Abstract
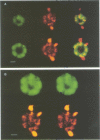
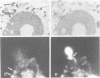
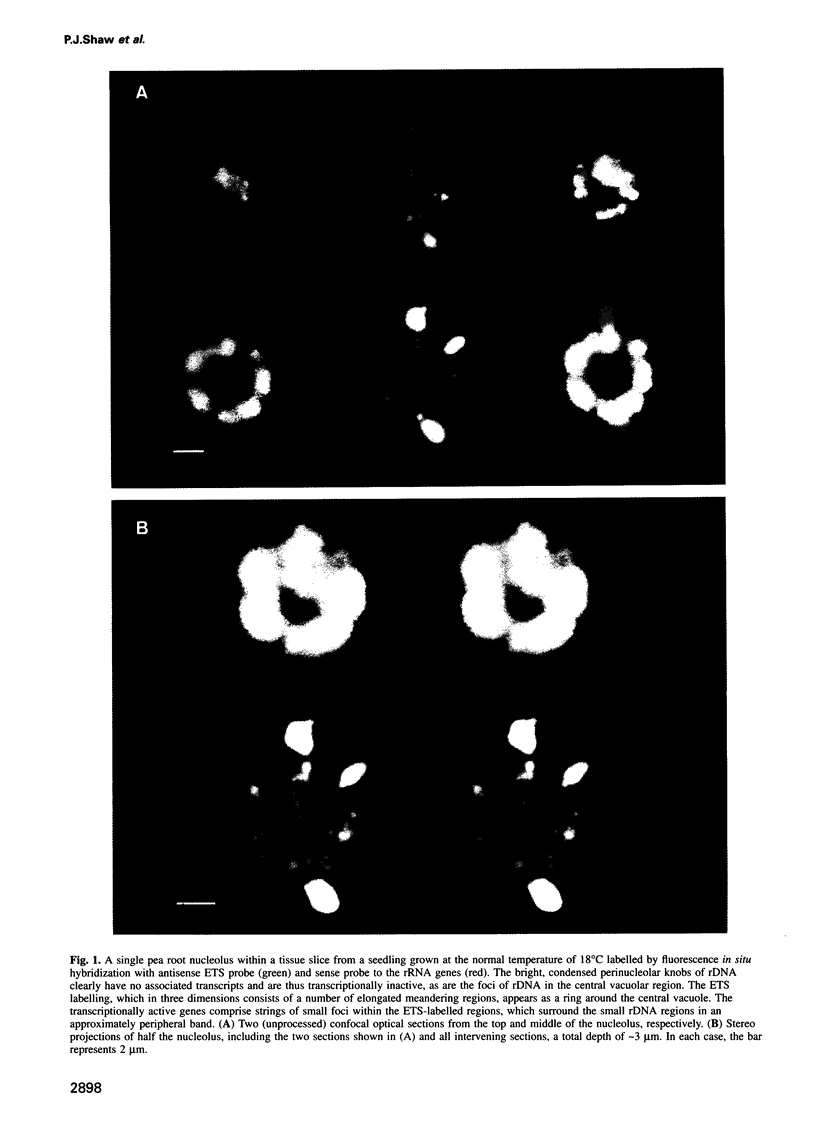
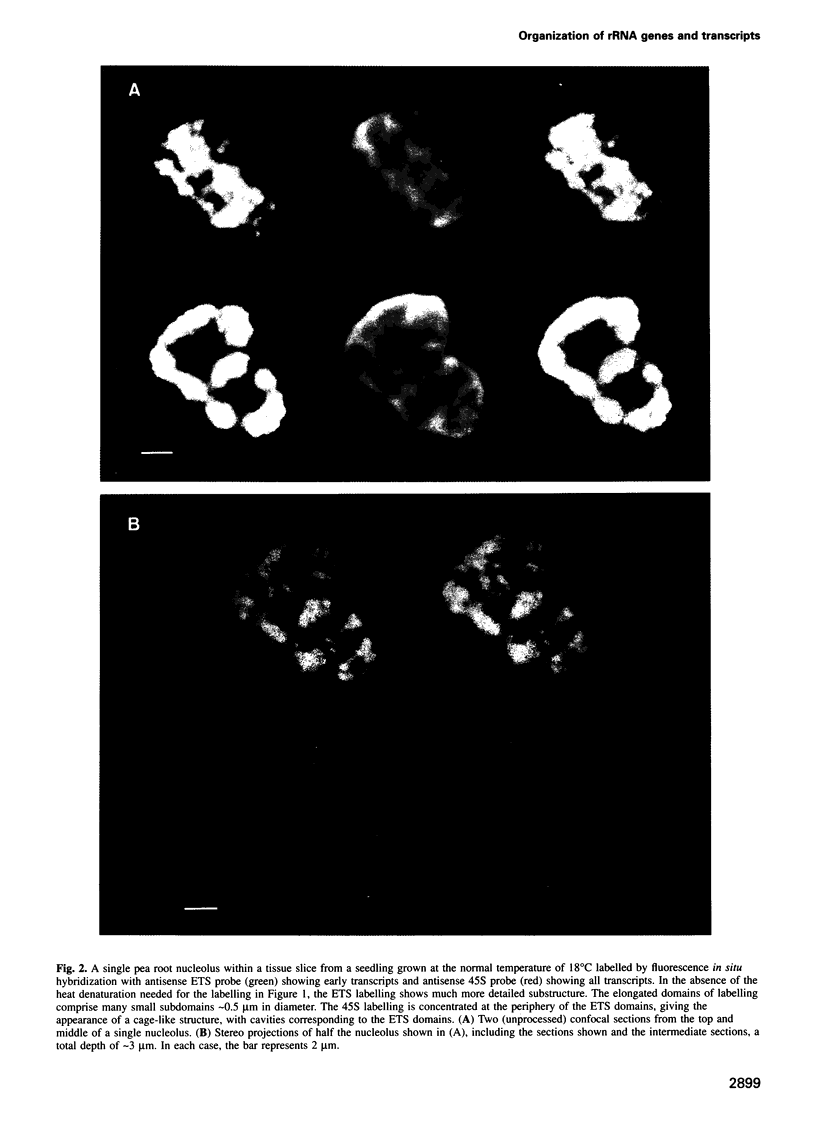
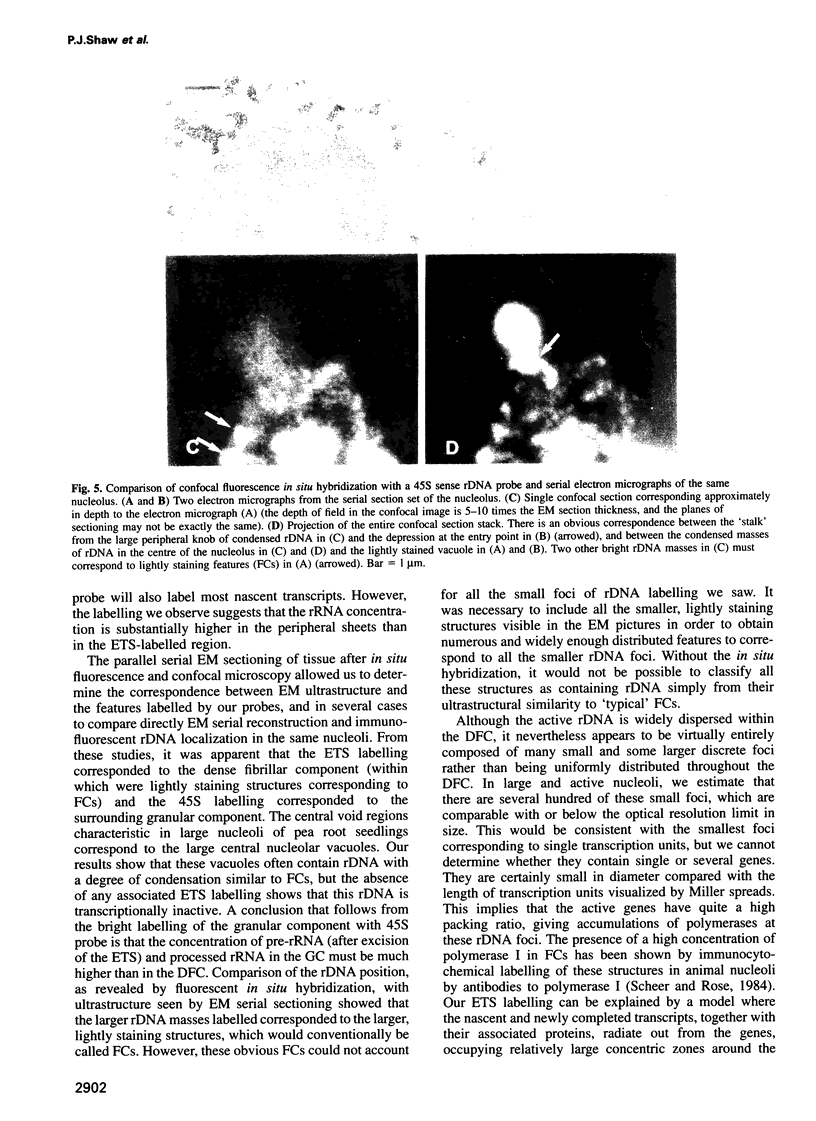
The nucleolus, the site of transcription and processing of the major ribosomal genes, generally reveals three distinct ultrastructural components in conventional thin-section electron micrographs (fibrillar centres, dense fibrillar component and granular component). We show here that different parts of the transcription and transcript processing pathway can be mapped to the different nucleolar components in pea root cells. This study shows the full three-dimensional arrangement of the different domains by in situ hybridization and confocal microscopy, and their correspondence with the major ultrastructural components of the nucleolus is revealed by parallel serial section electron microscopy. The active rDNA is widely dispersed in discrete foci, the larger of which, at least, correspond to well-defined fibrillar centres. A probe to the external transcribed spacer (ETS) sequence of the pre-rRNA transcripts labels clearly demarcated regions surrounding the foci of rDNA, and which we show correspond to the dense fibrillar component. Finally, a probe to the entire 45S transcript shows a higher concentration in regions corresponding to the granular component, surrounding the dense fibrillar component labelled by the ETS probe. The changes in structure that occur with heat shock show that nucleolar organization is dynamic and dependent upon transcriptional activity. These results show that the various RNA processing events are spatially highly organized and suggest a vectorial or radial model of transcription and transcript processing, where nascent and newly completed transcripts occupy zones surrounding the genes, which are in turn surrounded by regions containing the older more mature transcripts.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agard D. A., Hiraoka Y., Shaw P., Sedat J. W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Cox K. H., DeLeon D. V., Angerer L. M., Angerer R. C. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984 Feb;101(2):485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- Derenzini M., Farabegoli F., Trerè D. Localization of DNA in the fibrillar components of the nucleolus: a cytochemical and morphometric study. J Histochem Cytochem. 1993 Jun;41(6):829–836. doi: 10.1177/41.6.8315275. [DOI] [PubMed] [Google Scholar]
- Dundr M., Raska I. Nonisotopic ultrastructural mapping of transcription sites within the nucleolus. Exp Cell Res. 1993 Sep;208(1):275–281. doi: 10.1006/excr.1993.1247. [DOI] [PubMed] [Google Scholar]
- Geuskens M., Bernhard W. Cytochimie ultrastructurale du nucléole. 3. Action de l'actinomycine D sur le métabolisme du RNA nucléolaire. Exp Cell Res. 1966 Nov-Dec;44(2):579–598. doi: 10.1016/0014-4827(66)90462-9. [DOI] [PubMed] [Google Scholar]
- Goessens G. High resolution autoradiographic studies of ehrlich tumour cell nucleoli. Nucleolar labelling after [3H]actinomycin D binding to DNA or after [3H]TdR or [3H]uridine incorporation in nucleic acids. Exp Cell Res. 1976 Jun;100(1):88–94. doi: 10.1016/0014-4827(76)90330-x. [DOI] [PubMed] [Google Scholar]
- Highett M. I., Beven A. F., Shaw P. J. Localization of 5 S genes and transcripts in Pisum sativum nuclei. J Cell Sci. 1993 Aug;105(Pt 4):1151–1158. doi: 10.1242/jcs.105.4.1151. [DOI] [PubMed] [Google Scholar]
- Hozák P., Cook P. R., Schöfer C., Mosgöller W., Wachtler F. Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J Cell Sci. 1994 Feb;107(Pt 2):639–648. doi: 10.1242/jcs.107.2.639. [DOI] [PubMed] [Google Scholar]
- Hozák P., Hassan A. B., Jackson D. A., Cook P. R. Visualization of replication factories attached to nucleoskeleton. Cell. 1993 Apr 23;73(2):361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Hozák P., Schöfer C., Sylvester J., Wachtler F. A study on nucleolar DNA: isolation of DNA from fibrillar components and ultrastructural localization of different DNA probes. J Cell Sci. 1993 Apr;104(Pt 4):1199–1205. doi: 10.1242/jcs.104.4.1199. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Hassan A. B., Errington R. J., Cook P. R. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993 Mar;12(3):1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García L. F., Segura-Valdez M. L., Ochs R. L., Echeverría O. M., Vázquez-Nin G. H., Busch H. Electron microscopic localization of ribosomal DNA in rat liver nucleoli by nonisotopic in situ hybridization. Exp Cell Res. 1993 Aug;207(2):220–225. doi: 10.1006/excr.1993.1186. [DOI] [PubMed] [Google Scholar]
- Jordan E. G. Interpreting nucleolar structure: where are the transcribing genes? J Cell Sci. 1991 Apr;98(Pt 4):437–442. doi: 10.1242/jcs.98.4.437. [DOI] [PubMed] [Google Scholar]
- Jordan E. G. Nucleolar nomenclature. J Cell Sci. 1984 Apr;67:217–220. doi: 10.1242/jcs.67.1.217. [DOI] [PubMed] [Google Scholar]
- Kato A., Nakajima T., Yamashita J., Yakura K., Tanifuji S. The structure of the large spacer region of the rDNA in Vicia faba and Pisum sativum. Plant Mol Biol. 1990 Jun;14(6):983–993. doi: 10.1007/BF00019395. [DOI] [PubMed] [Google Scholar]
- Knibiehler B., Mirre C., Rosset R. Nucleolar organizer structure and activity in a nucleolus without fibrillar centres: the nucleolus in an established Drosophila cell line. J Cell Sci. 1982 Oct;57:351–364. doi: 10.1242/jcs.57.1.351. [DOI] [PubMed] [Google Scholar]
- Miller O. L., Jr, Beatty B. R. Visualization of nucleolar genes. Science. 1969 May 23;164(3882):955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- Mougey E. B., O'Reilly M., Osheim Y., Miller O. L., Jr, Beyer A., Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993 Aug;7(8):1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- Scheer U., Rose K. M. Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1431–1435. doi: 10.1073/pnas.81.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Thiry M., Goessens G. Structure, function and assembly of the nucleolus. Trends Cell Biol. 1993 Jul;3(7):236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- Scheer U., Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994 Jun;6(3):354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Schwarzacher H. G., Wachtler F. The functional significance of nucleolar structures. Ann Genet. 1991;34(3-4):151–160. [PubMed] [Google Scholar]
- Sollner-Webb B., Mougey E. B. News from the nucleolus: rRNA gene expression. Trends Biochem Sci. 1991 Feb;16(2):58–62. doi: 10.1016/0968-0004(91)90025-q. [DOI] [PubMed] [Google Scholar]
- Thiry M., Scheer U., Goessens G. Localization of DNA within Ehrlich tumour cell nucleoli by immunoelectron microscopy. Biol Cell. 1988;63(1):27–34. [PubMed] [Google Scholar]
- Thiry M. Ultrastructural distribution of DNA and RNA within the nucleolus of human Sertoli cells as seen by molecular immunocytochemistry. J Cell Sci. 1993 May;105(Pt 1):33–39. doi: 10.1242/jcs.105.1.33. [DOI] [PubMed] [Google Scholar]
- Vandelaer M., Thiry M., Goessens G. Ultrastructural distribution of DNA within the ring-shaped nucleolus of human resting T lymphocytes. Exp Cell Res. 1993 Apr;205(2):430–432. doi: 10.1006/excr.1993.1110. [DOI] [PubMed] [Google Scholar]
- Wachtler F., Hartung M., Devictor M., Wiegant J., Stahl A., Schwarzacher H. G. Ribosomal DNA is located and transcribed in the dense fibrillar component of human Sertoli cell nucleoli. Exp Cell Res. 1989 Sep;184(1):61–71. doi: 10.1016/0014-4827(89)90364-9. [DOI] [PubMed] [Google Scholar]
- Warner J. R. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990 Jun;2(3):521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Xing Y., Lawrence J. B. Nuclear RNA tracks: structural basis for transcription and splicing? Trends Cell Biol. 1993 Oct;3(10):346–353. doi: 10.1016/0962-8924(93)90105-a. [DOI] [PubMed] [Google Scholar]