Solitary fibrous tumors (SFTs) are rare mesenchymal tumors. Here, we describe the identification of a NAB2-STAT6 fusion from whole exome sequencing of 17 SFTs. Analysis in 53 tumors confirmed the presence of seven variants of this fusion transcript in 29 tumors (55%), a lower bound for fusion frequency at this locus, suggesting that the NAB2-STAT6 fusion is a distinct molecular feature of SFTs.
To better understand the molecular lesions contributing to SFT, we performed whole exome sequencing of DNA isolated from SFTs and matched blood from 17 patients (Supplementary Table 1). On average, we sequenced a median of 28.9 Mb per tumor, and 87.5% of the captured exons were covered to a depth of 20× or greater. An average of 22.5 non-synonymous somatic mutations per tumor were observed across the 17 samples (median: 19; range: 12-41; median rate of 0.66 mutations/Mb; Supplementary Fig. 1). This corresponded to 390 somatically mutated genes, of which one gene mutated in two samples (RBPJ) reached nominal statistical significance for recurrence (q < 0.1; Supplementary Tables 2 and 3; Supplementary Methods). Read count analysis of the tumor copy number profiles showed 11 tumors with structurally undisrupted genomes and no obvious gains or losses, while six tumors showed a broad loss of chromosome 13, including two samples with a concurrent broad gain of chromosome 8 (Supplementary Fig. 2).
Given that many soft tissue tumors are driven by genomic translocations,1we explored the possibility that gene fusions may be contributing to tumorigenesis in SFT. Using whole exome sequencing data, our fusion detection was limited to breaks that occurred within exons or near intronexon boundaries. However, we identified 19 potential fusion events in 10/17 tumor samples using algorithms designed for this application (Supplementary Table 4; Supplementary Methods).
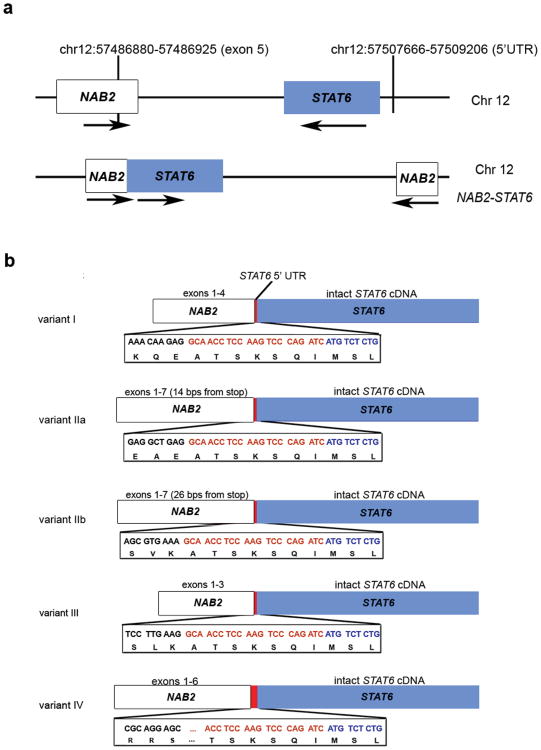
Rearrangements in 7/17 tumors represented in-frame fusions of the NAB2 and STAT6 genes both located on chromosome 12 (Supplementary Fig. 3a). Sequence review identified paired-end reads where one read mapped to an exonic region of NAB2 and its pair mate mapped 1.8 to 5.2 kilobases before the transcription start site of STAT6 (Supplementary Fig. 3b; Supplementary Table 4). Given that sequence mate pairs representing the two ends of a DNA fragment were mapped in opposite orientations on the same strand (Supplementary Fig. 3b), we concluded that this fusion was likely due to an intra-chromosomal inversion that juxtaposed NAB2 and STAT6 (Fig. 1a). Normally, NAB2 and STAT6 are in opposite orientations but the predicted inversions bring the two genes in close proximity in the same orientation (Fig. 1a). In all cases, we also identified at least one sequence that spanned the NAB2/STAT6 fusion boundary. We confirmed the genomic breakpoint in two tumors by sequencing of PCR products generated with breakpoint-spanning primers (Supplementary Fig. 4). The fusion was not observed in DNA from matched normal tissue (data not shown).
Figure 1. Structure of NAB2-STAT6.
(a) An inversion within chromosome 12 results in the juxtaposition of the NAB2 and STAT6 genes. (b) Schematic diagram of five representative fusion variants showing a conserved breakpoint within STAT6 and variable lengths of NAB2. White regions represent NAB2, red regions correspond to the 5′ untranslated region of STAT6, and blue regions show STAT6.
To analyze the expression and frequency of NAB2-STAT6 fusions, we performed RT-PCR using primers placed within exons 2-3 of NAB2 and within exon 1 of STAT6, and cDNA generated from 53 tumor samples representing 48 patients (Supplementary Table 1). An RT-PCR product was identified in 27 tumors (51%) from 23 patients. Sequencing of these products revealed multiple isoforms of the fusion that varied in the location of the fusion within NAB2 (Fig. 1b, Supplementary Fig. 4). The NAB2 sequence was followed by a sequence belonging to the 5′ untranslated region of STAT6. Comparing the cDNA structure to the inversion structure identified at the genomic level indicates that the region between exon 5 of NAB2 and the beginning of STAT6 (∼10kb) is spliced during transcription or that a large portion of this intronic region is deleted when the chromosome is rearranged. Additionally, we identified rare NAB2-STAT6 fusions that occurred in downstream exons of STAT6 in 2/17 cases from whole exome sequencing data (see Supplementary Table 4).
NAB2 encodes a transcriptional repressor of the zinc finger transcription factors EGR1 and EGR2, effectors of TGFβ signaling in smooth muscle.2,3 NAB2 contains two conserved domains that interact with EGR1 to mediate multimerization and repress transcription, respectively.2,3 Functional alterations of NAB2 have not been demonstrated in cancer but loss is reported in prostate cancer, lung cancer, non-Hodgkin's lymphoma, and neuroblastoma, and overexpression in melanoma, Ewing sarcoma, and rhabdomyosarcoma.4-6
STAT6 is a transcription factor that modulates signaling by IL-4 and IL-13 in the immune system.7 Activation of STAT family members in cancer underlies the hypothesis that these proteins are potential therapeutic targets.8 STAT6 in particular has been associated with increased proliferation and invasiveness in glioblastoma.9 Based on the properties of NAB2 and the oncogenic potential of STAT6, we hypothesize that the NAB2-STAT6 fusion dimerizes through the oligomerization domain of NAB2, translocates to the nucleus, and modulates STAT6-dependent gene expression. In experimental systems, fusion of STAT3 to a portion of the estrogen receptor results in a similar phenomenon.10
In summary, we have identified a novel NAB2-STAT6 fusion in at least half of SFT tumors from whole exome sequencing data. Although most fusions are identified from whole genome or whole transcriptome sequencing, this finding validates the use of exome data for the discovery of fusions that occur mid-exon. The NAB2-STAT6 fusion appears to be unique to SFT samples as fusion analysis of approximately 713 unique tumor-normal pairs from 5 tumor types analyzed by whole genome, exome, or transcriptome sequencing, or a combination of these techniques,11-15 failed to identify any fusions involving these genes. As such, this fusion represents the first molecular feature unique to SFTs. These data also suggest that small molecule inhibitors of STAT6 may be efficacious in this tumor type. Further experiments investigating the functional behavior of this fusion protein are ongoing. Our estimates of the frequency of NAB2-STAT6 fusions should be considered a lower bound because whole exome sequencing would fail to identify intronic breaks, and because our RT-PCR approach is limited to the regions covered.
Supplementary Material
Acknowledgments
We are grateful to the patients who contributed material for this study. We thank members of the Genomics Platform at the Broad Institute for their technical expertise, and Drs. Mohamed Abazeed and Austin Dulak for critically reviewing the manuscript. This work was conducted as part of the Slim Initiative for Genomic Medicine in the Americas (SIGMA), a project funded by the Carlos Slim Health Institute in Mexico. This work was supported by a SPORE grant in Soft Tissue Sarcoma (P50 CA140146) and a P01 grant (P01CA47179) to Samuel Singer (MSKCC). JC is supported by an American Cancer Society AstraZeneca Postdoctoral Fellowship; AMC is supported by a grant from the Kristen Ann Carr Foundation. MM is supported by generous donations from M.B. Zuckerman and Team Dragonfly of the Pan-Mass Challenge.
Footnotes
Accession codes: Sequence data used for this analysis are available in dbGaP under accession number phs000568.v1.p1, and are available at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000568.v1.p1.
Contributions: JC and MM conceived the project. JC, AMC, and MM wrote the manuscript with input from all co-authors. JC, AMC, and RO performed experiments. JC and MR performed computational analyses. AM contributed an unpublished algorithm to the analysis. JC, AMC, MR, SRW, RO, DF, and MM analyzed the results. AMC and MCH contributed samples. LA and DA facilitated transfer, sequencing, and analysis of samples.
Conflicts of interest: MM is a paid consultant for and equity holder in Foundation Medicine, a genomics-based oncology diagnostics company, and is a paid consultant for Novartis. MCH is a paid consultant for and equity holder in Molecular MD, a molecular diagnostics company, and is a paid consultant for Novartis.
References
- 1.Mertens F, et al. Semin Oncol. 2009;36:312–23. doi: 10.1053/j.seminoncol.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Svaren J, et al. Mol Cell Biol. 1996;16:3545–53. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya S, et al. PLoS One. 2009;4:e7620. doi: 10.1371/journal.pone.0007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimoyamada H, et al. Am J Pathol. 2010;177:70–83. doi: 10.2353/ajpath.2010.091164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdulkadir SA, et al. Hum Pathol. 2001;32:935–9. doi: 10.1053/hupa.2001.27102. [DOI] [PubMed] [Google Scholar]
- 6.Pal NR, Aguan K, Sharma A, Amari S. BMC Bioinformatics. 2007;8:5. doi: 10.1186/1471-2105-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. Cytokine Growth Factor Rev. 2006;17:173–88. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Nelson EA, Sharma SV, Settleman J, Frank DA. Oncotarget. 2011;2:518–24. doi: 10.18632/oncotarget.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merk BC, Owens JL, Lopes MB, Silva CM, Hussaini IM. BMC Cancer. 2011;11:184. doi: 10.1186/1471-2407-11-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He HJ, et al. J Biol Chem. 2006;281:31369–79. doi: 10.1074/jbc.M603762200. [DOI] [PubMed] [Google Scholar]
- 11.Imielinski M, et al. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammerman PS, et al. Nature. 2012 [Google Scholar]
- 13.Barbieri CE, et al. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugh TJ, et al. Nature. 2012;488:106–10. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerji S, et al. Nature. 2012;486:405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.