Abstract
Little is known about the genetic diversity of Sarcoptes scabiei mites in farm animals in Egypt. In this study, we characterized S. scabiei in 25 skin scrapes from water buffalo, cattle, sheep, and rabbits at the nuclear marker ITS2 and mitochondrial markers COX1 and 16S rRNA. Sequences of the ITS2 showed no host segregation or geographical isolation, whereas those of the mitochondrial COX1 and 16S rRNA genes indicated the presence of both host-adapted and geographically segregated populations of S. scabiei. Host adaptation may limit inter-species transmission of. S. scabiei, thus restrict gene flow among S. scabiei from different hosts. This is the first report on the molecular characterization of sarcoptic mites in Egypt. Further genetic studies involving larger numbers of specimens, especially those from humans and companion animals, are needed to understand the molecular epidemiology of sarcoptic mange in Egypt.
Introduction
The cosmopolitan mite Sarcoptes scabiei (Acari: Sarcoptidae) is an obligatory ectoparasite that infects the skin of a wide range of mammalian hosts, resulting in sarcoptic mange in companion animals, livestock, and wildlife, as well as scabies in humans [1]–[3]. The disease is highly contagious, characterized by pruritic dermatitis, alopecia, hyperkeratosis, and crust formation [4]–[6], and if left untreated, can lead to death due to dehydration, pneumonia, or bacterial septicemia [7]–[9]. In addition to its potential to cause huge economic losses due to weight loss and mortality in animals [10], [11], scabies imposes a global public health concerns as an emerging/re-emerging infectious disease [9], [12], [13]. Scabies outbreaks have been reported in industrialized countries [14]–[16], and the burden of the disease in developing countries is increasing [1], [17]–[19]. Drug residuals and toxicity due to extensive use of acaricides, especially in developing countries, and emergence of drug resistance are some other growing problems associated with sarcoptic mange and scabies [20]–[23].
Current knowledge suggests that humans and protohumans were most likely the initial source of animal scabies, first of dogs, and later of other species with further spread to wildlife [16]. Sarcoptes scabiei is taxonomically divided into different varieties based on host origin [24]. However, speciation in S. scabiei is a controversial issue due to the indistinguishable morphology of host-associated populations, evidence of apparent cross-species transmission during epizootics in sympatric wild animals [12], [25], limited or no cross-infestations between hosts in experimental studies [26], and presence of immunologically host-specific and cross-reactive epitopes [27], [28]. Characterizations of mitochondrial DNA (mtDNA) haplotypes and microsatellite allele frequencies have demonstrated significant associations between S. scabiei and host species or geographical locations [29], [30].
In Egypt, scabies has been reported in farm animals [31]–[33], wild games [34], and human [35]–[38]. However, there are no data on genetic diversity of S. scabiei. This preliminary study was conducted to examine the genetic characteristics of S. scabiei derived from different hosts in Egypt, including water buffalo, sheep, rabbits, and one cattle. Results of sequence characterization of the nuclear internal transcribed spacer 2 (ITS2) and mitochondrial cytochrome oxidase 1 (COX1) and 16S rRNA genes demonstrated the presence of host-adapted and geographically segregated S. scabiei populations in Egypt.
Materials and Methods
Ethics Statement
This study was carried out in strict compliance with the Guidelines of Animals Health Research Institute, Egypt. The study protocol was approved by the Committee on the Ethics of Animals Health Research Institute, Egypt (Permit Number 362 approved on August 31, 2010). All scrapings were collected by well trained and licensed veterinarians. This study was done on specimens from animals on private farms as part of the routine clinical examinations and care, with written consents from the owners. One of the co-investigator of the project, Dr. Abd El Naby Metwaly (Animal Health Research Institute, Kafr El Sheikh Provincial Lab, Kafr El Sheikh 33516, Egypt; e-mail: tahoon63@yahoo.com), should be contacted for permissions for future work on these farms. Efforts were made to minimize discomfort and stress to animals while performing skin scraping.
Specimens
Specimens of this study were collected during August 2010-April 2011 from buffalo, sheep, rabbits, and one cattle (Table 1) in Sheikh Province (130 km north of Cairo). Buffalo specimens were collected from animals on three farms at kafr El Sheikh District; specimens that had mixed infection with Psoroptes spp at the same infection site were excluded. Rabbit specimens were collected from three small rabbitries; rabbits were raised in wired cages. Sheep specimens were collected from two farms, with goats raised in the same herd on the second farm. The cattle specimen was from a sporadic case for veterinary consultation. Skin scrapings were collected in situ directly from infected animals into tightly closed plastic cups, transferred to the laboratory, and examined by microscopy. Positive samples were fixed in 75% ethyl alcohol and stored at 4°C for molecular biologic analyses.
Table 1. Sarcoptes scabiei isolates collected from four species of farm animals at Kafr El Sheikh Province, Egypt.
| Specimen ID | Host | Farm | Infection Site |
| 37025 | Water buffalo | Buffalo farm 1 | Perineal region |
| 37026 | Water buffalo | Buffalo farm 1 | Perineal region |
| 37027 | Water buffalo | Buffalo farm 1 | Perineal region |
| 37028 | Water buffalo | Buffalo farm 1 | Perineal region |
| 37031 | Water buffalo | Buffalo farm 2 | Perineal region |
| 37032 | Water buffalo | Buffalo farm 2 | Perineal region |
| 37033 | Water buffalo | Buffalo farm 2 | Perineal region |
| 37030 | Water buffalo | Buffalo farm 3 | Perineal region |
| 37073 | Cattle | Sporadic case | Body |
| 37061 | Sheep | Sheep farm 1 | Body |
| 37077 | Sheep | Sheep farm 1 | Body |
| 37078 | Sheep | Sheep farm 2 | Body |
| 37080 | Sheep | Sheep farm 2 | Body |
| 37081 | Sheep | Sheep farm 2 | Body |
| 37040 | Rabbit | Rabbitry 1 | Body |
| 37053 | Rabbit | Rabbitry 1 | Ear |
| 37054 | Rabbit | Rabbitry1 | Ear |
| 37056 | Rabbit | Rabbitry 1 | Ear |
| 37059 | Rabbit | Rabbitry1 | Foot |
| 37065 | Rabbit | Rabbitry 1 | Body |
| 37060 | Rabbit | Rabbitry 2 | Foot |
| 37063 | Rabbit | Rabbitry 2 | Ear |
| 37074 | Rabbit | Rabbitry 2 | Ear |
| 37064 | Rabbit | Rabbitry 3 | Foot |
| 37075 | Rabbit | Rabbitry 3 | Ear |
DNA extraction and PCR analysis
Ethyl alcohol was removed from microscopy-positive specimens by centrifugation and washing with distilled water. DNA was extracted from the specimens using the FastDNA SPIN Kit for Soil (MP Biomedicals, Colon, OH). PCR amplification of the ITS-2 was done using primers RIB-18 and RIB-3 as described by Zahler et al. [39]. PCR analyses of the mitochondrial COX1 and 16S rRNA genes were conducted as described by Walton et al. [30].
DNA sequence analyses
PCR products were sequenced directly using Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and an ABI 3130 Genetic Analyzer (Applied Biosystems). Sequences were assembled using the ChromasPro (version 1.5) software (http://www.technelysium.com.au/ChromasPro.html). The accuracy of data was confirmed by bi-directional sequencing. The obtained sequences were aligned with each other and reference sequences of each gene using ClustalX (ftp://ftp-igbmc.u-trasbg.fr/pub/ClustalX/) to confirm the identification of S. scabiei. A neighbor-joining (NJ) analysis implemented in the MEGA5 (http://www.megasoftware.net) was used to assess the phylogenetic relationship among different populations of S. scabiei. Unique nucleotide sequences generated in this study were deposited in GenBank under accession numbers AB778895 to AB778919 for ITS2, AB779564 to AB779587 for mitochondrial 16S rRNA, and AB779588 to AB779611 for mitochondrial COX1 genes.
Results
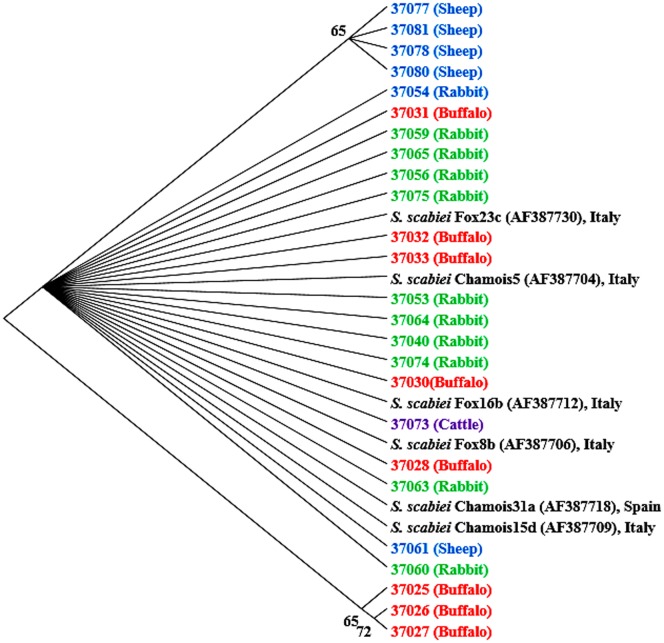
ITS2 sequence analysis of the Sarcoptes mites derived from different hosts from Egypt generated 7 sequence types. Despite the low number of polymorphic sites (5 sites), these sequences formed three groups on the NJ tree. All sequences derived from mites in rabbits, several sequences from mites in buffalo, and one sequence each from mites in cattle and sheep formed a cluster on the tree together with reference sequences from GenBank (Fig. 1), reflecting the broad host and geographical distribution of this S. scabiei population. In contrast, the other two groups were formed by sequences from this study, including one group containing the majority of sequences from mites of sheep (37077, 37078, 37080 and 37081), and one containing 3 sequences (37025–37027) from mites of buffalo.
Figure 1. Un-rooted NJ tree showing genetic relationship of Egyptian Sarcoptes mites to others in the GenBank database based on ITS2 sequences.
Evolutionary relationships of 31 taxa were inferred using the neighbor-joining method [46]. Numbers at the internodes correspond to percent bootstrap values from 2,000 replicates. Branches corresponding to partitions produced in less than 50% bootstrap replicates are collapsed. Sequences in bolded colors and with no GenBank accession numbers are generated from Egyptian specimens.
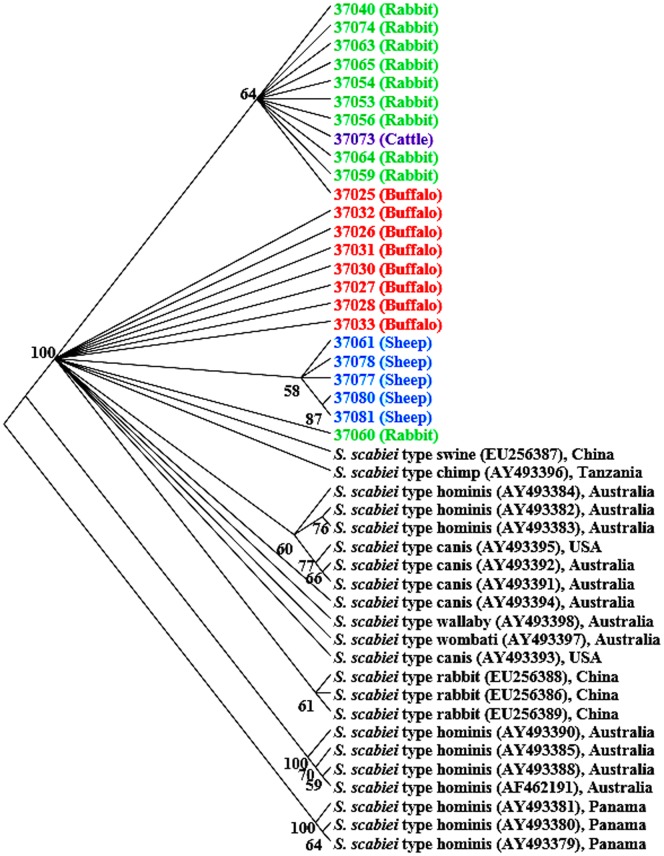
Sequence variability was greater at the mitochondrial COX1 and 16S rRNA genes. Altogether, 10 types of COX1 sequences and 5 types of 16S rRNA sequences were obtained, which differed from each other in the form of nucleotide substitutions and insertions or deletions. NJ analysis based on COX1 clearly showed the presence of 2 major clusters of S. scabiei in Egypt by host (Fig. 2). One cluster included all sequences from mites in rabbits, one cattle (37073), and one buffalo (37025). The other cluster had two branches, one of S. scabiei in sheep and one of S. scabiei in buffalo (Fig. 2). Comparing to S. scabiei isolates from other areas, the Egypt-derived sequences occupied unique positions in the NJ tree (Fig. 2). Phylogenetically, COX1 sequences from human isolates in Panama and some human isolates in Australia formed the two basal branches diverged from others containing sequences mostly from animal isolates. In the latter branches, sequences from different animals in different geographical locations showed host and geographical clustering, with sequences from Egyptian isolates separated from others. Sequences from several other human isolates in Australia formed a subgroup within the major cluster of largely animal isolates (Fig 2).
Figure 2. Un-rooted NJ tree showing genetic relationship of Egyptian Sarcoptes mites to others in the GenBank database based on COX1 sequences.
Evolutionary relationships of 46 taxa were inferred using the neighbor-joining method [46]. Numbers at the internodes correspond to percent bootstrap values from 2,000 replicates. Branches corresponding to partitions produced in less than 50% bootstrap replicates are collapsed. Sequences in bolded colors and with no GenBank accession numbers are generated from Egyptian specimens.
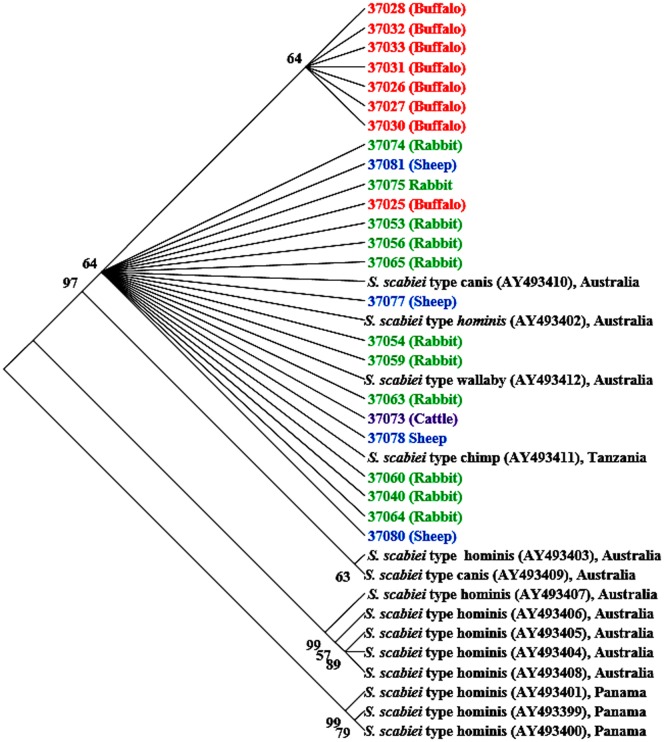
Sequences of the mitochondrial 16S rRNA gene divided the Egyptian S. scabiei isolates into two major groups, one containing most buffalo isolates and one containing all sheep and rabbit isolates and one each of buffalo and cattle isolates. In concordance with results of the COX1 sequence analysis, a NJ tree based on the 16S rRNA gene sequences placed sequences from all human isolates in Panama and some human isolates in Australia in the basal branches divergent from sequences from most animal isolates in various areas and some human isolates in Australia. However, there was less host and geographic segregation in the latter groups than seen at the COX1 locus, although the sequences from buffalo in Egypt clearly formed its own clade (Fig. 3). Sequences from isolates in rabbits, sheep, and one buffalo and cattle each were more related to each other and clustered together with sequences AY493410 from a dog in Australia, AY493402 from a human in Australia, AY493412 from a wallaby in Australia, and AY493411 from a chimpanzee in Tanzania. Thus, S. scabiei from sheep was genetically related to S. scabiei from buffalo at the COX locus, but was more related to S. scabiei from rabbits at the 16S rRNA locus, even though both loci are in the small mitochondrial genome. The relatively low bootstrap values of most branches in the phylogenetic trees were probably the result of limited sequence polymorphism and random distribution of some nucleotide substitutions at these loci.
Figure 3. Un-rooted NJ tree of showing genetic relationship of Egyptian Sarcoptes mites to others in the GenBank database based on mitochondrial 16S rRNA sequences.
Evolutionary relationships of 38 taxa were inferred using the neighbor-joining [46]. Numbers at the internodes correspond to percent bootstrap values from 2,000 replicates. Branches corresponding to partitions produced in less than 50% bootstrap replicates are collapsed. Sequences in bolded colors and with no GenBank accession numbers are generated from Egyptian specimens.
Discussion
In this study, we sequence-characterized S. scabiei isolates from buffalo, sheep, rabbits, and one cattle at three genetic loci. Results obtained showed that ITS2 sequences from Sarcoptes mites from these hosts are conserved with intra-sequence variability at only 5 positions. These results are in concordance with those of Zahler et al. [39] and Gu and Yang [40]. Thus, based on ITS2 sequence analysis, Zahler et al. [39] reported very little genetic variation in sarcoptic mites collected from different hosts and geographic locations, and, Gu and Yang [40] could not differentiate Sarcoptes mites from different hosts in China. Although Berrilli et al. [41] and Alasaad et al. [25] detected some gene variability between individual mites, the sequence variations were randomly distributed in different hosts from several locations, thus resulting in no distinct geographic or host-specific clustering.
In contrast to the nuclear ITS2 marker, mitochondrial markers analyzed in the present study showed clear sequence polymorphism related to the host species. COX1 sequence analysis showed the presence of 3 distinct groups by host species with additional geographic stratifications (Fig. 2). This was also supported by results of sequence analysis of the mitochondrial 16S rRNA gene (Fig 3). Using hypervariable microsatellite loci, Walton et al. [42] reported that S. scabiei from dogs and humans clustered by host species rather than by geographic location. In contrast, phylogenetic studies based on 16S rRNA and COX1 sequences demonstrated that clustering patterns of S. scabiei mites were under the impact of both host species and geographical locations [30], whereas sequence analysis of the mitochondrial 12S rRNA gene did not show any significant association between haplotypes and host species [43]. Thus, multilocus characterization of diverse isolates is needed to better understand host adaptation and geographic segregation in S. scabiei.
Host adaptation in S. scabiei has important implications in understanding the epidemiology and development of diagnostic tests and vaccines [30]. Previously, it was thought there was frequent inter-breeding among mites infecting distinct host species, increasing their genetic variability and allowing Sarcoptes to infect new species of animals [44]. In contrast, results of recent studies have shown the occurrence of host adaptation in S. scabiei [30], [42]. Data of the present study indicate that both host adaptation and geographic segregation also occur in S. scabiei in Egypt. Both host adaptation and geographic segregation would reduce the inter-species transmission of S. scabiei [12], [45], thus have important implications in our understanding of the epidemiology of S. scabiei and development of control strategies against mange in animals and scabies in humans.
The small number of specimens characterized is a major limitation of the current study. Thus, the conclusion on host-adaptation and geographic segregation in S. scabiei in Egypt needs support of multilocus genetic characterizations of parasites from a range of hosts, especially those of humans and companion animals. More advanced molecular biological tools, such as population genetics and comparative genomics, are also be needed to understanding the genetic mechanism responsible for host-adaptation and geographic segregation in S. scabiei.
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Funding Statement
This research was supported by the Arab Fund for Economic and Social Development (Zamalat Program), National Natural Science Foundation of China (Project No. 31110103901), and Centers for Disease Control and Prevention. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fuller LC (2013) Epidemiology of scabies. Curr Opin Infect Dis 26: 123–126. [DOI] [PubMed] [Google Scholar]
- 2. Holt DC, Fischer K (2013) Novel insights into an old disease: recent developments in scabies mite biology. Curr Opin Infect 26: 110–115. [DOI] [PubMed] [Google Scholar]
- 3. Pence D, Ueckermann E (2002) Sarcoptic mange in wildlife. Rev Sci Tech 21: 385–398. [PubMed] [Google Scholar]
- 4. Nimmervoll H, Hoby S, Robert N, Lommano E, Welle M, et al. (2013) Pathology of sarcoptic mange in red foxes (Vulpes vulpes): macroscopic and histologic characterization of three disease stages. J Wildl Dis 49: 91–102. [DOI] [PubMed] [Google Scholar]
- 5. Rafferty D, Gray J (1987) The feeding behaviour of Psoroptes spp. mites on rabbits and sheep. J Parasitol 73: 901–906. [PubMed] [Google Scholar]
- 6. Wells B, Burgess S, McNeilly T, Huntley J, Nisbet A (2012) Recent developments in the diagnosis of ectoparasite infections and disease through a better understanding of parasite biology and host responses. Mol Cell Probes 26: 47–53. [DOI] [PubMed] [Google Scholar]
- 7. Mounsey KE, McCarthy JS, Walton SF (2013) Scratching the itch: new tools to advance understanding of scabies. Trends Parasitol 29: 35–42. [DOI] [PubMed] [Google Scholar]
- 8. Tarry DW (1974) Sheep scab: its diagnosis and biology. Vet Record 95: 530–532. [DOI] [PubMed] [Google Scholar]
- 9. Walton S, Holt D, Currie B, Kemp D (2004) Scabies: new future for a neglected disease. Adv Parasitol 57: 309–376. [DOI] [PubMed] [Google Scholar]
- 10. Dagleish M, Ali Q, Powell R, Butz D, Woodford M (2007) Fatal Sarcoptes scabiei infection of blue sheep (Pseudois nayaur) in Pakistan. J Wildl Dis 43: 512–517. [DOI] [PubMed] [Google Scholar]
- 11. Damriyasa I, Failing K, Volmer R, Zahner H, Bauer C (2004) Prevalence, risk factors and economic importance of infestations with Sarcoptes scabiei and Haematopinus suis in sows of pig breeding farms in Hesse, Germany. Med Vet Entomol 18: 361–367. [DOI] [PubMed] [Google Scholar]
- 12. Alasaad S, Rossi L, Heukelbach J, Perez JM, Hamarsheh O, et al. (2013) The neglected navigating web of the incomprehensibly emerging and re-emerging Sarcoptes mite. Infect Genet Evol 17: 253–259. [DOI] [PubMed] [Google Scholar]
- 13. Fthenakis G, Karagiannidis A, Alexopoulos C, Brozos C, Papadopoulos E (2001) Effects of sarcoptic mange on the reproductive performance of ewes and transmission of Sarcoptes scabiei to newborn lambs. Vet Parasitol 95: 63–71. [DOI] [PubMed] [Google Scholar]
- 14. Achtari L, Erard P, Gueissaz F, Malinverni R (2007) An outbreak of scabies: a forgotten parasitic disease still present in Switzerland. Swiss Med Wkly 137: 695–699. [DOI] [PubMed] [Google Scholar]
- 15. Ariza L, Walter B, Worth C, Brockmann S, Weber ML, et al. (2012) Investigation of a scabies outbreak in a kindergarten in Constance, Germany. Eur J Clin Microbiol Infect Dis 32: 373–380. [DOI] [PubMed] [Google Scholar]
- 16. Currier R, Walton S, Currie B (2012) Scabies in animals and humans: history, evolutionary perspectives, and modern clinical management. Ann NY Acad Sci 1230: E50–60. [DOI] [PubMed] [Google Scholar]
- 17. Gilmore S (2011) Control strategies for endemic childhood scabies. PLoS One 6: e15990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hay R, Steer A, Engelman D, Walton S (2012) Scabies in the developing world-its prevalence, complications, and management. Clin Microbiol Infect 18: 313–23. [DOI] [PubMed] [Google Scholar]
- 19. Terry B, Kanjah F, Sahr F, Kortequee S, Dukulay I, et al. (2001) Sarcoptes scabiei infestation among children in a displacement camp in Sierra Leone. Public Health 115: 208–211. [DOI] [PubMed] [Google Scholar]
- 20. Bradberry S, Cage S, Proudfoot A, Vale J (2005) Poisoning due to pyrethroids. Toxicol Rev 24: 93–106. [DOI] [PubMed] [Google Scholar]
- 21. Currie B, Harumal P, McKinnon M, Walton S (2004) First documentation of in vivo and in vitro ivermectin resistance in Sarcoptes scabiei . Clin Infect Dis 39: 8–12. [DOI] [PubMed] [Google Scholar]
- 22. Mounsey K, Holt D, McCarthy J, Currie B, Walton S (2008) Scabies: molecular perspectives and therapeutic implications in the face of emerging drug resistance. Future Microbiol 3: 57–66. [DOI] [PubMed] [Google Scholar]
- 23. Sanderson H, Laird B, Pope L, Brain R, Wilson C, et al. (2007) Assessment of the environmental fate and effects of ivermectin in aquatic mesocosms. Aquat Toxicol 85: 229–240. [DOI] [PubMed] [Google Scholar]
- 24. Fain A (1978) Epidemiological problems of scabies. Int J Dermatol 17: 20–30. [DOI] [PubMed] [Google Scholar]
- 25. Alasaad S, Soglia D, Spalenza V, Maione S, Soriguer R, et al. (2009) Is ITS-2 rDNA suitable marker for genetic characterization of Sarcoptes mites from different wild animals in different geographic areas? Vet Parasitol 159: 181–185. [DOI] [PubMed] [Google Scholar]
- 26. Arlian L, Runyan R, Estes S (1984) Cross infestivity of Sarcoptes scabiei. J Am Acad Dermatol 10: 979–986. [DOI] [PubMed] [Google Scholar]
- 27. Arlian L, Morgan M, Arends J (1996) Immunologic cross-reactivity among various strains of Sarcoptes scabiei . J Parasitol 82: 66–72. [PubMed] [Google Scholar]
- 28. Haas N, Wagemann B, Hermes B, Henz B, Heile C, et al. (2005) Crossreacting IgG antibodies against fox mite antigens in human scabies. Arch Dermatol Res 296: 327–331. [DOI] [PubMed] [Google Scholar]
- 29. Alasaad S, Soglia D, Sarasa M, Soriguer R, Perez J, et al. (2008) Skin-scale genetic structure of Sarcoptes scabiei populations from individual hosts: empirical evidence from Iberian ibex-derived mites. Parasitol Res 104: 101–105. [DOI] [PubMed] [Google Scholar]
- 30. Walton S, Dougall A, Pizzutto S, Holt D, Taplin D, et al. (2004) Genetic epidemiology of Sarcoptes scabiei (Acari: Sarcoptidae) in northern Australia. Int J Parasitol 34: 839–849. [DOI] [PubMed] [Google Scholar]
- 31. Mazyad S, Sanad E, Morsy T (2001) Two types of scab mites infesting man and sheep in North Sinai. J Egypt Soc Parasitol 31: 213–222. [PubMed] [Google Scholar]
- 32. Osman S, Hanafy A, Amer S (2006) Clinical and therapeutic studies on mange in horses. Vet Parasitol 141: 191–195. [DOI] [PubMed] [Google Scholar]
- 33. Yassin M (2011) Mange mites causing scabies in Egyptian buffaloes at Giza Governorate. Egypt J Egypt Soc Parasitol 41: 55–64. [PubMed] [Google Scholar]
- 34. Felt S (2009) Acariasis in captive fat-tailed jirds (Pachyuromys duprasi). J Zoo Wildl Med 40: 217–219. [DOI] [PubMed] [Google Scholar]
- 35. Anbar T, El-Domyati M, Mansour H, Ahmad H (2007) Scaly scalp associated with crusted scabies: case series. Dermatol Online J 13: 18. [PubMed] [Google Scholar]
- 36. Kenawi M, Morsy T, Abdalla K, Nasr M, Awadalla R (1993) Clinical and parasitological aspects on human scabies in Qualyobia Governorate, Egypt. J Egypt Soc Parasitol 23: 247–253. [PubMed] [Google Scholar]
- 37. Morsy T, Rahem M, El-Sharkawy E, Shatat MA (2003) Eucalyptus globulus (camphor oil) against the zoonotic scabies, Sarcoptes scabiei. J. Egypt. Soc. Parasitol 33: 47–53. [PubMed] [Google Scholar]
- 38. Nofal A (2009) Variable response of crusted scabies to oral ivermectin: report on eight Egyptian patients. J Eur Acad Dermatol Venereol 23: 793–797. [DOI] [PubMed] [Google Scholar]
- 39. Zahler M, Essig A, Gothe R, Rinder H (1999) Molecular analyses suggest monospecificity of the genus Sarcoptes (Acari: Sarcoptidae). Int J Parasitol 29: 759–766. [DOI] [PubMed] [Google Scholar]
- 40. Gu X, Yang G (2008) A study on the genetic relationship of mites in the genus Sarcoptes (Acari: Sarcoptidae) in China. Int J Acarol 34: 183–190. [Google Scholar]
- 41. Berrilli F, D'Amelio S, Rossi L (2002) Ribosomal and mitochondrial DNA sequence variation in Sarcoptes mites from different hosts and geographical regions. Parasitol Res 88: 772–777. [DOI] [PubMed] [Google Scholar]
- 42. Walton S, Choy J, Bonson A, Valle A, McBroom J, et al. (1999) Genetically distinct dog-derived and human-derived Sarcoptes scabiei in scabies-endemic communities in northern Australia. Am J Trop Med Hyg 61: 542–547. [DOI] [PubMed] [Google Scholar]
- 43. Skerratt L, Campbell N, Murrell A, Walton S, Kemp D, et al. (2002) The mitochondrial 12S gene is a suitable marker of populations of Sarcoptes scabiei from wombats, dogs and humans in Australia. Parasitol Res 88: 376–379. [DOI] [PubMed] [Google Scholar]
- 44. Fain A (1994) Adaptation, specificity and host-parasite coevolution in mites (Acari). Int J Parasitol 24: 1273–1283. [DOI] [PubMed] [Google Scholar]
- 45. Rasero R, Rossi L, Maione S, Sacchi P, Rambozzi L, et al. (2010) Host taxon-derived Sarcoptes mites in European wildlife animals, revealed by microsatellite markers. Biol Cons 143: 1269–1277. [Google Scholar]
- 46. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]