Abstract
Objectives
This study was conducted to examine the development and molecular mechanisms of amphenicol resistance in Campylobacter jejuni by using in vitro selection with chloramphenicol and florfenicol. The impact of the resistance development on growth rates was also determined using in vitro culture.
Methods
Chloramphenicol and florfenicol were used as selection agents to perform in vitro stepwise selection. Mutants resistant to the selective agents were obtained from the selection process. The mutant strains were compared with the parent strain for changes in MICs and growth rates. The 23S rRNA gene and the L4 and L22 ribosomal protein genes in the mutant strains and the parent strain were amplified and sequenced to identify potential resistance-associated mutations.
Results
C. jejuni strains that were highly resistant to chloramphenicol and florfenicol were obtained from in vitro selection. A novel G2073A mutation in all three copies of the 23S rRNA gene was identified in all the resistant mutants examined, which showed resistance to both chloramphenicol and florfenicol. In addition, all the mutants selected by chloramphenicol also exhibited the G74D modification in ribosomal protein L4, which was previously shown to confer a low-level erythromycin resistance in Campylobacter species. The mutants selected by florfenicol did not have the G74D mutation in L4. Notably, the amphenicol-resistant mutants also exhibited reduced susceptibility to erythromycin, suggesting that the selection resulted in cross resistance to macrolides.
Conclusions
This study identifies a novel point mutation (G2073A) in 23S rRNA in amphenicol-selected mutants of C. jejuni. Development of amphenicol resistance in Campylobacter likely incurs a fitness cost as the mutant strains showed slower growth rates in antibiotic-free media.
Introduction
Campylobacter jejuni is a leading bacterial cause of acute gastroenteritis in humans [1]. Campylobacter infection is also the most common antecedent to Guillain-Barré syndrome (GBS), manifested as symmetric ascending paralysis [2], [3]. Usually, Campylobacter infections are self-limiting; however, clinical antimicrobial treatment is warranted in patients with severe or long-lasting infections, or with compromised immune systems [4], [5]. However, antimicrobial resistance in C. jejuni has increased significantly over the past decades, compromising clinical treatments and presenting a major public health threat [6]–[9].
Chloramphenicol (CHL) and florfenicol (FFC) are members of the amphenicol family, which are highly effective against a wide variety of Gram-positive and Gram-negative bacteria. Amphenicols were once widely applied in both human and veterinary practice for the prevention and treatment of many bacterial infections. Florfenicol, the fluorinated derivative of chloramphenicol, has been licensed for the control of bacterial respiratory tract infections in several food production animals, including cattle and pigs [10]. Nowadays, the use of chloramphenicol is limited to a small number of life-threatening infections in humans because of its adverse effects, which include bone-marrow depression, aplastic anaemia, and acute leukaemia [11]–[13]. The use of chloramphenicol in food-producing animals was banned in many countries; however, it is still widely used in pets and non-food-producing animals [14]–[16]. Chloramphenicol binds directly to the peptidyltransferase centre on the 50S ribosomal subunit, preventing peptide bond formation [17]. Acetylation of the drug by chloramphenicol acetyltransferase, which does not mediate resistance to FFC, is the most frequently encountered mechanism of bacteria resistance to CHL [10]. Other mechanisms of CHL resistance include efflux systems such as cmlA [18], floR [19], fexA [20], fexB [21], pexA [22]; point mutations in domain V of the 23S rRNA [23]; and methylation of A2503 in the 23S rRNA gene of the large ribosomal subunit, which is catalysed by cfr methyltransferase [24].
A series of studies showed that CHL resistance was very common in E. coli (40.6–79%) [25], [26] and Salmonella (28.42%) [27], while chloramphenicol resistance in Campylobacter rarely occurs. No chloramphenicol-resistant isolates were detected in studies performed in the United States [28], Spain [29], Japan [30], and Iran [31]. The chloramphenicol resistance detection rates were 0.8% in Korea [32]. In contrast to the low resistance rates in other countries, detection of chloramphenicol-resistant Campylobacter was high (37.5%) in Brazil [33]. In China, the florfenicol and chloramphenicol resistance rates of Campylobacter isolated from chicken [34] reached up to 61.7 and 24.5%, respectively. Chloramphenicol resistance in Campylobacter is mediated by chloramphenicol acetyltransferases [35]–[37]. To date, there have been no reports on FFC resistance mechanisms in Campylobacter. In this study, we examined the development of CHL and FFC resistance mechanisms in C. jejuni by in vitro selection and assessed the impact of the resistance on Campylobacter fitness.
Materials and Methods
Bacterial strains and culture conditions
The C. jejuni strain ATCC 33560, susceptible to CHL and FFC (MICCHL = 4 µg/mL and MICFFC = 2 µg/mL), was used as the parent strain for the selection studies with CHL and FFC. C. jejuni was cultured routinely on Mueller-Hinton agar (MHA, Sigma, St. Louis, MO) or in Mueller-Hinton broth (MHB, Sigma) for 24–48 hours at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2). If necessary, the MH media were supplemented with various concentrations of CHL or FFC. All strains were preserved in MH broth with 20% glycerol at −80°C.
CHL & FFC susceptibility testing
The MICs against CHL and FFC were determined using the agar dilution method as recommended by the Clinical and Laboratory Standards Institute [38]. According to the recommended breakpoints of NARMS [39], isolates were considered resistant to CHL and FFC if their MICs were ≥32 and ≥8 µg/mL, respectively. CHL, FFC, erythromycin, azithromycin, spiramycin, and clindamycin were purchased from the China Institute of Veterinary Drug Control, Beijing. Phenylalanine-arginine-β-naphthylamide (PAβN, Sigma-Aldrich, St. Louis, MO), an efflux pump inhibitor, was also used in this study to identify whether the antibiotic efflux systems of C. jejuni played a role in the resistance. The concentration of PAβN used in this study was 30 µg/mL [40]. A control plate of MH agar containing PAβN of the same concentration was also included to assess the effect of PAβN on the growth of the isolates investigated. The experiments mentioned above were repeated three times.
In vitro selection of CHL and FFC resistant strains
CHL- and FFC-resistant strains were selected in vitro with CHL and FFC as the selective agents, respectively. Briefly, the parent strain ATCC 33560 was first cultured on antimicrobial-free MHA media. For first round of selection, cultures of the parent strain were inoculated onto MHA media containing CHL or FFC at 0.5-fold MICs, and incubated under microaerobic conditions at 42°C for 3–5 days. For subsequent stepwise selection, cultures were collected and transferred to MHA containing a 2-fold higher concentration of antibiotics than the concentration used in the preceding round of selection. The highest concentrations of selective agents are 16-fold and 64-fold MIC for CHL and FFC, respectively. Single colonies (n = 10) were picked and tested at each stepwise selection. MICs of resistant clones against CHL and FFC and erythromycin, azithromycin, spiramycin, and clindamycin were determined accordingly.
Growth rates of the parent and mutant strains in MH broth
To compare the growth kinetics of selected resistant strains (C3 and F5) with the susceptible parent strain, a fresh culture of each strain was separately and equally inoculated into antimicrobial-free MH broth at an initial cell density of OD600 = 0.05. The cultures were incubated at 42°C under microaerobic conditions, and then aliquots of the samples were collected at 0, 4, 8, 12, 24, and 36 hours post-inoculation for OD600 determination. Three independent experiments were performed.
Sequence analysis of genes encoding 23S rRNA, L4 ribosomal protein, and L22 ribosomal protein
With respect to the 23S rRNA gene, we used the specific primer pair 23S-F and 23S-R to amplify all three copies of the 23S rRNA gene to detect potential CHL or FFC resistance-associated mutations (Table 1). To detect which specific copy of the gene contained the mutation, PCRs were performed to amplify each copy of the domain V of 23S rRNA gene. The three pairs of operon-specific PCR primers (FI/CJ copy-R; FII/CJ copy-R; and FIII/CJ copy-R) are listed in Table 1 [41]. The amplification reactions were performed with premixed LA Taq (TaKaRa Co. Ltd., Dalian, China), and the cycling parameters were 95°C for 5 min, followed by 30 cycles of 95°C for 30 sec for denaturation, 55°C for 30 sec for annealing, and 68°C for 5 min for extension. The PCR cycles were followed by a final extension period of 15 min at 68°C. The PCR products were purified by using TIANGEN DNA midi purification kit (TIANGEN, Beijing, China) and subsequently sequenced.
Table 1. Primers used in this article.
| Primers | Sequence (5′-3′) | References |
| 23S-F | 5′-AGCTACTAAGAGCGAATGGT-3′ | this study |
| 23S-R | 5′-AAAGATAAGCCAAACGCTCT-3′ | this study |
| FI | 5′-CCCTAAGTCAAGCCTTTCAATCC-3′ | [40] |
| FII | 5′-CGTTATAGATACGCTTAGCGGTTATG-3′ | [40] |
| FIII | 5′-CATCGAGCAAGAGTTTATGCAAGC-3′ | [40] |
| CJ copy-R | 5′-CTACCCACCAGACATTGTCCCAC-3′ | [40] |
| L4-F | 5′-GTAGTTAAAGGTGCAGTACCA-3′ | [40] |
| L4-R | 5′-GCGAAGTTTGAATAACTACG-3′ | [40] |
| L22-F | 5′-GAATTTGCTCCAACACGC-3′ | [40] |
| L22-R | 5′-ACCATCTTGATTCCCAGTTTC-3′ | [40] |
The sequences of the L4 and L22 ribosomal protein genes from the resistant clones were analysed in comparison with the parental strain using primers as previously described [42]. The PCR cycling conditions for the L4 and L22 genes were as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 45 sec, with a final extension at 72°C for 5 min. The PCR products were purified and sequenced for point mutations.
Results
Development of CHL and FFC resistance in C. jejuni by in vitro selection
Through independent stepwise in vitro selection experiments with CHL or FFC as the selective agents, we acquired C. jejuni strains of different resistance levels to the antibiotics. The selected resistant strains are listed in Table 2 . Three successive generations of CHL-resistant strains (MICC1 = 32 µg/mL; MICC2 = 64 µg/mL; and MICC3 = 128 µg/mL) and five successive generations of FFC-resistant strains (MICF1 = 16 µg/mL; MICF2 = 32 µg/mL; MICF3 = 64 µg/mL; MICF4 = 128 µg/mL; and MICF5 = 256 µg/mL) were chosen for further analysis. MICs of these strains were determined with or without PAβN in comparison with the parent strain ATCC 33560. As shown in Table 2, the selected mutants showed ≥8-fold increases in the MICs of CHL and FFC compared to ATCC33560. Addition of PAβN in the assay made little differences in the MICs (no change or 2-fold decrease; Table 2), and the PAβN alone did not affect the growth of the isolates investigated. As shown in Table 2, all CHL-resistant strains and high-level FFC-resistant strains (including F3, F4, and F5) showed decreased susceptibility to the both selective agents. Interestingly, the MICs of erythromycin against mutant strains C3 and F5 increased eight-fold (from 1 to 8 µg/mL), and the MICs of azithromycin increased four-fold (from 0.25 to 1 µg/mL), but were still lower than the resistance breakpoints (≥16 and ≥8 µg/mL, respectively), suggesting that selection with amphenicol led to a low-level cross-resistance to macrolides. However, the MICs of spiramycin and clindamycin only increased two-fold (data not shown).
Table 2. Target mutations identified in CHL- and FFC-resistant mutants.
| MIC (µg/mL)b | ||||||||||
| Strains a | CHL | CHL+PAβN | FFC | FFC+PAβN | Erythromicin | Azithromycin | Mutation in the 23S rRNA gene c | Change in protein L4 | Change in protein L22 | Selective Agents e |
| ATCC33560 | 4 | 4 | 2 | 2 | 1 | 0.25 | -d | - | - | NA |
| C1 | 32 | 32 | 16 | 16 | 8 | 1 | - | G74D | - | CHL |
| C2 | 64 | 32 | 32 | 16 | 8 | 1 | G2073A | G74D | - | CHL |
| C3 | 128 | 64 | 64 | 64 | 8 | 1 | G2073A | G74D | - | CHL |
| F1 | 8 | 4 | 16 | 8 | 2 | 0.5 | - | - | - | FFC |
| F2 | 16 | 8 | 32 | 32 | 8 | 1 | G2073A | - | - | FFC |
| F3 | 32 | 8 | 64 | 32 | 8 | 1 | G2073A | - | - | FFC |
| F4 | 64 | 16 | 128 | 64 | 8 | 1 | G2073A | - | - | FFC |
| F5 | 64 | 32 | 256 | 128 | 8 | 1 | G2073A | - | - | FFC |
C1-C3 were selected with CHL, while F1-F5 were selected with FFC.
Determined by the standard agar dilution method. ND, not determined.
The G2073A mutation was present in three copies of the domain V region of 23s rRNA gene.
no mutation detected.
CHL: Chloramphenicol; FFC: Florfenicol; NA: not applicable.
Molecular mechanism of Chloramphenicol and Florfenicol resistance
Analysis of domain V of the 23S rRNA gene
PCRs were performed with specific primers 23S-F and 23S-R (Table 1 ) to identify the potential mutations in domain V of the 23S rRNA gene in the selected resistant C. jejuni strains. Sequence analysis showed that a G-to-A single nucleotide change was detected at position 2073 of 23S rRNA (corresponding to position 2057 in the 23S rRNA gene of Escherichia coli) of all the mutants examined (Table 2). Further studies were performed with operon-specific primer pairs (FI/Cj copy R, FII/Cj copy R, and FIII/Cj copy R) to amplify and analyse the three copies of the 23S rRNA gene in the mutant strain. The results indicated that these mutants exhibited the G2073A mutation in all three copies of the gene (Table 2 ). When analysing the process of resistance development, we did not found the mutation in any of the three copies of the 23S rRNA gene in the FFC-selected clones with MICFFC ≤16 µg/mL and the CHL-selected clones with MICCHL ≤32 µg/mL.
Sequence analysis of L4 and L22 ribosomal protein genes
The selected mutant strains and the parent strain were examined for alterations in the L4 and L22 ribosomal protein genes. The G74D change, which was previously reported to confer low-level resistance to erythromycin in Campylobacter [42]–[44], was detected in the L4 protein in all the CHL-selected mutants (MICCHL≥16 µg/mL), but was absent in the FFC-selected strains, regardless of the resistance levels FFC (Table 2). No mutations were detected in the L22 ribosomal protein gene in either CHL-selected or FFC-selected mutants (Table 2).
Growth kinetics of the resistant strains
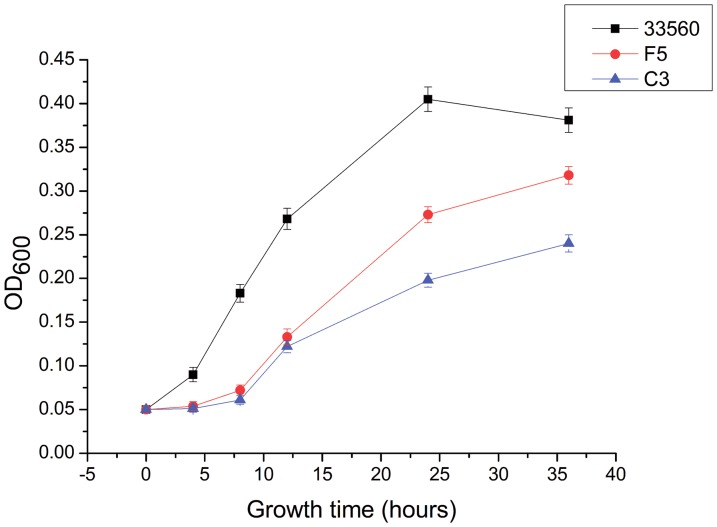
Two mutant strains (C3 and F5) and the parent strain were separately inoculated into antimicrobial-free MH broth and tested for growth rates. For the first 8 hours of incubation post-inoculation, the two mutant strains showed little growth (Figure 1). According to the OD values, the C3 and F5 strains showed apparent differences in growth kinetics between the wild-type parent strain and the mutants, suggesting that the C3 and F5 mutants might have impaired fitness.
Figure 1. Growth kinetics of the wild-type strain and mutant strains of C. jejuni in MH broth.
The OD600 were measured at 0, 4, 8, 12, 24 and 36 hours post-inoculation. Two mutant strains (C3 and F5) and the parent strain ATCC 33560 were included in the experiment. The experiment was repeated three times, and the results were shown as mean of three independent experiments.
Discussion
C. jejuni has become increasingly resistant to antimicrobials, which poses a significant threat to human health. There were numerous studies on antimicrobial resistance of Campylobacter species [45]-[47], but very few focused on the mechanism of CHL and FFC resistance [36]. In this study, C. jejuni strain ATCC 33560, which usually serves as a quality-control strain for the antimicrobial susceptibility testing of Campylobacter spp. [38], [48], was used as the parent strain to select for CHL- or FFC-resistant C. jejuni in vitro [49].
In this study, some unique features were revealed concerning the development of CHL and FFC resistance in C. jejuni. Firstly, irrespective of the selective agent, the resistant clones obtained in vitro exhibited dual resistance to both agents (Table 2 ). Similar cross-resistance was previously reported in E. coli and Salmonella [50], [51], which usually emerged under FFC selection. Combined resistance to CHL and FFC might be conferred by specific resistance genes, such as floR [19], fexA [20], fexB [21], pexA [22] and cfr [24]. Secondly, both the CHL- (MICCHL≥64 µg/mL) and FFC-selected (MICFFC≥32 µg/mL) resistant strains contained the G2073A mutation in three copies of the 23S rRNA gene (Table 2 ), which is the first reported mutation associated with amphenicol resistance in Campylobacter spp. The G2057A mutation of the E. coli 23S rRNA gene (corresponding to position 2073 in the 23S rRNA gene of Campylobacter spp.) was previously reported to confer resistance to chloramphenicol and intermediate-level resistance to 14-nunbered-ring macrolides. Indeed, the CHL- and FFC-selected C. jejuni mutants carrying the G2073A mutation also showed elevated MICs to erythromycin (8 µg/mL), consistent with the finding in E. coli that the G2057A conferred cross resistance to chloramphenicol and erythromycin [23]. Interestingly in propionibacteria [32], Mycoplasma hominis [33], and Francisella tularensis [31], the same G2057A mutation was associated with erythromycin resistance, but not with resistance to 16-membered macrolides or chloramphenicol. It was postulated that the G2057 mutation in the 23S rRNA gene might lead to conformational changes in the binding sites of CHL and 14-membered macrolides, attenuating the affinity of these antimicrobials to the ribosome [52].
The G74D modification was found in the L4 ribosomal protein of CHL-selected resistant strains (MICCHL≥16 µg/mL), while the modification was absent in FFC-selected mutants, suggesting that the structural differences between CHL and FFC might contribute to the difference in the mutant selection process. FFC is a synthetic derivative of CHL with a fluoro group substitution at position C3 and the replacement of a nitro group (-NO2) by a sulfomethyl group (-SO2CH3) [53]. It is also possible that there are differences in the binding sites for CHL and FFC in ribosomal protein L4, or differences in the mode of action between CHL and FFC. The G74D mutation is located in a conserved region of the L4 ribosomal protein, which is considered as the main anchoring site of this ribosomal protein to 23S rRNA [54]. Several previous reports also identified the G74D mutation in ribosomal protein L4 of Campylobacter spp. [33], [42], [55], and this mutation, by itself, usually conferred low-level resistance to erythromycin and contributed to higher-level resistance when combined with the efflux pump CmeABC [56] or a G57V mutation in L4 [43]. Furthermore, the result from the PAβN assay suggested that the efflux mechanism might not play a major role in resistance to CHL and FFC in the mutant strains that carried the G2073A mutation. Thus, the G2073A rRNA mutation alone may be sufficient to confer resistance to CHL and FFC in C. jejuni mutants. It was reported in E. coli that the G2057A change in combination with other mutations in the 23S rRNA gene (i.e., G2032A) conferred a higher level of chloramphenicol resistance [57].
Several studies investigated the fitness of antibiotic-resistant Campylobacter spp. with mutation-associated resistance [58]–[60]. In the study of Luo et al. [58], they reported that fluoroquinolone-resistant Campylobacter showed an enhanced fitness in vivo. However, macrolide-resistant Campylobacter demonstrated a severe defect in both in vitro and in vivo fitness [60]. These finding indicate that different target mutations have varied impact on Campylobacter fitness. The results from the growth experiment in this study (Figure 1) demonstrated that the G2073A alteration in the 23S rRNA gene had a negative effect on the growth rates of the mutant strains, which suggests that development of amphenicol resistance incurs a fitness cost in Campylobacter. It appeared that the CHL-selected strains exhibited the most significant fitness disadvantage (Figure 1 ).
In conclusion, we discovered a novel G2073A mutation in the 23S rRNA gene of C. jejuni that is associated with amphenicol resistance. This mutation was identified by in vitro selection using CHL or FFC. Notably, this G2073A mutation was also associated with reduced susceptibility to erythromicin, suggesting that it confers cross resistance to both amphenicols and macrolides. Considering that the amphenicol resistance rates in Campylobacter are rising in certain countries, it will be interesting to determine if this resistance-associated mutation is naturally present in clinical isolates and if the amphenicol-resistant mutants are able to continue to persist in the absence of antibiotic selection pressure.
Funding Statement
This work was supported by the National Natural Science Foundation of China (U1031004), Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2012BAK01B02), Special Fund for Agro-scientific Research in the Public Interest (201203040), and Specialized Research Fund for the Doctoral Program of Higher Education (SEFDP, 20100008120001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Samuel MC, Vugia DJ, Shallow S, Marcus R, Segler S, et al. (2004) Epidemiology of sporadic Campylobacter infection in the United States and declining trend in incidence, FoodNet 1996–1999. Clin Infect Dis 38: S165–S174. [DOI] [PubMed] [Google Scholar]
- 2. Zimmer J, Wierzba TF, Abdel-Messih IA, Gharib B, Baqar S, et al. (2008) Campylobacter Infection as a Trigger for Guillain-Barré Syndrome in Egypt. PLoS ONE 3: e3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Islam Z, Gilbert M, Mohammad QD, Klaij K, Li J, et al. (2012) Guillain-Barré Syndrome-Related Campylobacter jejuni in Bangladesh: Ganglioside Mimicry and Cross-Reactive Antibodies. PLoS ONE 7: e43976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allos BM (2001) Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis 32: 1201–1206. [DOI] [PubMed] [Google Scholar]
- 5. McGill K, Kelly L, Madden RH, Moran L, Carroll C, et al. (2009) Comparison of disc diffusion and epsilometer (E-test) testing techniques to determine antimicrobial susceptibility of Campylobacter isolates of food and human clinical origin. J Microbiol Methods 79: 238–241. [DOI] [PubMed] [Google Scholar]
- 6. Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, et al. (2009) Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol 4: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gupta A, Nelson JM, Barrett TJ, Tauxe RV, Rossiter SP, et al. (2004) Antimicrobial resistance among Campylobacter strains, United States, 1997–2001. Emerg Infect Dis 10: 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Asai T, Harada K, Ishihara K, Kojima A, Sameshima T, et al. (2007) Association of antimicrobial resistance in Campylobacter isolated from food-producing animals with antimicrobial use on farms. Jpn J Infect Dis 60: 290. [PubMed] [Google Scholar]
- 9. Bester L, Essack S (2008) Prevalence of antibiotic resistance in Campylobacter isolates from commercial poultry suppliers in KwaZulu-Natal, South Africa. J Antimicrob Chemother 62: 1298–1300. [DOI] [PubMed] [Google Scholar]
- 10. Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A (2004) Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28: 519–542. [DOI] [PubMed] [Google Scholar]
- 11.Mascaretti OA (2003) Bacteria Versus Antibacterial Agents: An Integrated Approach: ASM Press.
- 12. Xie YQ, Zhou ZW, Guo Y, Deng QL, Huang Y (2009) Investigation of Campylobacter jejuni infection in children with diarrhea in Guangzhou. Chinese Journal of Contemporary Pediatrics 11: 422–424. [PubMed] [Google Scholar]
- 13. Shi X, Wu A, Zheng S, Li R, Zhang D (2007) Molecularly imprinted polymer microspheres for solid-phase extraction of chloramphenicol residues in foods. J Chromatogr B Analyt Technol Biomed Life Sci 850: 24–30. [DOI] [PubMed] [Google Scholar]
- 14.China MoAo (2002) No. 235 Bulletin of the Ministry of Agriculture of the People's Republic of China. Beijing: Ministry of Agriculture of China.
- 15. Barreto F, Ribeiro C, Hoff RB, Costa TD (2012) Determination and confirmation of chloramphenicol in honey, fish and prawns by liquid chromatography–tandem mass spectrometry with minimum sample preparation: validation according to 2002/657/EC Directive. Food Additives & Contaminants Part A 29: 550–558. [DOI] [PubMed] [Google Scholar]
- 16. Stolker AA, Brinkman UA (2005) Analytical strategies for residue analysis of veterinary drugs and growth-promoting agents in food-producing animals—a review. J Chromatogr A 1067: 15–53. [DOI] [PubMed] [Google Scholar]
- 17. Bostan K, Aydin A, Ang MK (2009) Prevalence and antibiotic susceptibility of thermophilic Campylobacter species on beef, mutton, and chicken carcasses in Istanbul, Turkey. Microb Drug Resist 15: 143–149. [DOI] [PubMed] [Google Scholar]
- 18. Bischoff KM, White DG, Hume ME, Poole TL, Nisbet DJ (2005) The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli . FEMS Microbiol Lett 243: 285–291. [DOI] [PubMed] [Google Scholar]
- 19. Kim E, Aoki T (1996) Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscicida . Microbiol Immunol 40: 665–669. [DOI] [PubMed] [Google Scholar]
- 20. Kehrenberg C, Schwarz S (2004) fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob Agents Chemother 48: 615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H, Wang Y, Wu C, Schwarz S, Shen Z, et al. (2012) A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J Antimicrob Chemother 67: 322–325. [DOI] [PubMed] [Google Scholar]
- 22. Lang KS, Anderson JM, Schwarz S, Williamson L, Handelsman J, et al. (2010) Novel florfenicol and chloramphenicol resistance gene discovered in Alaskan soil by using functional metagenomics. Appl Environ Microbiol 76: 5321–5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ettayebi M, Prasad SM, Morgan EA (1985) Chloramphenicol-erythromycin resistance mutations in a 23S rRNA gene of Escherichia coli . J Bacteriol 162: 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwarz S, Werckenthin C, Kehrenberg C (2000) Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri . Antimicrob Agents Chemother 44: 2530–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harada K, Asai T, Kojima A, Ishihara K, Takahashi T (2006) Role of coresistance in the development of resistance to chloramphenicol in Escherichia coli isolated from sick cattle and pigs. Am J Vet Res 67: 230–235. [DOI] [PubMed] [Google Scholar]
- 26. Li XS, Wang GQ, Du XD, Cui BA, Zhang SM, et al. (2007) Antimicrobial susceptibility and molecular detection of chloramphenicol and florfenicol resistance among Escherichia coli isolates from diseased chickens. J Vet Sci 8: 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nagshetty K, Channappa ST, Gaddad SM (2010) Antimicrobial susceptibility of Salmonella Typhi in India. J Infect Dev Ctries 4: 70–73. [DOI] [PubMed] [Google Scholar]
- 28. Englen MD, Hill AE, Dargatz DA, Ladely SR, Fedorka-Cray PJ (2007) Prevalence and antimicrobial resistance of Campylobacter in US dairy cattle. J Appl Microbiol 102: 1570–1577. [DOI] [PubMed] [Google Scholar]
- 29. Perez-Boto D, Garcia-Pena FJ, Abad-Moreno JC, Echeita MA (2013) Antimicrobial Susceptibilities of Campylobacter jejuni and Campylobacter coli Strains Isolated from Two Early Stages of Poultry Production. Microb Drug Resist 19: 323–330. [DOI] [PubMed] [Google Scholar]
- 30. Ozawa M, Makita K, Tamura Y, Asai T (2012) Associations of antimicrobial use with antimicrobial resistance in Campylobacter coli from grow-finish pigs in Japan. Prev Vet Med 106: 295–300. [DOI] [PubMed] [Google Scholar]
- 31. Chakeri A, Foroushani MSH, Torki Z, Rahimi E, Ebadi AG (2012) Antimicrobial Resistance of Campylobacter Species Isolated from Fecal Samples from Cats and Dogs in Iran. J Pure Appl Microbiol 6: 1823–1827. [Google Scholar]
- 32. Shin E, Oh Y, Kim M, Jung J, Lee Y (2013) Antimicrobial resistance patterns and corresponding multilocus sequence types of the Campylobacter jejuni isolates from human diarrheal samples. Microb Drug Resist 19: 110–116. [DOI] [PubMed] [Google Scholar]
- 33. de Moura HM, Silva PR, da Silva PH, Souza NR, Racanicci AM, et al. (2013) Antimicrobial resistance of Campylobacter jejuni isolated from chicken carcasses in the Federal District, Brazil. J Food Prot 76: 691–693. [DOI] [PubMed] [Google Scholar]
- 34. Chen X, Naren GW, Wu CM, Wang Y, Dai L, et al. (2010) Prevalence and antimicrobial resistance of Campylobacter isolates in broilers from China. Vet Microbiol 144: 133–139. [DOI] [PubMed] [Google Scholar]
- 35. Adzitey F, Rusul G, Huda N, Cogan T, Corry J (2012) Prevalence, antibiotic resistance and RAPD typing of Campylobacter species isolated from ducks, their rearing and processing environments in Penang, Malaysia. Int J Food Microbiol 154: 197–205. [DOI] [PubMed] [Google Scholar]
- 36. Wang Y, Taylor DE (1990) Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94: 23–28. [DOI] [PubMed] [Google Scholar]
- 37. Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A (2004) Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28: 519–542. [DOI] [PubMed] [Google Scholar]
- 38.CLSI (2008) Performance Standards for Antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; Approved Standard-Third Edition. CLSI document M31-A3. Pennsylvania: Clinical and Laboratory Standards Institute.
- 39.Food and Drug Administration (2010) National Antimicrobial Resistance Monitoring System-Enteric Bacteria (NARMS): 2007 Executive Report. Rockville, MD, U.S.: Department of Health and Human Services, U.S. FDA.
- 40. Gibreel A, Wetsch NM, Taylor DE (2007) Contribution of the CmeABC efflux pump to macrolide and tetracycline resistance in Campylobacter jejuni . Antimicrob Agents Chemother 51: 3212–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gibreel A, Kos VN, Keelan M, Trieber CA, Levesque S, et al. (2005) Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob Agents Chemother 49: 2753–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cagliero C, Mouline C, Cloeckaert A, Payot S (2006) Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli . Antimicrob Agents Chemother 50: 3893–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Caldwell DB, Wang Y, Lin J (2008) Development, stability, and molecular mechanisms of macrolide resistance in Campylobacter jejuni . Antimicrob Agents Chemother 52: 3947–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hao H, Dai M, Wang Y, Peng D, Liu Z, et al. (2009) 23S rRNA mutation A2074C conferring high-level macrolide resistance and fitness cost in Campylobacter jejuni . Microb Drug Resist 15: 239–244. [DOI] [PubMed] [Google Scholar]
- 45. Rzewuska K, Korsak D, Mackiw E (2010) Antibiotic resistance of bacteria Campylobacter sp. Przegl Epidemiol 64: 63–68. [PubMed] [Google Scholar]
- 46. Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, et al. (2009) Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol 4: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alfredson DA, Korolik V (2007) Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli . FEMS Microbiol Lett 277: 123–32. [DOI] [PubMed] [Google Scholar]
- 48. McDermott PF, Bodeis SM, Aarestrup FM, Brown S, Traczewski M, et al. (2004) Development of a standardized susceptibility test for Campylobacter with quality-control ranges for ciprofloxacin, doxycycline, erythromycin, gentamicin, and meropenem. Microb Drug Resist 10: 124–131. [DOI] [PubMed] [Google Scholar]
- 49. Hänninen M-L, Hannula M (2007) Spontaneous mutation frequency and emergence of ciprofloxacin resistance in Campylobacter jejuni and Campylobacter coli . J Antimicrob Chemother 60: 1251–1257. [DOI] [PubMed] [Google Scholar]
- 50. Donaldson SC, Straley BA, Hegde NV, Sawant AA, DebRoy C, et al. (2006) Molecular Epidemiology of Ceftiofur-Resistant Escherichia coli Isolates from Dairy Calves. Appl Environ Microbiol 72: 3940–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arcangioli MA, Leroy-Setrin S, Martel JL, Chaslus-Dancla E (2000) Evolution of chloramphenicol resistance, with emergence of cross-resistance to florfenicol, in bovine Salmonella Typhimurium strains implicates definitive phage type (DT) 104. J Med Microbiol 49: 103–110. [DOI] [PubMed] [Google Scholar]
- 52. Noormohamed A, Fakhr MK (2012) Incidence and Antimicrobial Resistance Profiling of Campylobacter in Retail Chicken Livers and Gizzards. Foodborne Pathog Dis 9: 617–624. [DOI] [PubMed] [Google Scholar]
- 53. Switala M, Hrynyk R, Smutkiewicz A, Jaworski K, Pawlowski P, et al. (2007) Pharmacokinetics of florfenicol, thiamphenicol, and chloramphenicol in turkeys. J Vet Pharmacol Ther 30: 145–150. [DOI] [PubMed] [Google Scholar]
- 54. Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, et al. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688. [DOI] [PubMed] [Google Scholar]
- 55. Serraino A, Florio D, Giacometti F, Piva S, Mion D, et al. (2013) Presence of Campylobacter and Arcobacter species in in-line milk filters of farms authorized to produce and sell raw milk and of a water buffalo dairy farm in Italy. J Dairy Sci 96: 2801–2807. [DOI] [PubMed] [Google Scholar]
- 56. Oporto B, Juste RA, Hurtado A (2009) Phenotypic and Genotypic Antimicrobial Resistance Profiles of Campylobacter jejuni Isolated from Cattle, Sheep, and Free-Range Poultry Faeces. Int J Microbiol 2009: 456573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Douthwaite S (1992) Functional interactions within 23S rRNA involving the peptidyltransferase center. J Bacteriol 174: 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luo N, Pereira S, Sahin O, Lin J, Huang S, et al. (2005) Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci USA 102: 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Q, Sahin O, McDermott PF, Payot S (2006) Fitness of antimicrobial-resistant Campylobacter and Salmonella . Microbes Infect 8: 1972–1978. [DOI] [PubMed] [Google Scholar]
- 60. Luangtongkum T, Shen Z, Seng VW, Sahin O, Jeon B, et al. (2012) Impaired fitness and transmission of macrolide-resistant Campylobacter jejuni in its natural host. Antimicrob Agents Chemother 56: 1300–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]