activity of POMTs, Endo and co-workers used a radioactive sugar donor, Dol-P-[3H]Man, and a fragment of mouse α-DG as a protein acceptor. This fragment encompassed the amino acid sequence 313-483 of α-DG including mucin-type domain with multiple putative O-GalNAc attachment sites, based on the prediction by NetOGlyc program at (www.cbs.dtu.dk/services/netoglyc (Julenius et al., 2005)). Similar assays have been performed with microsomal fractions containing RT and TW while using as an acceptor the fragment of mammalian α-DG (Ichimiya et al., 2004), or mucin-type domain regions of Drosophila Dg-C and Dg-A isoforms (Nakamura et al., 2010b).
Critical Parameters and Troubleshooting
In vitro assay for O-mannosyltransferase activity
The assay appears to be sensitive to impurities that may arise from incomplete removal of solvent from Dol-P-[3H]Man (e.g., methanol:chlorophorm), or from some contaminants in protein acceptor preparations. Thus, careful preparation of both, donor and acceptor substrates for the assay can ensure their purity and improve the assay. Additionally, Dol-P-[3H]Man donor is not very stable at room temperature, and it should be used immediately once reconstituted in Assay Buffer, or quickly frozen and kept at −80°C. Moreover, Dol-P-[ H]Man can be absorbed by some plastic tubes and lost during reconstitution or the assay. Therefore, it is recommended to check the concentration of the donor sugar after reconstitution in Assay Buffer using a scintillation counter. Bacterial expression of some Dg-based substrates can be inefficient due to a problem with protein folding that leads to protein aggregation in inclusion bodies. This problem can be solved by decreasing temperature during protein expression to ≤25°C. Note that these conditions may require longer time of incubation after the induction of protein expression.
Expression of Drosophila O-mannosyltransferases and Dystroglycan in vivo
Crosses between UAS and GAL4 genotypes for ectopic expression can sometimes result in synthetic lethality (lethality caused by some combination of alleles that have little or no apparent disadvantage in other genetic backgrounds (Dobzhansky, 1946)) originating from a particular combination of several transgenic insertions. This lethality may not necessarily result from the overexpression of transgenic constructs, but rather be a consequence of mutations associated with transgene insertions that are combined together in the same genotype. In order to solve this problem, one can obtain other insertions of the constructs at the different genomic loci not causing synthetic lethality. Another potential problem can be associated with using a combination of a UAS construct and a GAL4 driver that may result in sick organisms due to exceedingly high level of expression causing toxicity. This situation can be revealed by a low number of progeny from a UAS x GAL4 cross, or dead pupae (or larvae) phenotype. To ensure the quality of tissue samples, it is important to avoid this situation and start with a different GAL4 and/or UAS transgene.
Purification of microsomal fractions with O-mannosyltransferase activity
The protein O-mannosyltransferase activity of microsomal membrane fraction is rather unstable, especially at temperatures above 15–20°C. Thus, it is important to carry out the protocol promptly, while keeping samples at ice-cold temperature throughout purification. Once the microsomal fraction is isolated, it should be used in assays right away for best results.
Analysis of Drosophila Dystroglycan O-mannosylation using lectin blots
Lectin blots can be prone to a high background and non-specific bands. To troubleshoot these problems, one can decrease lectin and/or ABC reagent concentration during incubation with blots. Additionally, it’s imperative to perform control incubation in the presence of inhibiting sugar. Finally, the specificity of the staining can be confirmed by treating the sample with a glycosidase that is expected to remove the structure recognized by the lectin. In this case, lectin blot analysis should confirm that specific bands disappear upon this treatment.
Analysis of Drosophila Dystroglycan O-mannosylation using glycosidase treatment and western blots
Some glycosidase isolates can be contaminated with a protease activity that could destroy glycoprotein during glycosidase treatment. Thus, it is important to compare the amount of glycoprotein before and after the treatment (e.g., by western blot). A different lot or source of glycosidase can be used to troubleshoot this problem. Additionally, some glycan chains can be more resistant to glycosidases due to their location, overall composition and presence of additional modifications. If incomplete digestion or glycosidase resistance is suspected, an alternative way to confirm the structure should be used, such as a different glycosidase with similar or overlapping activity, lectin blot analysis, or mass spectrometry approach.
Analysis of O-glycosylated peptides via beta-elimination
One must take caution in the mapping of O-glycoslyation sites with this method since other post-translational modifications, such as phosphorylation, are susceptible to beta-elimination. One possible strategy to address this shortcoming is to treat a portion of the sample with a phosphatase or a cocktail of glycosidases before the beta-elimination (Vosseller et al., 2005; Wells et al., 2002).
Anticipated Results
In vitro assay for protein O-mannosyltransferase activity
The assay can be useful for comaparing protein O-mannosyltransferase activity toward various protein acceptors, in different types of cells or in organisms of different genotypes. For example, this approach has been used to analyze this activity at its endogenous level, as well as upon transgenic overexpression of POMT1 and POMT2, in HEK293 cell cultures (Manya et al., 2004), in wildtype, mutant or transgenic Drosophila (Ichimiya et al., 2004; Nakamura et al., 2010b), to compare activity of mutant POMT enzymes (Manya et al., 2009), and to compare protein O-mannosyltransferase activity toward different Dg-derived peptide and protein substrates (Manya et al., 2007; Nakamura et al., 2010b). As an example of anticipated results for protein O-mannosyltransferase assays, Figure 3A illustrates the analysis of endogeneous RT-TW in Drosophila larvae with or without knockdown of rt and tw, demonstrating that both POMT genes are required for protein O-mannosylation (Ichimiya et al., 2004). A typical result of expression and purification of O-mannosyltransferase substrates from bacterial cells (Support Protocol 1.1) is shown in Figure 3B as a Coomassie staining of purified mucin-type domain of Drosophila Dg-A (Nakamura et al., 2010b).
Figure 3. Examples of protein O-mannosyltransferase assays (A) and a purified protein substrate for O-mannosylation (B).
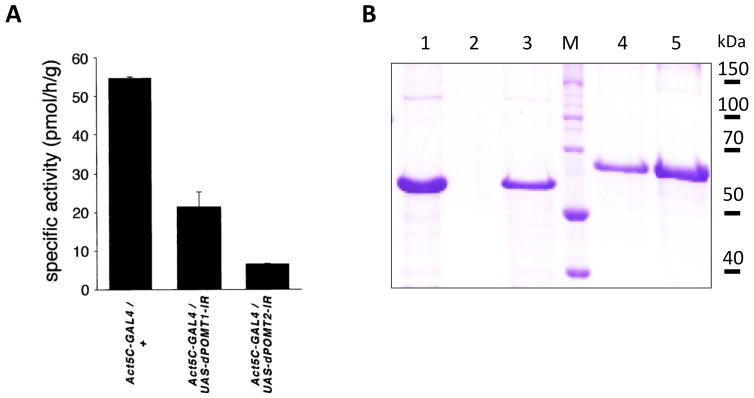
A, The activity of microsomal fraction isolated from Drosophila larvae was assayed using a fragment of mucin-type domain of mouse α-DG protein. The activity was assayed for 3 genotypes: Act5C-GAL4/+ (having wild-type level of rt and tw expression), Act5C-GAL4/UAS-dPOMT1-IR (RNAi-mediated knockdown of rt) and Act5C-GAL4/UAS-dPOMT2-IR (RNAi-mediated knockdown of tw). Figure adapted, with permission, from (Ichimiya et al., 2004). B, Coomassie staining of SDS-PAGE gel with purified fragment of Drosophila DG-A-GST protein expressed in E.coli cells (Nakamura et al., 2010b). Lane 1, DG-A-GST released from 10 μl of GST beads; Lane 2, blank; Lane 3, 1 μl of purified DG-A-GST eluted from after dialysis and concentration (estimated amount ~1.5 μg); M, protein molecular mass standards; Lane 4–5, BSA control samples for protein amount quantification, 1 and 2 μg, respectively.
Analysis of Drosophila Dystroglycan O-mannosylation using lectin blots
In vivo expression of Drosophila Dystroglycan protein can be induced at relatively high levels in larvae or pupae using act-GAL4 or hs-GAL4 drivers (Nakamura et al., 2010b). A purification experiment using about twenty larvae expressing UAS-ExDg-FLAG under control of act-GAL4 is expected to generate the amount of ExDg-FLAG protein sufficient for several western and lectin blot analyses. As little amount of material as 0.8 larvae/lane of SDS-PAGE can suffice to reliably detect ExDG-FLAG by western blot (Fig. 4A). Combining glycosidase treatments with western and lectin blot analyses can provide important information on the presence of certain glycan structures on purified protein. Figures 4B shows an example of such analysis performed with ExDg purified from different genetic backgrounds.
Figure 4. Western and lectin blot analyses of O-mannosylated forms of Drosophila ExDg-FLAG expressed in vivo.
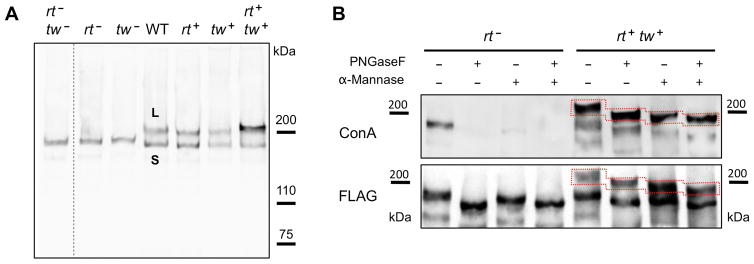
A, Western blot detection of ExDG expressed in rt-tw double mutants (rt− tw−), rt mutants (rt−), tw mutants (tw−), wildtype background (WT), and backgrounds with ubiquitous ectopic expression of RT (rt+), TW (tw+), or RT-TW co-expression (rt+ tw+). L band represents a highly O-mannosylated glycoform, while S band corresponds to a glycoform without significant O-mannosylation. B, Analysis of ExDg glycosylation by glycosidase treatments. The top panel shows Con A reactivity of purified ExDG after treatments with PNGaseF and α-mannosidase. The S glycoform purified from rt mutant background (left side) loses its Con A reactivity either after the removal of N-linked glycans by PNGaseF or after treatment with α-mannosidase removing α-linked mannose residues, suggesting the absence of O-mannose modifications and efficient removal of oligomannose structures either by trimming N-linked branches with α-mannosidase or by complete elimination of N-linked glycans with PNGaseF. The L glycoform purified from RT-TW co-expression background (right side) retains Con A reactivity after treatment with PNGaseF, α-mannosidase, or both glycosidases, suggesting that L glycoform is O-mannosylated, and that α-mannosidase does not remove O-mannose completely. The bottom panel shows anti-FLAG western blot control corresponding to the lectin blot shown in the top panel. Dashed outline shows the region of the L glycoform on the blots. Figure adapted with permission from (Nakamura et al., 2010b).
O-linked glycomics
Direct glycomics experiments as outlined can provide much more detailed results as to the exact O-glycans present in a sample and direct glycoproteomics analysis can define sites of attachment. As an example of direct glycopeptide analysis, see Figure 5. Using collision-induced dissociation, one can see that the major fragment of a singly O-mannose modified peptide is the neutral loss of the hexose (M.W. 162, loss of 81 m/z due to doubly charged state of the peptide) (Nakamura et al., 2010b). This type of neutral loss signature indicates the presence of a hexose modified peptide and can be used to trigger the instrument to perform an electron-transfer dissociation fragmentation in which the modification is not as labile that facilitates exact site mapping (Zhao et al., 2011).
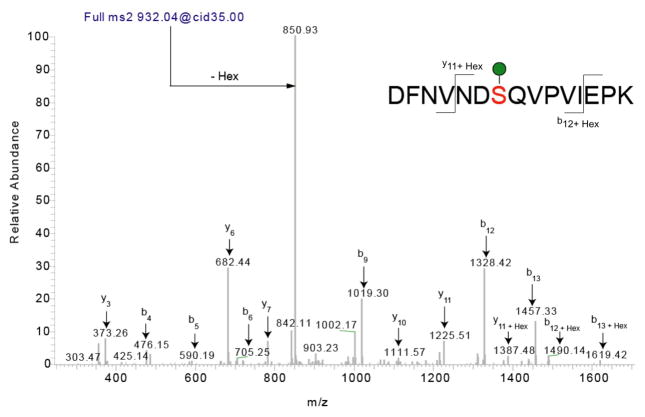
Figure 5. Identification of O-linked mannose sites using mass spectrometry.
Collision induced MS2 fragmentation of a singly O-mannosylated peptide derived from Drosophila Dystroglycan with m/z of 932.04 results in a predominant neutral loss ion at 850.93. This loss represents a loss of a hexose from the doubly charged peptide. In this example, there are sufficient fragments (b and y) to assign the peptide confidently and there is only one possible site of attachment. For peptides with multiple potential sites, the neutral loss observed upon collision induced fragmentation could be used to trigger an electron transfer dissociation fragmentation for confident site mapping. Figure adapted with permission from (Nakamura et al., 2010b).
Time Consideration
In vitro assay for O-mannosyltransferase activity (Basic Protocol 1) requires a couple of hours to complete and analyze the results. Expressing in bacterial cells and purification of protein acceptor substrates usually takes 2.5–3 days, starting from cell transformations and finishing with the analysis of purified proteins by gel staining. Expression of Drosophila O- mannosyltransferases and Dystroglycan in vivo requires two-three weeks, starting from setting up crosses till collecting progeny of genotypes of interest. This time is approximate and can vary significantly depending on the vigor of genotypes used in the experiments and the scale of the experiment. For large-scale expression experiments and when working with difficult genotypes, this time can be as along as a couple of months. Purification of microsomal fractions with O- mannosyltransferase activity takes 2–3 hours. A typical lectin blot procedure takes 1.5–2 days. Glycosidase treatment followed by western blot analysis normally requires 2–3 days. Isolation of glycoproteins, release and permethylation takes 2–3 days, and analysis by mass spectrometry takes 1–2 hours. Data interpretation can take up to a week depending on expertise and availability of software to assist. For analysis of glycopeptides, generation of peptides takes one day and desalting and LC-MS/MS analysis takes 1–2 hours. Data interpretation can take up to two days depending on expertise and availability of software to assist.
Acknowledgments
We would like to thank Naosuke Nakamura, Dmitry Lyalin, Stephanie Stalnaker and all members of the Panin’s and Wells’ laboratories for helpful discussions and contributions to the development of protocols described in this publication. This article is based on research supported in part by NIH grants GM069952 and NS075534 to V.M.P. and by a P41 grant from NCRR (P41RR018502, L.W., senior investigator) and grant from Muscular Dystrophy Association (MDA4074) to L.W.
Abbreviations
- Ser
serine
- Thr
theronine
- α-Dg
α-Dystroglycan (protein)
- Gal
galactose
- GlcNAc
N-Acetylglucosamine
- GalNAc
N-Acetylgalactosamine
- Man
mannose
- PMT
Protein O-mannosyltransferase
- POMT1(2)
Protein O-mannosyltransferase 1(2)
- rt
rotated abdomen (gene)
- tw
twisted (gene)
- Dol-P-Man
dolichol-phosphate-activated mannose
- POMGnT1
- FKRP
Fukutin-related protein (gene)
- ECM
Extracellular Matrix
- WWS
Walker Warburg Syndrome
- MEB
Muscle-Eye-Brain disease
- FCMD
Fukuyama congenital muscular dystrophy
- CMD
congenital muscular dystrophy
- MDDG
Muscular Dystrophy-Dystroglycanopathy
- DGC
dystrophin-associated glycoprotein complex
- NOS
nitric oxide synthase
Contributor Information
Vladislav M. Panin, Email: panin@tamu.edu.
Lance Wells, Email: lwells@ccrc.uga.edu.
Literature Cited
- Abbott KL, Matthews RT, Pierce M. Receptor tyrosine phosphatase beta (RPTPbeta) activity and signaling are attenuated by glycosylation and subsequent cell surface galectin-1 binding. J Biol Chem. 2008;283:33026–33035. doi: 10.1074/jbc.M803646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka-Manya K, Manya H, Nakajima A, Kawakita M, Endo T. Physical and functional association of human protein o-mannosyltransferases 1 and 2. J Biol Chem. 2006;281:19339–19345. doi: 10.1074/jbc.M601091200. [DOI] [PubMed] [Google Scholar]
- Anumula KR, Taylor PB. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Analytical biochemistry. 1992;203:101–108. doi: 10.1016/0003-2697(92)90048-c. [DOI] [PubMed] [Google Scholar]
- Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M. The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem. 2008;283:30385–30400. doi: 10.1074/jbc.M804925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone R, Aiello C, Race V, Morava E, Foulquier F, Riemersma M, Passarelli C, Concolino D, Carella M, Santorelli F, Vleugels W, Mercuri E, Garozzo D, Sturiale L, Messina S, Jaeken J, Fiumara A, Wevers RA, Bertini E, Matthijs G, Lefeber DJ. DPM2-CDG: A muscular dystrophy-dystroglycanopathy syndrome with severe epilepsy. Ann Neurol. 2012;72:550–558. doi: 10.1002/ana.23632. [DOI] [PubMed] [Google Scholar]
- Barresi R, Michele DE, Kanagawa M, Harper HA, Dovico SA, Satz JS, Moore SA, Zhang W, Schachter H, Dumanski JP, Cohn RD, Nishino I, Campbell KP. LARGE can functionally bypass alpha-dystroglycan glycosylation defects in distinct congenital muscular dystrophies. Nat Med. 2004;10:696–703. doi: 10.1038/nm1059. [DOI] [PubMed] [Google Scholar]
- Beedle AM, Turner AJ, Saito Y, Lueck JD, Foltz SJ, Fortunato MJ, Nienaber PM, Campbell KP. Mouse fukutin deletion impairs dystroglycan processing and recapitulates muscular dystrophy. J Clin Invest. 2012;122:3330–3342. doi: 10.1172/JCI63004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran-Valero de Bernabe D, Currier S, Steinbrecher A, Celli J, van Beusekom E, van der Zwaag B, Kayserili H, Merlini L, Chitayat D, Dobyns WB, Cormand B, Lehesjoki AE, Cruces J, Voit T, Walsh CA, van Bokhoven H, Brunner HG. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am J Hum Genet. 2002;71:1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann C, Geyer H, Lieberoth A, Splittstoesser F, Liu Y, Feizi T, Schachner M, Kleene R, Reinhold V, Geyer R. O-glycosylation pattern of CD24 from mouse brain. Biol Chem. 2009;390:627–645. doi: 10.1515/BC.2009.044. [DOI] [PubMed] [Google Scholar]
- Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- Breloy I, Schwientek T, Gries B, Razawi H, Macht M, Albers C, Hanisch FG. Initiation of mammalian O-mannosylation in vivo is independent of a consensus sequence and controlled by peptide regions within and upstream of the alpha-dystroglycan mucin domain. J Biol Chem. 2008;283:18832–18840. doi: 10.1074/jbc.M802834200. [DOI] [PubMed] [Google Scholar]
- Brockington M, Blake DJ, Prandini P, Brown SC, Torelli S, Benson MA, Ponting CP, Estournet B, Romero NB, Mercuri E, Voit T, Sewry CA, Guicheney P, Muntoni F. Mutations in the fukutin-related protein gene (FKRP) cause a form of congenital muscular dystrophy with secondary laminin alpha2 deficiency and abnormal glycosylation of alpha-dystroglycan. Am J Hum Genet. 2001a;69:1198–1209. doi: 10.1086/324412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockington M, Torelli S, Prandini P, Boito C, Dolatshad NF, Longman C, Brown SC, Muntoni F. Localization and functional analysis of the LARGE family of glycosyltransferases: significance for muscular dystrophy. Hum Mol Genet. 2005;14:657–665. doi: 10.1093/hmg/ddi062. [DOI] [PubMed] [Google Scholar]
- Brockington M, Yuva Y, Prandini P, Brown SC, Torelli S, Benson MA, Herrmann R, Anderson LV, Bashir R, Burgunder JM, Fallet S, Romero N, Fardeau M, Straub V, Storey G, Pollitt C, Richard I, Sewry CA, Bushby K, Voit T, Blake DJ, Muntoni F. Mutations in the fukutin-related protein gene (FKRP) identify limb girdle muscular dystrophy 2I as a milder allelic variant of congenital muscular dystrophy MDC1C. Hum Mol Genet. 2001b;10:2851–2859. doi: 10.1093/hmg/10.25.2851. [DOI] [PubMed] [Google Scholar]
- Chiba A, Matsumura K, Yamada H, Inazu T, Shimizu T, Kusunoki S, Kanazawa I, Kobata A, Endo T. Structures of sialylated O-linked oligosaccharides of bovine peripheral nerve alpha-dystroglycan. The role of a novel O-mannosyl-type oligosaccharide in the binding of alpha-dystroglycan with laminin. J Biol Chem. 1997;272:2156–2162. doi: 10.1074/jbc.272.4.2156. [DOI] [PubMed] [Google Scholar]
- de Bernabe DB, Inamori K, Yoshida-Moriguchi T, Weydert CJ, Harper HA, Willer T, Henry MD, Campbell KP. Loss of alpha-dystroglycan laminin binding in epithelium-derived cancers is caused by silencing of LARGE. J Biol Chem. 2009;284:11279–11284. doi: 10.1074/jbc.C900007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of Natural Populations. Xiii. Recombination and Variability in Populations of Drosophila Pseudoobscura. Genetics. 1946;31:269–290. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Manya H. O-mannosylation in mammalian cells. Methods Mol Biol. 2006;347:43–56. doi: 10.1385/1-59745-167-3:43. [DOI] [PubMed] [Google Scholar]
- Finne J, Krusius T, Margolis RK, Margolis RU. Novel mannitol-containing oligosaccharides obtained by mild alkaline borohydride treatment of a chondroitin sulfate proteoglycan from brain. J Biol Chem. 1979;254:10295–10300. [PubMed] [Google Scholar]
- Fujimura K, Sawaki H, Sakai T, Hiruma T, Nakanishi N, Sato T, Ohkura T, Narimatsu H. LARGE2 facilitates the maturation of alpha-dystroglycan more effectively than LARGE. Biochem Biophys Res Commun. 2005;329:1162–1171. doi: 10.1016/j.bbrc.2005.02.082. [DOI] [PubMed] [Google Scholar]
- Greis KD, Hayes BK, Comer FI, Kirk M, Barnes S, Lowary TL, Hart GW. Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by beta-elimination and tandem electrospray mass spectrometry. Analytical biochemistry. 1996;234:38–49. doi: 10.1006/abio.1996.0047. [DOI] [PubMed] [Google Scholar]
- Grewal PK, McLaughlan JM, Moore CJ, Browning CA, Hewitt JE. Characterization of the LARGE family of putative glycosyltransferases associated with dystroglycanopathies. Glycobiology. 2005;15:912–923. doi: 10.1093/glycob/cwi094. [DOI] [PubMed] [Google Scholar]
- Haines N, Seabrooke S, Stewart BA. Dystroglycan and protein O-mannosyltransferases 1 and 2 are required to maintain integrity of Drosophila larval muscles. Mol Biol Cell. 2007;18:4721–4730. doi: 10.1091/mbc.E07-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R, Hitchen PG, Panico M, Morris HR, Mekhaiel D, Pleass RJ, Dell A, Hewitt JE, Haslam SM. Glycoproteomic characterization of recombinant mouse alpha-dystroglycan. Glycobiology. 2012;22:662–675. doi: 10.1093/glycob/cws002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam SM, North SJ, Dell A. Mass spectrometric analysis of N- and O-glycosylation of tissues and cells. Current opinion in structural biology. 2006;16:584–591. doi: 10.1016/j.sbi.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Hutzler J, Schmid M, Bernard T, Henrissat B, Strahl S. Membrane association is a determinant for substrate recognition by PMT4 protein O-mannosyltransferases. Proc Natl Acad Sci U S A. 2007;104:7827–7832. doi: 10.1073/pnas.0700374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimiya T, Manya H, Ohmae Y, Yoshida H, Takahashi K, Ueda R, Endo T, Nishihara S. The twisted abdomen phenotype of Drosophila POMT1 and POMT2 mutants coincides with their heterophilic protein O-mannosyltransferase activity. J Biol Chem. 2004;279:42638–42647. doi: 10.1074/jbc.M404900200. [DOI] [PubMed] [Google Scholar]
- Inamori K, Yoshida-Moriguchi T, Hara Y, Anderson ME, Yu L, Campbell KP. Dystroglycan function requires xylosyl- and glucuronyltransferase activities of LARGE. Science. 2012a;335:93–96. doi: 10.1126/science.1214115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamori KI, Hara Y, Willer T, Anderson ME, Zhu Z, Yoshida-Moriguchi T, Campbell KP. Xylosyl- and glucuronyltransferase functions of LARGE in alpha-dystroglycan modification are conserved in LARGE2. Glycobiology. 2012b doi: 10.1093/glycob/cws152. Epub ahead of print Nov. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang-Lee J, Curwen RS, Ashton PD, Tissot B, Mathieson W, Panico M, Dell A, Wilson RA, Haslam SM. Glycomics analysis of Schistosoma mansoni egg and cercarial secretions. Molecular & cellular proteomics : MCP. 2007;6:1485–1499. doi: 10.1074/mcp.M700004-MCP200. [DOI] [PubMed] [Google Scholar]
- Jiang X, Han G, Feng S, Jiang X, Ye M, Yao X, Zou H. Automatic validation of phosphopeptide identifications by the MS2/MS3 target-decoy search strategy. Journal of proteome research. 2008;7:1640–1649. doi: 10.1021/pr700675j. [DOI] [PubMed] [Google Scholar]
- Jiao J, Zhang H, Reinhold VN. High Performance IT-MS Sequencing of Glycans (Spatial Resolution of Ovalbumin Isomers) International journal of mass spectrometry. 2011;303:109–117. doi: 10.1016/j.ijms.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Kobata A. Use of endo- and exoglycosidases for structural studies of glycoconjugates. Analytical biochemistry. 1979;100:1–14. doi: 10.1016/0003-2697(79)90102-7. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa M, Hamano K, Sakakihara Y, Nonaka I, Nakagome Y, Kanazawa I, Nakamura Y, Tokunaga K, Toda T. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- Krusius T, Finne J, Margolis RK, Margolis RU. Identification of an O-glycosidic mannose-linked sialylated tetrasaccharide and keratan sulfate oligosaccharides in the chondroitin sulfate proteoglycan of brain. J Biol Chem. 1986;261:8237–8242. [PubMed] [Google Scholar]
- Kunz S, Rojek JM, Kanagawa M, Spiropoulou CF, Barresi R, Campbell KP, Oldstone MB. Posttranslational modification of alpha-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J Virol. 2005;79:14282–14296. doi: 10.1128/JVI.79.22.14282-14296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeber DJ, Schonberger J, Morava E, Guillard M, Huyben KM, Verrijp K, Grafakou O, Evangeliou A, Preijers FW, Manta P, Yildiz J, Grunewald S, Spilioti M, van den Elzen C, Klein D, Hess D, Ashida H, Hofsteenge J, Maeda Y, van den Heuvel L, Lammens M, Lehle L, Wevers RA. Deficiency of Dol-P-Man synthase subunit DPM3 bridges the congenital disorders of glycosylation with the dystroglycanopathies. Am J Hum Genet. 2009;85:76–86. doi: 10.1016/j.ajhg.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel M, Strahl S. Protein O-mannosylation: conserved from bacteria to humans. Glycobiology. 2009;19:816–828. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- Longman C, Brockington M, Torelli S, Jimenez-Mallebrera C, Kennedy C, Khalil N, Feng L, Saran RK, Voit T, Merlini L, Sewry CA, Brown SC, Muntoni F. Mutations in the human LARGE gene cause MDC1D, a novel form of congenital muscular dystrophy with severe mental retardation and abnormal glycosylation of alpha-dystroglycan. Hum Mol Genet. 2003;12:2853–2861. doi: 10.1093/hmg/ddg307. [DOI] [PubMed] [Google Scholar]
- Lyalin D, Koles K, Roosendaal SD, Repnikova E, Van Wechel L, Panin VM. The twisted gene encodes Drosophila protein O-mannosyltransferase 2 and genetically interacts with the rotated abdomen gene encoding Drosophila protein O-mannosyltransferase 1. Genetics. 2006;172:343–353. doi: 10.1534/genetics.105.049650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manya H, Akasaka-Manya K, Nakajima A, Kawakita M, Endo T. Role of N-glycans in maintaining the activity of protein O-mannosyltransferases POMT1 and POMT2. J Biochem. 2009 doi: 10.1093/jb/mvp170. [DOI] [PubMed] [Google Scholar]
- Manya H, Bouchet C, Yanagisawa A, Vuillaumier-Barrot S, Quijano-Roy S, Suzuki Y, Maugenre S, Richard P, Inazu T, Merlini L, Romero NB, Leturcq F, Bezier I, Topaloglu H, Estournet B, Seta N, Endo T, Guicheney P. Protein O-mannosyltransferase activities in lymphoblasts from patients with alpha-dystroglycanopathies. Neuromuscul Disord. 2008;18:45–51. doi: 10.1016/j.nmd.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Manya H, Chiba A, Yoshida A, Wang X, Chiba Y, Jigami Y, Margolis RU, Endo T. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc Natl Acad Sci U S A. 2004;101:500–505. doi: 10.1073/pnas.0307228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manya H, Suzuki T, Akasaka-Manya K, Ishida HK, Mizuno M, Suzuki Y, Inazu T, Dohmae N, Endo T. Regulation of mammalian protein O-mannosylation: preferential amino acid sequence for O-mannose modification. J Biol Chem. 2007;282:20200–20206. doi: 10.1074/jbc.M702369200. [DOI] [PubMed] [Google Scholar]
- Manzini MC, Tambunan DE, Hill RS, Yu TW, Maynard TM, Heinzen EL, Shianna KV, Stevens CR, Partlow JN, Barry BJ, Rodriguez J, Gupta VA, Al-Qudah AK, Eyaid WM, Friedman JM, Salih MA, Clark R, Moroni I, Mora M, Beggs AH, Gabriel SB, Walsh CA. Exome Sequencing and Functional Validation in Zebrafish Identify GTDC2 Mutations as a Cause of Walker-Warburg Syndrome. Am J Hum Genet. 2012;91:541–547. doi: 10.1016/j.ajhg.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez T, Pace D, Brady L, Gerhart M, Balland A. Characterization of a novel modification on IgG2 light chain. Evidence for the presence of O-linked mannosylation. J Chromatogr A. 2007;1156:183–187. doi: 10.1016/j.chroma.2007.04.050. [DOI] [PubMed] [Google Scholar]
- Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochimica et biophysica acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni F, Torelli S, Wells DJ, Brown SC. Muscular dystrophies due to glycosylation defects: diagnosis and therapeutic strategies. Curr Opin Neurol. 2011;24:437–442. doi: 10.1097/WCO.0b013e32834a95e3. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Lyalin D, Panin VM. Protein O-mannosylation in animal development and physiology: from human disorders to Drosophila phenotypes. Semin Cell Dev Biol. 2010a;21:622–630. doi: 10.1016/j.semcdb.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Stalnaker SH, Lyalin D, Lavrova O, Wells L, Panin VM. Drosophila Dystroglycan is a target of O-mannosyltransferase activity of two protein O-mannosyltransferases, Rotated Abdomen and Twisted. Glycobiology. 2009 doi: 10.1093/glycob/cwp189. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Stalnaker SH, Lyalin D, Lavrova O, Wells L, Panin VM. Drosophila Dystroglycan is a target of O-mannosyltransferase activity of two protein O-mannosyltransferases, Rotated Abdomen and Twisted. Glycobiology. 2010b;20:381–394. doi: 10.1093/glycob/cwp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMIM. Online Mendelian Inheritance in Man. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; Baltimore, MD: 2012. World Wide Web URL: http://omim.org/ [Google Scholar]
- Pacharra S, Hanisch FG, Breloy I. Neurofascin 186 is O-mannosylated within and outside of the mucin domain. J Proteome Res. 2012;11:3955–3964. doi: 10.1021/pr200996y. [DOI] [PubMed] [Google Scholar]
- Rademaker GJ, Pergantis SA, Blok-Tip L, Langridge JI, Kleen A, Thomas-Oates JE. Mass spectrometric determination of the sites of O-glycan attachment with low picomolar sensitivity. Analytical biochemistry. 1998;257:149–160. doi: 10.1006/abio.1997.2548. [DOI] [PubMed] [Google Scholar]
- Roberts DB. Drosophila: A Practical Approach. 2. Oxford University Press; New York: 1998. [Google Scholar]
- Sharma RC, Schimke RT. Preparation of Electro-Competent E. coli Using Salt-free Growth Medium. BioTechniques. 1996;20:42–44. doi: 10.2144/96201bm08. [DOI] [PubMed] [Google Scholar]
- Shcherbata HR, Yatsenko AS, Patterson L, Sood VD, Nudel U, Yaffe D, Baker D, Ruohola-Baker H. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. Embo J. 2007;26:481–493. doi: 10.1038/sj.emboj.7601503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Stalnaker SH, Aoki K, Lim JM, Porterfield M, Liu M, Satz JS, Buskirk S, Xiong Y, Zhang P, Campbell KP, Hu H, Live D, Tiemeyer M, Wells L. Glycomic analyses of mouse models of congenital muscular dystrophy. J Biol Chem. 2011a;286:21180–21190. doi: 10.1074/jbc.M110.203281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalnaker SH, Hashmi S, Lim JM, Aoki K, Porterfield M, Gutierrez-Sanchez G, Wheeler J, Ervasti JM, Bergmann C, Tiemeyer M, Wells L. Site mapping and characterization of O-glycan structures on alpha-dystroglycan isolated from rabbit skeletal muscle. J Biol Chem. 2010;285:24882–24891. doi: 10.1074/jbc.M110.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalnaker SH, Stuart R, Wells L. Mammalian O-mannosylation: unsolved questions of structure/function. Curr Opin Struct Biol. 2011b;21:603–609. doi: 10.1016/j.sbi.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DT, Lim JM, Liu M, Stalnaker SH, Wells L, Ten Hagen KG, Live D. Glycosylation of alpha-dystroglycan: O-mannosylation influences the subsequent addition of GalNAc by UDP-GalNAc polypeptide N-acetylgalactosaminyltransferases. J Biol Chem. 2012;287:20967–20974. doi: 10.1074/jbc.M112.370387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Reeuwijk J, Janssen M, van den Elzen C, Beltran-Valero de Bernabe D, Sabatelli P, Merlini L, Boon M, Scheffer H, Brockington M, Muntoni F, Huynen MA, Verrips A, Walsh CA, Barth PG, Brunner HG, van Bokhoven H. POMT2 mutations cause alpha-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J Med Genet. 2005;42:907–912. doi: 10.1136/jmg.2005.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosseller K, Hansen KC, Chalkley RJ, Trinidad JC, Wells L, Hart GW, Burlingame AL. Quantitative analysis of both protein expression and serine/threonine post-translational modifications through stable isotope labeling with dithiothreitol. Proteomics. 2005;5:388–398. doi: 10.1002/pmic.200401066. [DOI] [PubMed] [Google Scholar]
- Wada Y, Dell A, Haslam SM, Tissot B, Canis K, Azadi P, Backstrom M, Costello CE, Hansson GC, Hiki Y, Ishihara M, Ito H, Kakehi K, Karlsson N, Hayes CE, Kato K, Kawasaki N, Khoo KH, Kobayashi K, Kolarich D, Kondo A, Lebrilla C, Nakano M, Narimatsu H, Novak J, Novotny MV, Ohno E, Packer NH, Palaima E, Renfrow MB, Tajiri M, Thomsson KA, Yagi H, Yu SY, Taniguchi N. Comparison of methods for profiling O-glycosylation: Human Proteome Organisation Human Disease Glycomics/Proteome Initiative multi-institutional study of IgA1. Molecular & cellular proteomics : MCP. 2010;9:719–727. doi: 10.1074/mcp.M900450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wairkar YP, Fradkin LG, Noordermeer JN, DiAntonio A. Synaptic defects in a Drosophila model of congenital muscular dystrophy. J Neurosci. 2008;28:3781–3789. doi: 10.1523/JNEUROSCI.0478-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- Willer T, Lee H, Lommel M, Yoshida-Moriguchi T, de Bernabe DB, Venzke D, Cirak S, Schachter H, Vajsar J, Voit T, Muntoni F, Loder AS, Dobyns WB, Winder TL, Strahl S, Mathews KD, Nelson SF, Moore SA, Campbell KP. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat Genet. 2012;44:575–580. doi: 10.1038/ng.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing DR, Rademacher TW, Schmitz B, Schachner M, Dwek RA. Comparative glycosylation in neural adhesion molecules. Biochem Soc Trans. 1992;20:386–390. doi: 10.1042/bst0200386. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Kobayashi K, Manya H, Taniguchi K, Kano H, Mizuno M, Inazu T, Mitsuhashi H, Takahashi S, Takeuchi M, Herrmann R, Straub V, Talim B, Voit T, Topaloglu H, Toda T, Endo T. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev Cell. 2001;1:717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, Oldstone MB, Schachter H, Wells L, Campbell KP. O-mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science. 2010;327:88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen CT, Chai W, Loveless RW, Lawson AM, Margolis RU, Feizi T. Brain contains HNK-1 immunoreactive O-glycans of the sulfoglucuronyl lactosamine series that terminate in 2-linked or 2,6-linked hexose (mannose) J Biol Chem. 1997;272:8924–8931. doi: 10.1074/jbc.272.14.8924. [DOI] [PubMed] [Google Scholar]
- Zhao P, Viner R, Teo CF, Boons GJ, Horn D, Wells L. Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. Journal of proteome research. 2011;10:4088–4104. doi: 10.1021/pr2002726. [DOI] [PMC free article] [PubMed] [Google Scholar]