Abstract
Background: Environmental endocrine disruptors (EEDs) are exogenous chemicals that mimic endogenous hormones such as estrogens. Previous studies using a zebrafish transgenic reporter demonstrated that the EEDs bisphenol A and genistein preferentially activate estrogen receptors (ERs) in the larval heart compared with the liver. However, it was not known whether the transgenic zebrafish reporter was sensitive enough to detect estrogens from environmental samples, whether environmental estrogens would exhibit tissue-specific effects similar to those of BPA and genistein, or why some compounds preferentially target receptors in the heart.
Methods: We tested surface water samples using a transgenic zebrafish reporter with tandem estrogen response elements driving green fluorescent protein expression (5xERE:GFP). Reporter activation was colocalized with tissue-specific expression of ER genes by RNA in situ hybridization.
Results: We observed selective patterns of ER activation in transgenic fish exposed to river water samples from the Mid-Atlantic United States, with several samples preferentially activating receptors in embryonic and larval heart valves. We discovered that tissue specificity in ER activation was due to differences in the expression of ER subtypes. ERα was expressed in developing heart valves but not in the liver, whereas ERβ2 had the opposite profile. Accordingly, subtype-specific ER agonists activated the reporter in either the heart valves or the liver.
Conclusion: The use of 5xERE:GFP transgenic zebrafish revealed an unexpected tissue-specific difference in the response to environmentally relevant estrogenic compounds. Exposure to estrogenic EEDs in utero was associated with adverse health effects, with the potentially unanticipated consequence of targeting developing heart valves.
Citation: Gorelick DA, Iwanowicz LR, Hung AL, Blazer VS, Halpern ME. 2014. Transgenic zebrafish reveal tissue-specific differences in estrogen signaling in response to environmental water samples. Environ Health Perspect 122:356–362; http://dx.doi.org/10.1289/ehp.1307329
Introduction
Estrogens are small molecules that influence organ formation and function (Deroo and Korach 2006). Estrogens bind to and activate receptors in the cytosol, which then travel to the nucleus and directly regulate gene expression. Multiple estrogen receptor (ER) genes are present in vertebrates, such as the Esr1 and Esr2 genes in mice (coding for ERα and ERβ proteins, respectively) and the esr1, esr2a, and esr2b genes in zebrafish (coding for ERα, ERβ1, and ERβ2 proteins, respectively). Exposure to environmental endocrine disruptors (EEDs) that bind to ERs are associated with increased risk of cancers and abnormal reproductive tract formation in mammals and fish (Ma 2009). Because ERs are expressed widely in many tissues (Kuiper et al. 1997), exposure to estrogenic EEDs may also influence the development of nonreproductive tissues (Meeker 2012). Therefore, detecting environmental estrogens and identifying their sites and mechanism of action during organismal development is of paramount importance.
Standard methods to detect ER activity use yeast and mammalian cell culture assays (Legler et al. 1999; Leskinen et al. 2005; Routledge and Sumpter 1996; Sanseverino et al. 2005) that are limited in their utility because they are not representative of tissue diversity. In addition, although these methods can demonstrate the presence of estrogenic chemicals in environmental samples, they do not address whether chemicals are being absorbed and producing an effect at the organismal level. ER activity assays have been developed for fish and mice; however, most reporter constructs are designed to act in certain tissues exclusively (such as liver or brain) (Brion et al. 2012; Kurauchi et al. 2005) or have used a bioluminescent reporter (such as luciferase) that has limited spatial resolution (Ciana et al. 2003; Legler et al. 2000).
Previously, we developed transgenic zebrafish that specifically report ER transcriptional activity in all tissues of embryos and larvae with single cell resolution (Gorelick and Halpern 2011). The reporter line (5xERE:GFP) contains tandem estrogen response element (ERE) DNA sequences (Gruber et al. 2004) driving green fluorescent protein (GFP) expression. The 5xERE:GFP line serves as a tissue-specific reporter of ER-mediated transcriptional activity following exposure of zebrafish embryos to estrogenic compounds. Exposure to certain purified compounds results in preferential activation of GFP in heart valves, whereas other compounds activate the reporter only in the liver (Gorelick and Halpern 2011). Similar results were reported independently using a 3xERE zebrafish reporter (Lee et al. 2012). We sought to determine whether the ER reporter zebrafish would also be useful in detecting the presence of environmental estrogens and in discovering the basis for the tissue-specific differences in response to estrogens.
Materials and Methods
Chemicals. Estradiol (purity ≥ 98%), bisphenol A (BPA; purity ≥ 99%) and dimethyl sulfoxide (DMSO; purity ≥ 99.9%) were purchased from Sigma-Aldrich (St. Louis, MO). ICI 182,780 (ICI), an ER antagonist; 2,3-bis(4-hydroxyphenyl)-propionitrile (DPN) and 4,4´,4´´-(4-propyl-[1H]-pyrazole-1,3,5-triyl)trisphenol (PPT), synthetic ER agonists with affinity for human ERα and ERβ, respectively; and 1,3-bis(4-hydroxyphenyl)-4-methyl-5-[4-(2-piperidinylethoxy)phenol]-1H-pyrazole dihydrochloride (MPP) and 4-[2-phenyl-5,7-bis(trifluoromethyl)pyrazolo[1,5-a]pyrimidin-3-yl]phenol (PHTPP), antagonists for ERα and ERβ, respectively, were obtained from Tocris Biosciences (Bristol, UK), with purity of > 99%, except for MPP (purity > 98%). All chemicals were dissolved in DMSO and diluted into dechlorinated fish water for a final DMSO concentration of 0.1%.
Zebrafish. Water used to house zebrafish was ultraviolet-light sterilized and circulated through a fluidized bed filtration system (Aquaneering Inc., San Diego, CA). We used three zebrafish strains: the wild-type AB laboratory strain (Walker 1999) and the transgenic strains Tg(5xERE:GFP)c262/c262 and Tg(5xERE:GFP)c263 (Gorelick and Halpern 2011). All work was approved by the Institutional Animal Care and Use Committee of the Carnegie Institution for Science. All animals were treated humanely and with regard for alleviation of suffering.
RNA in situ hybridization. We used antisense RNA probes corresponding to esr1 (ERα), esr2a (ERβ1), and esr2b (ERβ2) as described previously (Gorelick and Halpern 2011). [Note that some previous publications (e.g., Bertrand et al. 2007) referred to the esr2b gene as esr2a and the esr2a gene as esr2b.] We also assayed sense RNA probes, but they did not produce signals above background (data not shown). Colorimetric whole-mount in situ hybridization was performed on zebrafish embryos and larvae as described previously (Gorelick and Halpern 2011), except 5% dextran (final) was included in the hybridization solution (Lauter et al. 2011). Images were collected using a Zeiss Axioskop microscope equipped with an AxioCam HRc digital camera (Carl Zeiss Microimaging, Thornwood, NJ). Image adjustments and cropping were performed using Photoshop CS5 and InDesign CS5 (both from Adobe Systems Inc., San Jose, CA).
Water sampling. To concentrate estrogens over time, passive sampling devices [Polar Organic Chemical Integrative Sampler (POCIS), fabricated at the U.S. Geological Survey (USGS) Columbia Environmental Research Center (Columbia, MO) as described by Alvarez et al. (2004)] were deployed in rivers and streams at 19 locations in the Shenandoah watershed and the Allegheny, Delaware, and Susquehanna Rivers in Virginia and Pennsylvania in April 2010 and remained in place for 31–45 days (see Supplemental Material, Table S1). The Shenandoah and Susquehanna sites are part of an ongoing monitoring and research program to determine the factors involved in fish lesions and mortalities and to assess signs of reproductive endocrine disruption (testicular oocytes and plasma vitellogenin in male bass) observed in these watersheds (Blazer et al. 2010; Reif et al. 2012). The Allegheny and Delaware sites were used as comparisons for the Susquehanna sites in the Pennsylvania emerging contaminants project (Reif et al. 2012). The POCIS devices were deployed during April and May because these months were previously identified as periods of high estrogenicity in the Virginia watershed (Ciparis et al. 2012). After 31–45 days, the sampling devices were retrieved as described by Alvarez (2010), and POCIS membranes were shipped to the USGS Columbia Environmental Research Center for analyte recovery as previously described (Alvarez et al. 2009). Briefly, the POCIS membranes were extracted using 50 mL of 1:1:8 (vol:vol:vol) methanol:toluene:dichloromethane followed by 20 mL ethyl acetate. Extracts were reduced by rotary evaporation, filtered, and composited into two equivalent POCIS samples, thereby increasing the amount of chemical present in each sample to aid in detection.
Samples were resuspended in DMSO and diluted into fish water between 1:100 and 1:4,000 (vol:vol). At 1 day postfertilization (dpf), Tg(5xERE:GFP)c262/c262 embryos were exposed to treated water; they were examined for fluorescent labeling at 3 or 4 dpf. Four embryos were exposed per treatment (see Supplemental Material, Table S2). Embryos were incubated in 24- or 96-well plates at a density of no more than four and two embryos per well, respectively. Exposure occurred under static water conditions, with no water changes during exposure. Embryos were incubated at 28°C under an 18-hr light/6-hr dark cycle.
For discrete water sampling, two sites from the POCIS deployment were selected for follow-up analysis based on results from the initial zebrafish assay. Muddy Creek was selected because samples from that site preferentially activated the reporter in heart valves. Hawksbill Creek was selected because samples from that site exhibited the most intense fluorescence. Water was collected from the Muddy Creek and Hawksbill Creek locations (corresponding to samples 7 and 16 from the POCIS study; see Supplemental Material, Table S1) approximately 1 year after passive sampling to minimize seasonal effects. Samples were extracted with OASIS HLB glass cartridges (Waters Corporation, Milford, MA) as described by Ciparis et al. (2012). The methanol/methanol:dichloromethane eluate was dried under a continuous flow of atmospheric air, resuspended in DMSO, and serially diluted into fish water from 1:500 to 1:10,000 (vol:vol; equivalent to exposing larvae to 5–100 times the concentration found at sampling sites).
For negative controls, a field blank was prepared for each POCIS site and treated identically to POCIS extractions (Alvarez 2010). Briefly, field blanks were stored in airtight containers and transported to the field locations in insulated coolers. During both deployment and retrieval of the passive samplers the lids of the field blank containers were opened and exposed to the surrounding air; this simulated possible exposure to airborne contaminants of the actual deployed sampler. The field blanks were then extracted using the same method as for the deployed sampler. For POCIS samples diluted 1:100 into fish water, the vehicle control was zebrafish incubated in fish water containing 1% DMSO. For other conditions, the vehicle control was zebrafish incubated in fish water containing 0.1% DMSO. For positive controls, zebrafish were incubated in water containing 100 ng/mL estradiol.
Samples were randomly coded so that researchers were blinded to sample identity during zebrafish testing. For the initial screening, GFP fluorescence within live embryos and larvae was visualized using an Olympus MVX10 fluorescent stereomicroscope (Olympus, Center Valley, PA) equipped with a Leica DCF500 digital camera (Leica Microsystems Inc., Buffalo Grove, IL). Images were captured using identical microscope and camera settings. For secondary imaging at higher magnification, embryos and larvae were mounted on bridged coverslips and examined on a Zeiss Axio Imager microscope equipped with an AxioCam HRm digital camera (Carl Zeiss Microimaging, Thornwood, NJ).
Morpholinos. To reduce levels of ER protein, 1-cell-stage Tg(5xERE:GFP)c262/c262 embryos were injected with antisense morpholino oligonucleotides targeting the translation start sites of either esr2a (5´-ACATGGTGAAGGCGGATGAGTTCAG) or esr2b (5´-AGCTCATGCTGGAGAACACAAGAGA) (Gene Tools, Philomath, OR). Morpholinos were resuspended in water at 30 μM, and 1–2 nL was injected into each embryo as described by Nasevicius and Ekker (2000). Beginning at 2 dpf, embryos were incubated in 10 μM BPA or vehicle control (0.1% ethanol). At 3 dpf, fluorescence was assayed as described above.
Yeast ER reporter assay. To measure estrogen equivalents (relative to 17β-estradiol) of the analytes present in the POCIS extracts, we performed a bioluminescent yeast estrogen screen (BLYES) (Sanseverino et al. 2005) as described previously (Ciparis et al. 2012). All assay plates included a 12-point standard curve consisting of estradiol (2.3 × 10–11 to 5.0 × 10–7 M) and sample blanks containing minimal media only. Samples, standards, and blanks were run in triplicate. Luminescence was quantified using a SpectraFluor Plus plate reader (Tecan Group Ltd., Durham, NC). A linear calibration curve was created using log10 transformations of the five lowest standards (2.3 × 10–11 to 2.1 × 10–10 M estradiol) and their associated mean luminescence. Concentrations in samples with luminescence above this range were quantified using four points from the linear portion of the dose–response curve (log10[estradiol] vs. mean luminescence; 1.2 × 10–10 to 1.9 × 10–9 M estradiol), extrapolated from these standards, and reported as ng/POCIS estradiol equivalents (E2Eq).
Results
Environmental estrogens preferentially activate receptors in heart valves. At 1 dpf, groups of 5xERE:GFP transgenic zebrafish embryos were exposed for 3–4 days to POCIS extracts collected from 19 locations in the Shenandoah River watershed and the Allegheny, Delaware, and Susquehanna Rivers (Figure 1; see also Supplemental Material, Table S1). A surprisingly large number of samples (16) activated the ER reporter in transgenic zebrafish, with 5 samples preferentially inducing GFP labeling of the heart valves (Figure 1; see also Supplemental Material, Table S2 and Movie S1). In embryos exposed to samples 3 (Delaware River; diluted 1:1,000) and 6 (Naked Creek, VA; diluted 1:500), the ER reporter was activated in the heart valves but not the liver (see Supplemental Material, Table S2). Embryos exposed to sample 7 (Muddy Creek, VA) showed activation in both tissues, but with increased sensitivity in the heart valves (1:1,000 dilution) compared with the liver (1:500 dilution).
Figure 1.

Sites of sample collection (n = 19) in April and May of 2010. Each circle represents a sampling site, and the color of the circle indicates the presence or absence of ER activity in the 5xERE:GFP zebrafish reporter after incubation in water containing extracts from sampled water. Abbreviations: DE, Delaware; MD, Maryland; NJ, New Jersey; PA, Pennsylvania; VA, Virginia; WV, West Virginia.
To confirm that reporter activity was specific for ERs, we exposed embryos to water samples that either activated the reporter in the heart valves alone (samples 3 or 7, diluted 1:1,000) or together with the liver (samples 16 or 18, diluted 1:1,000) in the presence of the ER antagonist ICI (Robertson 2001) (Figure 2). Co-treatment with 10 μM ICI abolished fluorescence in all embryos (Figure 2B,D; see also Supplemental Material, Table S2), indicating that the chemicals in the water were either ER agonists or led to the production of ER agonists in zebrafish. Embryos treated with 100 ng/mL estradiol exhibited robust fluorescence in the heart and liver (Figure 2G), whereas embryos treated with 10 μM ICI alone did not exhibit fluorescence (data not shown), consistent with previous studies (Gorelick and Halpern 2011). Thus, 5xERE:GFP transgenic zebrafish larvae were able to report tissue-specific ER activation of unknown estrogens from passively sampled water.
Figure 2.
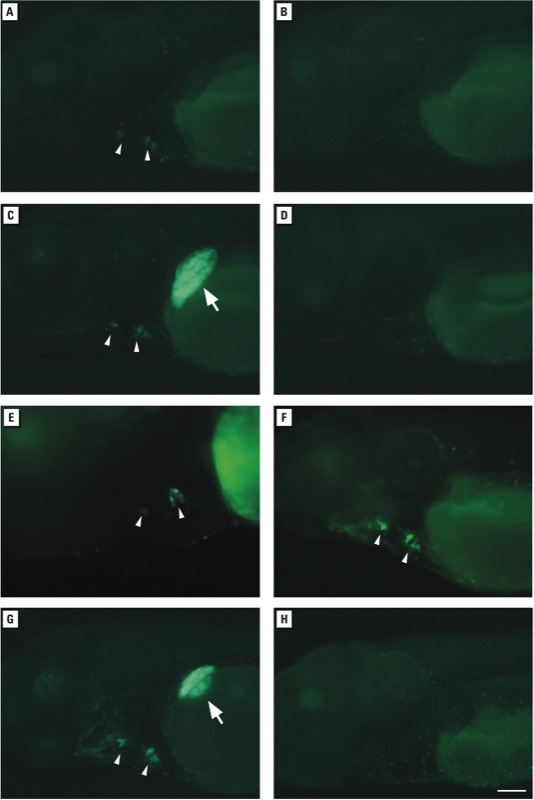
Tissue-specific responses of Tg(5xERE:GFP)c262 zebrafish embryos to environmental estrogens after incubation in water containing extracts from water sampled from the Shenandoah Watershed and nearby rivers. Samples shown in (A–E) were collected in 2010 using passive sampling. (A) Delaware River, Pennsylvania, sample 3 (diluted 1:1,000). (B) Delaware River, sample 3 plus the estrogen receptor antagonist ICI (10 μM). (C) Hawksbill Creek, Virginia, sample 16 (diluted 1:1,000). (D) Hawksbill Creek, sample 16 plus 10 μM ICI. (E) Naked Creek, Virginia, sample 6 (diluted 1:500). (F) Hawksbill Creek sample (discrete sampling) collected in 2011 (diluted 1:500). (G) Positive control water containing 100 ng/mL estradiol. (H) Negative control field blank. Fluorescence was visualized in the liver (arrows) and heart valves (arrow heads) of live larvae at 3 dpf (F) or 4 dpf (A–E,G,H). All images are lateral views, with anterior to the left and dorsal to the top. Bar = 100 μm.
POCIS sampling provides a time-weighted average of chemical exposure over several weeks, whereas discrete sampling provides a snapshot of chemical exposure at a single point in time. We examined whether the zebrafish reporter was sensitive enough to detect environmental estrogens from single pass collections at the Muddy Creek and Hawksbill Creek locations (samples 7 and 16 from the passive sampling study; see Supplemental Material, Table S1). Approximately 1 year after passive sampling, we collected and concentrated 1 L of water from the same locations. As in the previous findings, water from Hawksbill Creek diluted 1:500 or 1:1,000 activated the reporter in the heart valves (Figure 2F), whereas fluorescence was not observed at greater dilutions (1:5000, 1:10,000) (data not shown; n = 20 embryos per dilution per sample). Thus, 5xERE:GFP embryos were able to detect environmental estrogens from water samples collected from the same sites at different times using passive or discrete sampling methods.
To assess the sensitivity of the 5xERE:GFP zebrafish reporter, we compared the responses in zebrafish with those measured using a widely used yeast reporter assay (Balsiger et al. 2010; Leskinen et al. 2005; Sanseverino et al. 2005). Passively sampled water was tested using the BLYES (Sanseverino et al. 2005), which utilizes a yeast strain containing the human ESR1 (ERα) gene and a tandem ERE that drives an inducible luxAB reporter. Every water sample that activated the zebrafish reporter was readily detected using the yeast system (E2Eq between 0.76 and 7.98 ng/POCIS; Supplemental Material, Table S2). Moreover, the three water samples that failed to activate the zebrafish reporter exhibited the lowest levels of activity in the yeast assay (< 0.8 E2Eq; see Supplemental Material, Table S2).
Tissue specificity in ER gene expression. A plausible explanation for the tissue-specific differences in activation of the transgenic reporter by known estrogenic compounds and environmental samples is diversity in ERs. Zebrafish express three ER subtypes, ERα, ERβ1, and ERβ2 (encoded by the genes esr1, esr2a, and esr2b), which, similar to their mammalian orthologues, bind ligands with different affinities in vitro (Cosnefroy et al. 2012; Menuet et al. 2002). Previous studies demonstrated that esr2b is expressed in the embryonic and larval liver (Bertrand et al. 2007; Gorelick and Halpern 2011). However, there has been no report of ER transcripts or proteins in the developing heart of zebrafish.
To determine the presence of ER transcripts in the zebrafish heart, we reexamined expression of ER genes in 3–5 dpf zebrafish using a method for whole-mount in situ hybridization with enhanced sensitivity (Lauter et al. 2011). In 5 dpf zebrafish larvae, we observed robust expression of esr2b in the liver, consistent with previous studies (Bertrand et al. 2007; Gorelick and Halpern 2011) and discovered that esr1 is selectively transcribed in the developing valves of the heart (Figure 3). We did not detect esr2a transcripts at this stage (Figure 3B). The results demonstrate that different ER subtypes are specifically expressed in the heart and liver. In a previous study we found that the ER ligands BPA and genistein preferentially activated receptors in zebrafish heart valves compared with the liver (Gorelick and Halpern 2011). The differences in ER subtype localization reported here support the idea that BPA and genistein preferentially activate ERα in the heart because they have a higher affinity for this ER subtype.
Figure 3.

Expression of esr1, esr2a, and esr2b genes in heart and liver detected using whole mount in situ hybridization larvae at 5 dpf. (A) esr1 transcripts are present in heart valves (arrow heads) but not in liver. Inset, high magnification ventral view of heart showing labeling of atrioventricular valve leaflets. (B) esr2a transcripts are not present in heart or liver. (C) esr2b transcripts are present in liver (arrow) but not in heart valves. All views are lateral views with anterior to the left. Bar = 50 μm.
Selective ER modulation in the heart and liver. To corroborate the findings of differential gene expression, we used genetic and pharmacological approaches to activate or inhibit ERα, ERβ1, or ERβ2 selectively in 3–4 dpf transgenic zebrafish. On the basis of gene expression, reducing ERβ2 protein levels should reduce ER activity in the liver but not in heart valves. We injected 5xERE:GFP embryos with antisense morpholino oligonucleotides targeting esr2a or esr2b genes (1–2 nL of 30 μM solution), incubated embryos in 10 μM BPA, and assayed fluorescence. Fluorescence in the liver was reduced in esr2b-morphant embryos (Figure 4B) compared with esr2a-morphant embryos (Figure 4A), whereas robust labeling of the heart valves was observed in all morphant embryos (Figure 4A,B and Table 1). Embryos exposed to the vehicle alone (0.1% ethanol) exhibited no fluorescence in the liver or heart (data not shown). Attempts to reduce ERα levels using three different morpholinos targeting esr1 gene translation and RNA splicing were ineffective because esr1-morphant embryos exhibited pleiotropic developmental defects such as cardiac edema, a small head, and curved tail (data not shown), suggesting a nonspecific response.
Figure 4.

ER subtype-specific activity and tissue-specific response of 5xERE:GFP embryos that were injected with esr2a (A) or esr2b (B) antisense morpholino oligonucleotides (MO) to inhibit translation of ERβ1 or ERβ2 proteins. Embryos were exposed to 10 μM BPA at 2 dpf, and fluorescence was visualized a day later. Fluorescence was visualized in the liver (arrows) and heart valves (arrow heads) of live larvae. Embryos injected with esr2a‑MO (A) exhibited fluorescence in the liver and heart valves, whereas those injected with esr2b‑MO (B) exhibited fluorescence in heart valves but not liver. (C–E) Embryos were exposed to 10 μM BPA or one of the ER subtype-specific agonists (PPT, 100 μM; DPN, 1 μM) at 3 dpf, and fluorescence was visualized a day later. PPT and DPN selectively activated the reporter in liver or heart valves. All images are lateral views, with anterior to the left and dorsal to the top. Bar = 100 μm.
Table 1.
Results of esr morpholino treatment.
| Morpholino target | Morpholino dose of 30 μM solution | GFP+ heart valves only (%) | GFP+ liver only (%) | GFP+ heart valves and liver (%) | Embryos (n) |
|---|---|---|---|---|---|
| esr2a | 2 nL | 0 | 0 | 100 | 18 |
| esr2b | 1 nL | 56 | 0 | 44 | 18 |
| esr2b | 2 nL | 94 | 0 | 6 | 18 |
| One-cell stage 5xERE:GFP embryos were injected with translation blocking morpholinos to reduce ER levels. At 2 dpf, embryos were incubated in 10 μM BPA. Fluorescence was assayed at 3 dpf, and data are presented as the percentage of GFP-positive embryos (GFP+) in the indicated tissues. | |||||
To activate ER subtypes selectively, we used the synthetic ER ligands PPT and DPN (Meyers et al. 2001; Stauffer et al. 2000). PPT has higher affinity for human ERα than for ERβ, but DPN has higher affinity for ERβ. We found, however, that 5xERE:GFP zebrafish embryos exposed to 100 μM PPT showed GFP labeling of only the liver, where ERβ2 is produced (n = 10; Figure 4D); those exposed to 1 μM DPN showed GFP-labeling of the heart valves, which synthesize ERα (n = 20; Figure 4E). Embryos exposed to 10 μM BPA, the positive control, showed GPF labeling of the heart valves and liver (Figure 4C). To inhibit ER subtypes selectively, we exposed zebrafish to selective antagonists designed against human ER subtypes. Treatment of 5xERE:GFP embryos with either the ERα or ERβ antagonists MPP (Sun et al. 2002) or PHTPP (Compton et al. 2004) failed to inhibit reporter activity in any tissue (data not shown). Thus, different ER agonists selectively activated receptors in either the heart valves or the liver of zebrafish larvae, but in a manner opposite to what we expected based on their activation of human ER receptors.
Discussion
In the present study we observed that 5xERE:GFP reporter zebrafish can detect the tissue-specific effects of environmental estrogens. This represents a significant improvement over traditional detection assays using yeast (Routledge and Sumpter 1996) or cultured cells (Legler et al. 1999), which do not allow comparisons between multiple tissues. Furthermore, testing compounds for ER activity in zebrafish larvae involves the physiologically relevant parameters of absorption, distribution, metabolism, and excretion.
We found a high concordance between responses in zebrafish and in a bioluminescent yeast assay for detection of estrogens from the same environmental samples. This indicates that the whole embryo assay of transgenic zebrafish correlates well with an established and sensitive method (Bergamasco et al. 2011; Ciparis et al. 2012; Sanseverino et al. 2005) for measuring estrogenic compounds in water samples. In addition, the zebrafish reporter revealed a previously unknown tissue and developmental stage for ER signaling, the newly formed heart valves. With the genetic and pharmacological tools available to manipulate zebrafish, transgenic models can be readily applied to detect tissue-specific environmental estrogens and identify their mode of action. Future studies will broaden this approach to report the activity of other environmentally relevant small molecules such as androgens and dioxins.
Although estrogens in discrete and time-integrated passively collected water samples were detectable in 5xERE:GFP zebrafish, our results suggest that estrogen levels vary depending on the sampling method. For example, POCIS extracts prepared from water collected from Muddy Creek in June 2010 activated the reporter preferentially in heart valves, whereas discrete water samples collected the following year did not. Similarly, POCIS extracts from Hawksbill Creek collected in June 2010 activated the reporter in the heart valves and liver, but discrete water samples collected the following year preferentially activated the reporter only in heart valves. These differences are not surprising, however, given the likely daily and seasonal variations in the concentration of environmental estrogens (Ciparis et al. 2012; Martinovic et al. 2008).
An unexpected finding is that that DPN and PPT appear to activate zebrafish ERα and ERβ2, respectively, the opposite of what has been observed for the human ER subtypes (Meyers et al. 2001; Stauffer et al. 2000). One possibility is that zebrafish ERα has greater functional homology to human ERβ. Although zebrafish ERα is most similar to human ERα when comparing the entire protein sequence, similarities between functional domains within each protein are more relevant for predicting functional homology. For example, in the N-terminal AF-1 domain that regulates transcriptional activation (also referred to as the A/B domain) (Metzger et al. 1995), zebrafish ERα is more similar to human ERβ (13.2%) than to human ERα (8.4%) (Menuet et al. 2002). Low sequence homology (< 15% identity) between the AF-1 domains from human and zebrafish ERs makes it difficult to predict functional homology between subtypes with accuracy. Furthermore, studies using chimeric ER proteins from rainbow trout and humans suggest that, despite low sequence homology, ER domains from different species may function similarly and interact with the same transcription factors (Petit et al. 2000). It is therefore not surprising that agonists might show altered affinities for ERs in species as diverse as fish and humans.
Although ER-subtype–selective agonists (DPN and PPT) designed against human ERs were effective in zebrafish, selective antagonists designed against human ER subtypes were not. These in vivo results are consistent with those obtained in cultured cells expressing zebrafish ERs, where MPP and PHTPP also failed to inhibit ERE-dependent reporter activity induced by 17α-ethynylestradiol (Notch and Mayer 2011). Together, these data suggest that MPP and PHTPP do inhibit zebrafish ERs.
The environmental estrogenic compound(s) capable of activating the zebrafish reporter with tissue specificity remains to be identified. The low levels of known estrogens in water samples make this a challenging endeavor, requiring sequential rounds of HPLC fractionation for purification and mass spectrometry for identification. However, the small size and transparency of zebrafish embryos are advantageous for rapid, high-throughput screening of fractions for tissue-specific ER activity. Ultimately, it will be possible to identify unknown EEDs that affect estrogen signaling, their sites of action, and effects on embryonic development.
The activation of ERs in heart valves during development leads to the intriguing hypothesis that estrogen signaling influences valve formation. In humans, the occurrence of heart valve abnormalities differs between the sexes, which could be due to sex differences in estrogen levels. Bicuspid aortic valve defects, where the aortic valve develops two leaflets instead of three, are four times more prevalent in men than in women (Warnes 2008). Because ERs are ligand-dependent transcription factors, it will be important to identify which genes are directly regulated by estrogens and to test whether they are important for cell migration or proliferation of valve precursors. Exposure to environmental endocrine-disrupting compounds that mimic or inhibit endogenous estrogens in utero is associated with adverse health effects (Soto and Sonnenschein 2010), with the potentially unanticipated consequence of causing heart valve malformations.
Supplemental Material
Acknowledgments
We thank M. Macurak, T. McDonough, and A. Sperry for technical assistance; W. Goessling and T. North for sharing morpholino sequences; J. Sanseverino for providing the BLYES yeast strain; and R. Braham for assistance generating the map in Figure 1.
Footnotes
D.A.G. was funded by a National Institutes of Health National Research Service Award Postdoctoral Fellowship from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (5F32HD061119-02). Partial funding for the project was provided by the U.S. Geological Survey Chesapeake Bay Priority Ecosystems program.
Use of trade names is for identification purposes only and does not imply endorsement by the U.S. government.
The authors declare they have no actual or potential competing financial interests.
References
- Alvarez DA. Guidelines for the Use of the Semipermeable Membrane Device (SPMD) and the Polar Organic Chemical Integrative Sampler (POCIS) in Environmental Monitoring Studies. Techniques and Methods 1–D4. Reston, VA:U.S. Geological Survey. 2010. Available: http://pubs.usgs.gov/tm/tm1d4/pdf/tm1d4.pdf [accessed 19 February 2014]
- Alvarez DA, Cranor WL, Perkins SD, Schroeder VL, Iwanowicz LR, Clark RC, et al. Reproductive health of bass in the Potomac, USA, drainage: part 2. Seasonal occurrence of persistent and emerging organic contaminants. Environ Toxicol Chem. 2009;28(5):1084–1095. doi: 10.1897/08-417.1. [DOI] [PubMed] [Google Scholar]
- Alvarez DA, Petty JD, Huckins JN, Jones-Lepp TL, Getting DT, Goddard JP, Manahan SE. Development of a passive, in situ, integrative sampler for hydrophilic organic contaminants in aquatic environments. Environ Toxicol Chem. 2004;23:1640–1648. doi: 10.1897/03-603. [DOI] [PubMed] [Google Scholar]
- Balsiger HA, de la Torre R, Lee WY, Cox MB. A four-hour yeast bioassay for the direct measure of estrogenic activity in wastewater without sample extraction, concentration, or sterilization. Sci Total Environ. 2010;408(6):1422–1429. doi: 10.1016/j.scitotenv.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamasco AM, Eldridge M, Sanseverino J, Sodre FF, Montagner CC, Pescara IC, et al. Bioluminescent yeast estrogen assay (BLYES) as a sensitive tool to monitor surface and drinking water for estrogenicity. J Environ Monit. 2011;13(11):3288–3293. doi: 10.1039/c1em10464k. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PL, et al. 2007Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genetics 311e188; 10.1371/journal.pgen.0030188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazer VS, Iwanowicz LR, Starliper CE, Iwanowicz DD, Barbash P, Hedrick JD, et al. Mortality of centrarchid fishes in the Potomac drainage: survey results and overview of potential contributing factors. J Aquat Anim Health. 2010;22:190–218. doi: 10.1577/H10-002.1. [DOI] [PubMed] [Google Scholar]
- Brion F, Le Page Y, Piccini B, Cardoso O, Tong SK, Chung BC, et al. 2012Screening estrogenic activities of chemicals or mixtures in vivo using transgenic (cyp19a1b-GFP) zebrafish embryos. PLoS ONE 75e36069; 10.1371/journal.pone.0036069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciana P, Raviscioni M, Mussi P, Vegeto E, Que I, Parker MG, et al. In vivo imaging of transcriptionally active estrogen receptors. Nat Med. 2003;9(1):82–86. doi: 10.1038/nm809. [DOI] [PubMed] [Google Scholar]
- Ciparis S, Iwanowicz LR, Voshell JR. Effects of watershed densities of animal feeding operations on nutrient concentrations and estrogenic activity in agricultural streams. Sci Total Environ. 2012;414:268–276. doi: 10.1016/j.scitotenv.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Compton DR, Sheng S, Carlson KE, Rebacz NA, Lee IY, Katzenellenbogen BS, et al. Pyrazolo[1,5-a]pyrimidines: estrogen receptor ligands possessing estrogen receptor β antagonist activity. J Med Chem. 2004;47(24):5872–5893. doi: 10.1021/jm049631k. [DOI] [PubMed] [Google Scholar]
- Cosnefroy A, Brion F, Maillot-Maréchal E, Porcher JM, Pakdel F, Balaguer P, et al. Selective activation of zebrafish estrogen receptor subtypes by chemicals by using stable reporter gene assay developed in a zebrafish liver cell line. Toxicol Sci. 2012;125(2):439–449. doi: 10.1093/toxsci/kfr297. [DOI] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116(3):561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick DA, Halpern ME. Visualization of estrogen receptor transcriptional activation in zebrafish. Endocrinology. 2011;152(7):2690–2703. doi: 10.1210/en.2010-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber CJ, Gruber DM, Gruber IM, Wieser F, Huber JC. Anatomy of the estrogen response element. Trends Endocrinol Metab. 2004;15(2):73–78. doi: 10.1016/j.tem.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Kuiper GGJM, Carlsson B, Grandien K, Enmark E, Häggblad J, Nilsson S, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology. 1997;138(3):863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- Kurauchi K, Nakaguchi Y, Tsutsumi M, Hori H, Kurihara R, Hashimoto S, et al. In vivo visual reporter system for detection of estrogen-like substances by transgenic medaka. Environ Sci Technol. 2005;39(8):2762–2768. doi: 10.1021/es0486465. [DOI] [PubMed] [Google Scholar]
- Lauter G, Söll I, Hauptmann G.2011Multicolor fluorescent in situ hybridization to define abutting and overlapping gene expression in the embryonic zebrafish brain. Neural Dev 610; 10.1186/1749-8104-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee O, Takesono A, Tada M, Tyler CR, Kudoh T.2012Biosensor zebrafish provide new insights into potential health effects of environmental estrogens. Environ Health Perspect 1207990–996.; 10.1289/ehp.1104433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legler J, Broekhof JLM, Brouwer A, Lanser PH, Murk AJ, van der Saag PT, et al. A novel in vivo bioassay for (xeno-)estrogens using transgenic zebrafish. Environ Sci Technol. 2000;34(20):4439–4444. [Google Scholar]
- Legler J, van den Brink CE, Brouwer A, Murk AJ, van der Saag PT, Vethaak AD, et al. Development of a stably transfected estrogen receptor-mediated luciferase reporter gene assay in the human T47D breast cancer cell line. Toxicol Sci. 1999;48(1):55–66. doi: 10.1093/toxsci/48.1.55. [DOI] [PubMed] [Google Scholar]
- Leskinen P, Michelini E, Picard D, Karp M, Virta M. Bioluminescent yeast assays for detecting estrogenic and androgenic activity in different matrices. Chemosphere. 2005;61(2):259–266. doi: 10.1016/j.chemosphere.2005.01.080. [DOI] [PubMed] [Google Scholar]
- Ma L. Endocrine disruptors in female reproductive tract development and carcinogenesis. Trends Endocrinol Metab. 2009;20(7):357–363. doi: 10.1016/j.tem.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinovic D, Denny JS, Schmieder PK, Ankley GT, Sorensen PW. Temporal variation in the estrogenicity of a sewage treatment plant effluent and its biological significance. Environ Sci Technol. 2008;42(9):3421–3427. doi: 10.1021/es0708013. [DOI] [PubMed] [Google Scholar]
- Meeker JD. Exposure to environmental endocrine disruptors and child development. Arch Pediatr Adolesc Med. 2012;166(10):952–958. [PubMed] [Google Scholar]
- Menuet A, Pellegrini E, Anglade I, Blaise O, Laudet V, Kah O, et al. Molecular characterization of three estrogen receptor forms in zebrafish: binding characteristics, transactivation properties, and tissue distributions. Biol Reprod. 2002;66(6):1881–1892. doi: 10.1095/biolreprod66.6.1881. [DOI] [PubMed] [Google Scholar]
- Metzger D, Ali S, Bornert JM, Chambon P. Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J Biol Chem. 1995;270(16):9535–9542. doi: 10.1074/jbc.270.16.9535. [DOI] [PubMed] [Google Scholar]
- Meyers MJ, Sun J, Carlson KE, Marriner GA, Katzenellenbogen BS, Katzenellenbogen JA. Estrogen receptor-β potency-selective ligands: structure-activity relationship studies of diarylpropionitriles and their acetylene and polar analogues. J Med Chem. 2001;44(24):4230–4251. doi: 10.1021/jm010254a. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Gen. 2000;26(2):216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Notch EG, Mayer GD. Efficacy of pharmacological estrogen receptor antagonists in blocking activation of zebrafish estrogen receptors. Gen Comp Endocrinol. 2011;173(1):183–189. doi: 10.1016/j.ygcen.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Petit FG, Valotaire Y, Pakdel F. The analysis of chimeric human/rainbow trout estrogen receptors reveals amino acid residues outside of P- and D-boxes important for the transactivation function. Nucleic Acids Res. 2000;28(14):2634–2642. doi: 10.1093/nar/28.14.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif AG, Crawford JK, Loper CA, Proctor A, Manning R, Titler R. Occurrence of Pharmaceuticals, Hormones, and Organic Wastewater Compounds in Pennsylvania Waters, 2006–09. Scientific Investigations Report 2012–5106. Reston, VA:U.S. Geological Survey. 2012. Available: http://pubs.usgs.gov/sir/2012/5106/pdf/sir2012-5106.pdf [accessed 19 February 2014]
- Robertson JF. ICI 182,780 (Fulvestrant™)—the first oestrogen receptor down-regulator—current clinical data. Br J Cancer. 2001;85(suppl 2):11–14. doi: 10.1054/bjoc.2001.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routledge EJ, Sumpter JP. Estrogenic activity of surfactants and some of their degradation products assessed using a recombinant yeast screen. Environ Toxicol Chem. 1996;15(3):241–248. [Google Scholar]
- Sanseverino J, Gupta RK, Layton AC, Patterson SS, Ripp SA, Saidak L, et al. Use of Saccharomyces cerevisiae BLYES expressing bacterial bioluminescence for rapid, sensitive detection of estrogenic compounds. Appl Environ Microbiol. 2005;71(8):4455–4460. doi: 10.1128/AEM.71.8.4455-4460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto AM, Sonnenschein C. Environmental causes of cancer: endocrine disruptors as carcinogens. Nat Rev Endocrinol. 2010;6(7):363–370. doi: 10.1038/nrendo.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer SR, Coletta CJ, Tedesco R, Nishiguchi G, Carlson K, Sun J, et al. Pyrazole ligands: structure-affinity/activity relationships and estrogen receptor-α-selective agonists. J Med Chem. 2000;43(26):4934–4947. doi: 10.1021/jm000170m. [DOI] [PubMed] [Google Scholar]
- Sun J, Huang YR, Harrington WR, Sheng S, Katzenellenbogen JA, Katzenellenbogen BS. Antagonists selective for estrogen receptor α. Endocrinology. 2002;143(3):941–947. doi: 10.1210/endo.143.3.8704. [DOI] [PubMed] [Google Scholar]
- Walker C. Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 1999;60:43–70. doi: 10.1016/s0091-679x(08)61893-2. [DOI] [PubMed] [Google Scholar]
- Warnes CA. Sex differences in congenital heart disease: Should a woman be more like a man? Circulation. 2008;118(1):3–5. doi: 10.1161/CIRCULATIONAHA.108.785899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


