Abstract
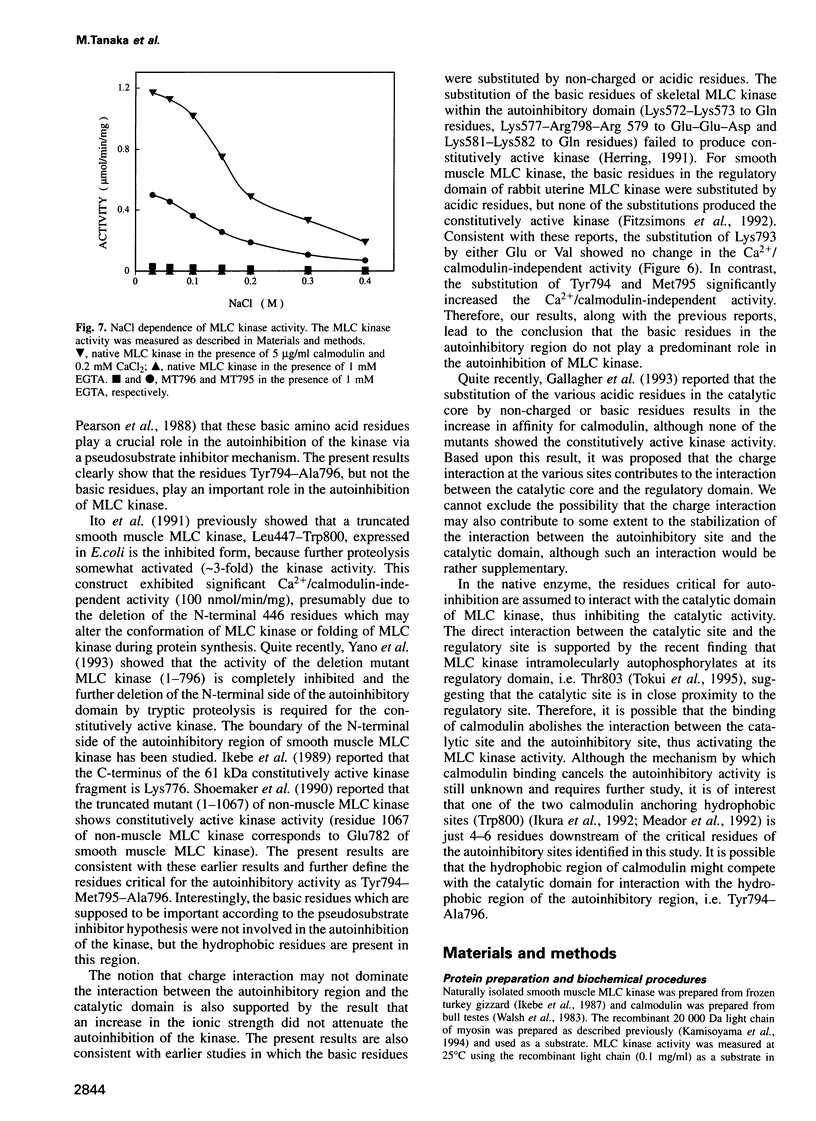
It has been hypothesized that basic residues in the autoinhibitory region of myosin light chain (MLC) kinase, which resemble the substrate sequence, interact with the catalytic core via charge interaction and thus inhibit the kinase activity (pseudosubstrate inhibitory hypothesis). In the present study, we produced seven MLC kinase mutants in which the residues in the autoinhibitory region are deleted to various extents, and determined the residues crucial for the autoinhibition of the kinase activity. The activities of MT799 (1-799) and MT796 (1-796) were completely inhibited, whereas MT793 (1-793), MT791 (1-791), MT787 (1-787) and MT783 (1-783) were constitutively active. The tryptic proteolysis of MT799 and MT796 activated the kinase activity, presumably due to the removal of the residues essential for autoinhibition. The mutants which showed the constitutively active kinase activity were not further activated by tryptic proteolysis, suggesting that the residues crucial for autoinhibition were already deleted. On the other hand, MT795 (1-795) was partially constitutively active (33% of maximum activity) and the tryptic proteolysis further activated the enzyme activity, suggesting that MT795 loses part of the residues essential for autoinhibition. The substitution of the residues Tyr794-Met795 but not Lys793 of untruncated MLC kinase significantly increased the Ca2+/calmodulin-independent kinase activity. These results clearly show that the region Tyr794-Met795-Ala796 is critical for autoinhibition. This study shows that the pseudosubstrate sequence is not critical for the autoinhibition mechanism of MLC kinase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Araki Y., Ikebe M. Activation of smooth muscle myosin light chain kinase activity by a monoclonal antibody which recognizes the calmodulin-binding region. Biochem J. 1991 May 1;275(Pt 3):679–684. doi: 10.1042/bj2750679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi I. C., Kemp B. E., Means A. R. Myosin light chain kinase structure function analysis using bacterial expression. J Biol Chem. 1989 Sep 25;264(27):15843–15849. [PubMed] [Google Scholar]
- Fitzsimons D. P., Herring B. P., Stull J. T., Gallagher P. J. Identification of basic residues involved in activation and calmodulin binding of rabbit smooth muscle myosin light chain kinase. J Biol Chem. 1992 Nov 25;267(33):23903–23909. [PMC free article] [PubMed] [Google Scholar]
- Gallagher P. J., Herring B. P., Griffin S. A., Stull J. T. Molecular characterization of a mammalian smooth muscle myosin light chain kinase. J Biol Chem. 1991 Dec 15;266(35):23936–23944. [PMC free article] [PubMed] [Google Scholar]
- Gallagher P. J., Herring B. P., Trafny A., Sowadski J., Stull J. T. A molecular mechanism for autoinhibition of myosin light chain kinases. J Biol Chem. 1993 Dec 15;268(35):26578–26582. [PMC free article] [PubMed] [Google Scholar]
- Guerriero V., Jr, Russo M. A., Olson N. J., Putkey J. A., Means A. R. Domain organization of chicken gizzard myosin light chain kinase deduced from a cloned cDNA. Biochemistry. 1986 Dec 30;25(26):8372–8381. doi: 10.1021/bi00374a007. [DOI] [PubMed] [Google Scholar]
- Herring B. P. Basic residues are important for Ca2+/calmodulin binding and activation but not autoinhibition of rabbit skeletal muscle myosin light chain kinase. J Biol Chem. 1991 Jun 25;266(18):11838–11841. [PubMed] [Google Scholar]
- Herring B. P., Stull J. T., Gallagher P. J. Domain characterization of rabbit skeletal muscle myosin light chain kinase. J Biol Chem. 1990 Jan 25;265(3):1724–1730. [PMC free article] [PubMed] [Google Scholar]
- Highashihara M., Frado L. L., Craig R., Ikebe M. Inhibition of conformational change in smooth muscle myosin by a monoclonal antibody against the 17-kDa light chain. J Biol Chem. 1989 Mar 25;264(9):5218–5225. [PubMed] [Google Scholar]
- Ikebe M., Maruta S., Reardon S. Location of the inhibitory region of smooth muscle myosin light chain kinase. J Biol Chem. 1989 Apr 25;264(12):6967–6971. [PubMed] [Google Scholar]
- Ikebe M., Stepinska M., Kemp B. E., Means A. R., Hartshorne D. J. Proteolysis of smooth muscle myosin light chain kinase. Formation of inactive and calmodulin-independent fragments. J Biol Chem. 1987 Oct 5;262(28):13828–13834. [PubMed] [Google Scholar]
- Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992 May 1;256(5057):632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- Ito M., Guerriero V., Jr, Chen X. M., Hartshorne D. J. Definition of the inhibitory domain of smooth muscle myosin light chain kinase by site-directed mutagenesis. Biochemistry. 1991 Apr 9;30(14):3498–3503. doi: 10.1021/bi00228a021. [DOI] [PubMed] [Google Scholar]
- Kamisoyama H., Araki Y., Ikebe M. Mutagenesis of the phosphorylation site (serine 19) of smooth muscle myosin regulatory light chain and its effects on the properties of myosin. Biochemistry. 1994 Jan 25;33(3):840–847. doi: 10.1021/bi00169a027. [DOI] [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B., Guerriero V., Jr, Bagchi I. C., Means A. R. The calmodulin binding domain of chicken smooth muscle myosin light chain kinase contains a pseudosubstrate sequence. J Biol Chem. 1987 Feb 25;262(6):2542–2548. [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B., House C. Role of basic residues in the phosphorylation of synthetic peptides by myosin light chain kinase. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7471–7475. doi: 10.1073/pnas.80.24.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B. Spatial requirements for location of basic residues in peptide substrates for smooth muscle myosin light chain kinase. J Biol Chem. 1985 Mar 25;260(6):3355–3359. [PubMed] [Google Scholar]
- Knighton D. R., Pearson R. B., Sowadski J. M., Means A. R., Ten Eyck L. F., Taylor S. S., Kemp B. E. Structural basis of the intrasteric regulation of myosin light chain kinases. Science. 1992 Oct 2;258(5079):130–135. doi: 10.1126/science.1439761. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Meador W. E., Means A. R., Quiocho F. A. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992 Aug 28;257(5074):1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- Olson N. J., Pearson R. B., Needleman D. S., Hurwitz M. Y., Kemp B. E., Means A. R. Regulatory and structural motifs of chicken gizzard myosin light chain kinase. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2284–2288. doi: 10.1073/pnas.87.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R. B., Ito M., Morrice N. A., Smith A. J., Condron R., Wettenhall R. E., Kemp B. E., Hartshorne D. J. Proteolytic cleavage sites in smooth muscle myosin-light-chain kinase and their relation to structural and regulatory domains. Eur J Biochem. 1991 Sep 15;200(3):723–730. doi: 10.1111/j.1432-1033.1991.tb16237.x. [DOI] [PubMed] [Google Scholar]
- Pearson R. B., Wettenhall R. E., Means A. R., Hartshorne D. J., Kemp B. E. Autoregulation of enzymes by pseudosubstrate prototopes: myosin light chain kinase. Science. 1988 Aug 19;241(4868):970–973. doi: 10.1126/science.3406746. [DOI] [PubMed] [Google Scholar]
- Roush C. L., Kennelly P. J., Glaccum M. B., Helfman D. M., Scott J. D., Krebs E. G. Isolation of the cDNA encoding rat skeletal muscle myosin light chain kinase. Sequence and tissue distribution. J Biol Chem. 1988 Jul 25;263(21):10510–10516. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker M. O., Lau W., Shattuck R. L., Kwiatkowski A. P., Matrisian P. E., Guerra-Santos L., Wilson E., Lukas T. J., Van Eldik L. J., Watterson D. M. Use of DNA sequence and mutant analyses and antisense oligodeoxynucleotides to examine the molecular basis of nonmuscle myosin light chain kinase autoinhibition, calmodulin recognition, and activity. J Cell Biol. 1990 Sep;111(3):1107–1125. doi: 10.1083/jcb.111.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takio K., Blumenthal D. K., Walsh K. A., Titani K., Krebs E. G. Amino acid sequence of rabbit skeletal muscle myosin light chain kinase. Biochemistry. 1986 Dec 2;25(24):8049–8057. doi: 10.1021/bi00372a038. [DOI] [PubMed] [Google Scholar]
- Walsh M. P., Hinkins S., Dabrowska R., Hartshorne D. J. Smooth muscle myosin light chain kinase. Methods Enzymol. 1983;99:279–288. doi: 10.1016/0076-6879(83)99063-8. [DOI] [PubMed] [Google Scholar]
- Yano K., Araki Y., Hales S. J., Tanaka M., Ikebe M. Boundary of the autoinhibitory region of smooth muscle myosin light-chain kinase. Biochemistry. 1993 Nov 16;32(45):12054–12061. doi: 10.1021/bi00096a016. [DOI] [PubMed] [Google Scholar]