Abstract
Aberrant hypo-methylation of DNA is evident in a range of human diseases including cancer and diabetes. Development of sensitive assays capable of detecting traces of un-methylated DNA within methylated samples can be useful in several situations. Here we describe a new approach, fast-COLD-MS-PCR, which amplifies preferentially un-methylated DNA sequences. By employing an appropriate denaturation temperature during PCR of bi-sulfite converted DNA, fast-COLD-MS-PCR enriches un-methylated DNA and enables differential melting analysis or bisulfite sequencing. Using methylation on the MGMT gene promoter as a model, it is shown that serial dilutions of controlled methylation samples lead to the reliable sequencing of un-methylated sequences down to 0.05% un-methylated-to-methylated DNA. Screening of clinical glioma tumor and infant blood samples demonstrated that the degree of enrichment of un-methylated over methylated DNA can be modulated by the choice of denaturation temperature, providing a convenient method for analysis of partially methylated DNA or for revealing and sequencing traces of un-methylated DNA. Fast-COLD-MS-PCR can be useful for the detection of loss of methylation/imprinting in cancer, diabetes or diet-related methylation changes.
Introduction
The role of DNA promoter methylation in cancer genetics has been widely studied and some of its mechanisms elucidated, however, the implication of DNA hypomethylation in oncogenesis is less well understood. Studies show a decrease in 5-methylcytosine levels in cancer tissues compared to their surrounding normal tissue in colon adenocarcinomas, Wilms' tumors and ovarian epithelial carcinomas, among others. The relationship of DNA hypomethylation to tumorigenesis is important to consider in view of cancer therapies that operate by decreasing DNA methylation. [1]. Furthermore, during gonad development gene imprinting takes place through parent of origin allele-specific methylation processes. The loss of imprinting (i.e. un-methylation of a previously methylated allele) may result in placental defects as well as in a series of genetic conditions that could increase the risk of cancer [2], [3]. Loss of methylation is also important in diabetes, where differential methylation occurs in genes that encode pre-proinsulin where β-cells are mainly un-methylated compared to other cell types. Detection of un-methylated pre-proinsulin DNA in serum has been associated to pancreatic β-cell death. Thus detecting DNA un-methylation at early stages of disease progression may provide actionable interventions in pre-diabetic stages [4]. Given such possibilities, the identification of small traces of un-methylated DNA within methylated DNA is of significance and is clinically desirable in cancer, diabetes and in other fields where epigenetics provides a potential link between lifestyle (diet, alcohol, smoking) with diabetes, obesity and cancer [5], [6].
A range of methods currently exist for detecting methylated or un-methylated DNA including methylation-specific PCR [7]–[10], differential enzymatic restriction methods [11], isothermal amplification methods [12], high resolution melting [13] or bisulfite sequencing [14] carried out as traditional Sanger sequencing or using next generation sequencing technologies. By revealing the status of individual CpG di-nucleotides, sequencing provides the most complete information on the methylation status of a target region. On the other hand real-time methylation-specific PCR is able to detect the presence of rare un-methylated alleles among an excess of methylated alleles or vice versa, something which is difficult to achieve with bisulfite sequencing alone. Below we demonstrate that Co-amplification at Lower Denaturation temperature PCR, COLD-PCR, [15]–[17], can be adapted for detection and sequencing of traces of un-methylated alleles within a population of methylated alleles. This approach relies on the substitution of the denaturation temperature (typically 95–98°C) by a decreased denaturation temperature (critical denaturation temperature, Tc). This allows the preferential amplification of sequences that have a lower melting temperature. Detection can be performed using an intercalating dye and melting analysis, followed by direct sequencing of the resulting amplicons without the need for specific reagents or use of special instrumentation.
We chose methylation of the gene encoding for O6 methylguanine methyl transferase (MGMT) as an example to validate COLD Methylation-specific (MS)-PCR in DNA from glioblastoma or infant blood samples. MGMT is of particular importance in glioblastoma. It encodes an evolutionarily conserved protein whose primary function is to repair guanine nucleotides that are alkylated at the O6 position [18]. As O6-methyl guanine constitutes the major cytotoxic lesion related to temozolomide, the only efficacious chemotherapy against glioblastoma, MGMT serves as the primary determinant of therapeutic response. MGMT promoter methylation leads to decreased expression of MGMT, and this methylation pattern have been associated with improved therapeutic response by three independent, well-conducted clinical trials, including the EORTIC-NCIC, NOA-8, and the Nordic trials [19]. As such, methodologies for sensitive and reproducible detection MGMT promoter methylation would better facilitate clinical management of glioblastoma patients. Using COLD-MS-PCR we demonstrate that we are able to amplify and sequence 0.05% of un-methylated MGMT DNA within an excess of 99.95% of methylated MGMT DNA.
Materials and Methods
DNA, clinical samples and bisulfite treatment
Un-methylated and enzymatically methylated DNA (Millipore Corporation, Billerica, MA, USA) were obtained and subjected to bisulfite treatment using the EpiTect Bisulfite kit following manufacturer's recommendations (Qiagen Inc.). Additional Epitect bisulfite-converted un-methylated and methylated controls were obtained from Qiagen. Serial dilutions of un-methylated and methylated DNA were prepared.
Two sets of clinical samples were tested, chosen for their potential to harbor methylation (or lack of methylation) on the MGMT gene. Glioma tumor tissue specimens (N = 20), obtained from the Division of Neurosurgery at the Beth Israel – Deaconess Medical Center; and blood samples (N = 10) isolated from blood of infants at the Department of Pediatrics, Boston Medical Center. Infant samples were obtained after written informed consent by the mothers and following approval by the IRB committee of Boston Medical Center. Glioma samples were also obtained following written consent prior to surgery (BIDMC Division of Neurosurgery) and following approval by the IRB committee of DFCI/BIDMC. These samples were processed for DNA isolation using the DNAeasy Blood & Tissue Kit (Qiagen) as described [20], followed by bisulfite sequencing.
Methylation-specific fast-COLD-PCR (fast-COLD-MS-PCR)
Fast-COLD-PCR is the simplest version of COLD-PCR [15]. It leads to the preferential amplification of sequences containing genomic alterations that result to a reduction of the melting temperature of the PCR amplicon and combines well with downstream detection formats including high resolution melting [21]–[24]. This property is exploited here for sensitive detection of un-methylated DNA. During sodium bisulfite conversion of DNA, deamination of un-methylated cytosines generates uracils whereas 5-methylcytosines remain unaffected. In a subsequent PCR reaction uracil residues are amplified as thymines and 5-methyl-cytosines as cytosines [14]. This results in a large difference in GC content and melting temperature differential changes that can then be exploited by COLD-PCR for selectively amplifying un-methylated DNA.
PCR reactions, specific for bisulfite-converted DNA, were designed and optimized to amplify a region of MGMT (O-6-methyl-guanine-DNA methyltransferase) gene. A pre-amplification of 379 bp using primers that bind target regions without CpG sites 5′-TTTGTTTTTTTTAGGTTTT-3′ and 5′-CCAAAAACCCCAAACCCGA-3′ with an annealing temperature of 48°C was performed as reported [25]. A second, nested PCR reaction was designed using 5′-GGATATGTTGGGATAGTT-3′ [25] and 5′-CACCTAAAAAACACTTAAAAC-3′ with an annealing temperature of 52°C to produce a 255 bp amplicon. The second reaction was performed in a fast-COLD-PCR format to favor the amplification of un-methylated DNA. Alternatively a nested conventional PCR reaction was performed for comparison. PCR reagents for both first and nested PCR reactions comprised 1× Colorless Gotaq reaction buffer, 2 mM MgCl2, 0.2 mmol/L each dNTP, 0.25 µM each primer, 0.1× LCGreen+ dye (Idaho Technologies Inc.), and 1 unit/ul GoTaq polymerase (Promega, Madison, WI, USA). Fast-COLD-MS-PCR thermocycling protocol for amplification on the SmartCycler II (Cepheid, Inc.) was (5 cycles of conventional PCR followed by 35 cycles at the selected critical denaturation temperature (Tc) for 10 s, 52°C for 30 s and 72°C for 20 s with a final extension at 72°C). Conventional PCR protocol was (initial denaturation at 95°C for 2 min, then 40 cycles at 95°C for 30 s, 52°C for 30 s and 72°C for 20 s with a final extension at 72°C for 5 min). Immediately following the extension step of the nested reaction, PCR amplicons were subjected to melting curve analysis on the Smartcycler II. Amplicons were processed for Sanger sequencing. Experiments were repeated three independent times.
Results and Discussion
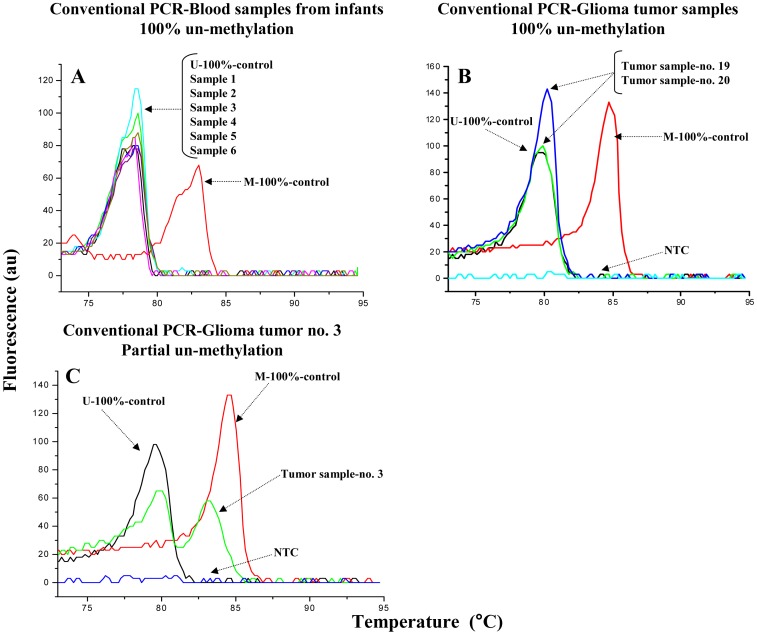
Melting profiles of MGMT target gene regions using conventional PCR-amplification of bisulfite-converted DNA from glioma tumor samples and infant blood samples, along with fully un-methylated and fully methylated DNA controls are depicted on Figure 1, A-C . All the samples from infant blood generated amplicons with melting profiles matching the 100% un-methylated control DNA at the MGMT promoter region, and six representative samples are depicted on Figure 1A . The fully un-methylated and fully methylated DNA produced amplicons with melting temperatures (Tm) that differed by approximately 5°C ( Figure 1A ). Out of 20 glioma samples tested, 19 samples generated amplicons with melting profiles consistent with 100% un-methylated control DNA. Figure 1B depicts two such un-methylated glioma samples. A single glioma sample (no. 3) generated a melting profile consistent with a mix of methylated plus un-methylated DNA sequences at the MGMT promoter region, which was different from the other glioma tumor samples used to explain the reliability and stability of the approach ( Figure 1C ).
Figure 1. Melting profiles of bisulfite-converted DNA from clinical samples following conventional PCR.
Post-PCR melting profiles of the 255 bp bisulfite-converted MGMT gene amplicon after conventional PCR. Examples of fully un-methylated DNA samples isolated from infant blood (Panel A) and glioma samples (Panel B) are depicted. 100% methylated (M) and 100% un-methylated (U) DNA controls are used as reference standards, demonstrating a ∼5°C melting temperature difference among the two. Panel C. A glioma sample with mixed methylation/unmethylation pattern is shown.
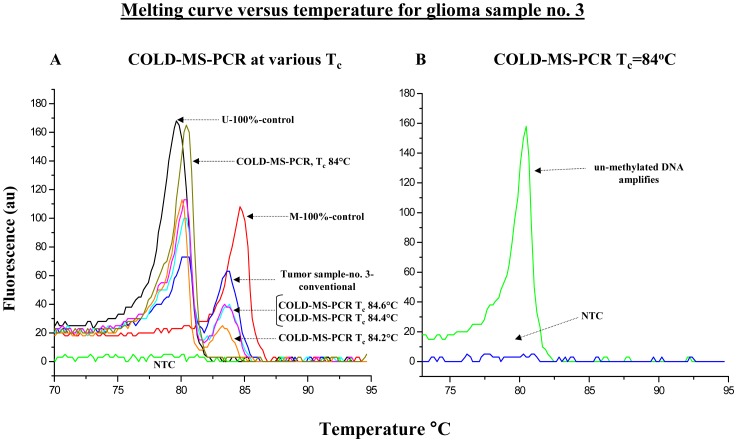
Next we demonstrated that by modulating the denaturation temperature during PCR, we can select with different efficiency the un-methylated DNA fraction of a give sample. In Figure 2 , Panel A, we analyzed the effect of gradually lowering the denaturation temperature during PCR, using for this purpose glioma sample no. 3 with a mixture of un-methylated and methylated DNA. By gradully shifting the Tc during PCR, a gradual shifting of the un-methylated and methylated DNA portions is observed. When a Tc of 84°C was used during fast- COLD-MS-PCR, the methylated peak disappeared in the subsequent melting profiles, consistent with an almost exclusive amplification of un-methylated DNA ( Figure 2B ).
Figure 2. Melting profiles of bisulfite-converted DNA from clinical samples following fast-COLD-MS-PCR.
The effect of lowering the denaturation temperature in PCR is depicted. Panel A. Glioma sample no. 3 was subjected to different critical denaturation temperature-Tc during fast-COLD-MS-PCR. The modulation of the preferential amplification of the un-methylated DNA fraction is shown. Panel B. fast-COLD-MS-PCR performed at a Tc of 84°C demonstrates that the amplification of the methylated DNA fraction is completely inhibited.
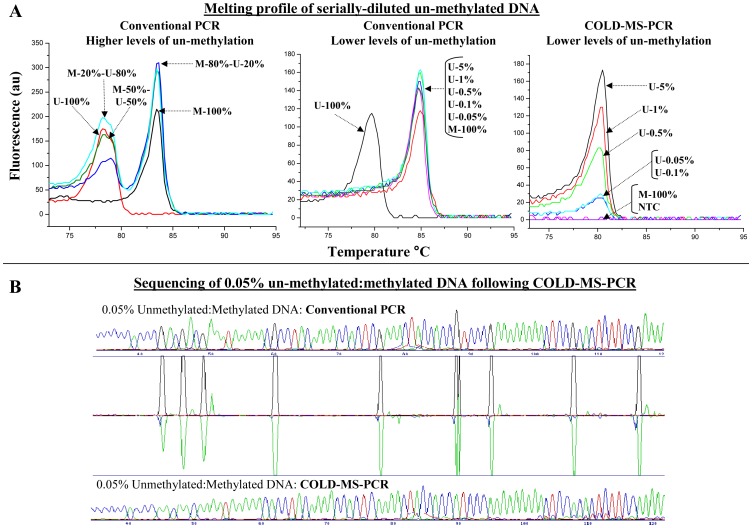
To examine the lowest fraction of un-methylated MGMT DNA within an excess of methylated MGMT DNA that can be preferentially amplified, detected and sequenced using fast- COLD-MS-PCR we tested serial dilutions of un-methylated to methylated DNA ( Figure 3A ). Using conventional MS-PCR, fractions of un-methylated-to-methylated DNA of 20–80% generate the anticipated double peaks in their melting profiles. However, in fractions below 5% the methylated peak cannot be observed in either the melting profiles or the sequencing chromatograms. In contrast, fast-COLD-MS-PCR can enrich and detect the un-methylated DNA down to 0.05%. Furthermore, direct sequencing of the product from an initial 0.05% un-methylated DNA reveals un-methylated C at individual CpG sequence positions Figure 3B .
Figure 3. Detection of un-methylated DNA within different abundances of methylated DNA background by conventional or fast-COLD-MS-PCR.
Panel A. Post-PCR melting profile of the 255 bp bisulfite-converted-specific amplicon after conventional PCR or COLD-PCR. Serial dilutions of un-methylated (U) to methylated (M) genomic DNA are depicted (top half). Higher abundances of un-methylated DNA can be discriminated from methylated DNA by the melt peak, whereas lower abundances are only detectable if fast-COLD-MS-PCR replaces conventional PCR. Panel B. Sanger sequencing results of the 0.05% un-methylated (U): methylated (M) DNA sample as amplified by conventional and fast-COLD-MS-PCR are shown (bottom half). Chromatograms are aligned and compared using SeqDoc, and the CpG methylation positions are revealed in the middle panel.
Fast-COLD-MS-PCR utilizes primers that bind sequence positions that do not contain CpG sites, i.e. they are neutral in regards to CpG methylation. This is a potential advantage over approaches that utilize methylation-specific primers, such as MS-PCR or methyl-Light [7], [8], as the methylation status of the primer binding sites ideally should not pre-determine which fraction of the target amplicons can be studied. Approaches like COBRA [11] or methylation sensitive high resolution melting [13] employ CpG-neutral primer binding sites and detect low levels of un-methylation, but they do not enable bisulfite sequencing of the product. The ability of fast-COLD-MS-PCR to modulate the enrichment effect using graded denaturation temperatures provides a convenient method for analysis of partially methylated DNA or for revealing and sequencing traces of un-methylated DNA. Using fast-COLD-MS-PCR enabled the detection of traces of un-methylated sequences by virtue of the Tm-lowering effect of C>T transversions. On the other hand using different versions of COLD-PCR (full-COLD-PCR [26]; ice-COLD-PCR [27]) it should also be possible to reverse the process, i.e. enrich small amounts of methylated DNA within an excess of un-methylated DNA.
In summary, COLD-PCR technology can be adapted to reveal traces of un-methylated DNA within a high excess of methylated DNA.
When fast-COLD-MS-PCR was used to analyze bisulfite-converted clinical samples and serially-diluted un-methylated DNA in methylated DNA, low abundances of un-methylated MGMT DNA, down to 0.05%, could be preferentially enriched and sequenced. Thereby one can enrich and sequence traces of un-methylated DNA which can precede disease progression in cancer and diabetes, can be predictive of chemotherapy-caused remission and are linked to diet and lifestyle-induced epigenetic changes.
Funding Statement
This work was supported by NCI grants 1R03CA139406 (AM); R21CA-155615 and R21CA-175542 (GMM). The contents of this manuscript do not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ehrlich M (2002) DNA methylation in cancer: too much, but also too little. Oncogene 21: 5400–5413. [DOI] [PubMed] [Google Scholar]
- 2. McGraw S, Oakes CC, Martel J, Cirio MC, de Zeeuw P, et al. (2013) Loss of DNMT1o Disrupts Imprinted X Chromosome Inactivation and Accentuates Placental Defects in Females. PLoS Genet 9: e1003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jelinic P, Shaw P (2007) Loss of imprinting and cancer. J Pathol 211: 261–268. [DOI] [PubMed] [Google Scholar]
- 4. Fisher MM, Perez Chumbiauca CN, Mather KJ, Mirmira RG, Tersey SA (2013) Detection of islet beta-cell death in vivo by multiplex PCR analysis of differentially methylated DNA. Endocrinology 154: 3476–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waterland RA, Jirtle RL (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23: 5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waterland RA (2009) Is epigenetics an important link between early life events and adult disease? Horm Res 71 Suppl 1: 13–16. [DOI] [PubMed] [Google Scholar]
- 7. Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, et al. (2000) MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 28: E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 93: 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonanno C, Shehi E, Adlerstein D, Makrigiorgos GM (2007) MS-FLAG, a novel real-time signal generation method for methylation-specific PCR. Clin Chem 53: 2119–2127. [DOI] [PubMed] [Google Scholar]
- 10. Roy K, Wang L, Makrigiorgos GM, Price BD (2006) Methylation of the ATM promoter in glioma cells alters ionizing radiation sensitivity. Biochem Biophys Res Commun 344: 821–826. [DOI] [PubMed] [Google Scholar]
- 11. Xiong Z, Laird PW (1997) COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 25: 2532–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zerilli F, Bonanno C, Shehi E, Amicarelli G, Adlerstein D, et al. (2010) Methylation-specific loop-mediated isothermal amplification for detecting hypermethylated DNA in simplex and multiplex formats. Clin Chem 56: 1287–1296. [DOI] [PubMed] [Google Scholar]
- 13. Wojdacz TK, Dobrovic A (2007) Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res 35: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, et al. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A 89: 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J, Wang L, Mamon H, Kulke MH, Berbeco R, et al. (2008) Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med 14: 579–584. [DOI] [PubMed] [Google Scholar]
- 16. Li J, Makrigiorgos GM (2009) COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing. Biochem Soc Trans 37: 427–432. [DOI] [PubMed] [Google Scholar]
- 17. Milbury CA, Li J, Liu P, Makrigiorgos GM (2011) COLD-PCR: improving the sensitivity of molecular diagnostics assays. Expert Rev Mol Diagn 11: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Y, Hu F, Zhou Y, Chen W, Shao H, et al. (2013) MGMT promoter methylation and glioblastoma prognosis: a systematic review and meta-analysis. Arch Med Res 44: 281–290. [DOI] [PubMed] [Google Scholar]
- 19. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, et al. (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10: 459–466. [DOI] [PubMed] [Google Scholar]
- 20. Milbury CA, Chen CC, Mamon H, Liu P, Santagata S, et al. (2011) Multiplex amplification coupled with COLD-PCR and high resolution melting enables identification of low-abundance mutations in cancer samples with low DNA content. J Mol Diagn 13: 220–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milbury CA, Li J, Makrigiorgos GM (2009) COLD-PCR-enhanced high-resolution melting enables rapid and selective identification of low-level unknown mutations. Clin Chem 55: 2130–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kristensen LS, Daugaard IL, Christensen M, Hamilton-Dutoit S, Hager H, et al. (2010) Increased sensitivity of KRAS mutation detection by high-resolution melting analysis of COLD-PCR products. Hum Mutat 31: 1366–1373. [DOI] [PubMed] [Google Scholar]
- 23. Mancini I, Santucci C, Sestini R, Simi L, Pratesi N, et al. (2010) The use of COLD-PCR and high-resolution melting analysis improves the limit of detection of KRAS and BRAF mutations in colorectal cancer. J Mol Diagn 12: 705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pritchard CC, Akagi L, Reddy PL, Joseph L, Tait JF (2010) COLD-PCR enhanced melting curve analysis improves diagnostic accuracy for KRAS mutations in colorectal carcinoma. BMC Clin Pathol 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, et al. (2000) Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 60: 5954–5958. [PubMed] [Google Scholar]
- 26. Li J, Milbury CA, Li C, Makrigiorgos GM (2009) Two-round coamplification at lower denaturation temperature-PCR (COLD-PCR)-based sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma. Hum Mutat 30: 1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Milbury CA, Li J, Makrigiorgos GM (2011) Ice-COLD-PCR enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations. Nucleic Acids Res 39: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]