Abstract
The SCAR/WAVE complex drives the actin polymerisation that underlies protrusion of the front of the cell and thus drives migration. However, it is not understood how the activity of SCAR/WAVE is regulated to generate the infinite range of cellular shape changes observed during cell motility. What are the relative roles of the subunits of the SCAR/WAVE complex? What signaling molecules do they interact with? And how does the complex integrate all this information in order to control the temporal and spatial polymerisation of actin during protrusion formation? Unfortunately, the interdependence of SCAR complex members has made genetic dissection hard. In our recent paper,1 we describe stabilization of the Dictyostelium SCAR complex by a small fragment of Abi. Here we summarize the main findings and discuss how this approach can help reveal the inner workings of this impenetrable complex.
Keywords: SCAR, WAVE, Abi, Arp 2/3, actin, pseudopod, chemotaxis, motility
The SCAR/WAVE complex and its inputs
The highly conserved SCAR complex (also called the “WAVE complex”) causes actin-based protrusion during cell migration in a diverse range of eukaryotes including the amoeba Dictyostelium discoideum, Drosophila melanogaster, and mammalian cells.2-4 SCAR/WAVE is a WASP family member that induces actin nucleation via recruitment and activation of the Arp2/3 complex.5,6 In vivo, SCAR activity is regulated by its inclusion within a ~400 kDa regulatory complex consisting of PIR121, Nap1, HSPC300, and Abi.7-9 This regulatory complex undoubtedly acts as a signaling hub, where competing inputs are integrated and coupled to the activation of SCAR. However, we still lack even a basic knowledge of what the complex interacts with, never mind how such possible interactions are interpreted and translated into actin polymerisation, motility, and chemotaxis.10 Confusingly, WASP, which lacks the regulatory complex, can respond to many of the same inputs.2
The different roles of SCAR complex members
The recent resolution of the human SCAR complex crystal structure has greatly advanced our understanding of how these proteins interact with SCAR and one another.11,12 However, the relative contribution of the individual complex members to the activity of the whole remains poorly understood and still awaits elucidation.
Thus far, it has been established that the SCAR complex interacts with its best-known activator, Rac, via PIR121.11,13 As highlighted in Figure 1, Abi has been implicated in the recruitment of the SCAR complex to signaling complexes containing Rac.14 Furthermore, multiple, stimulus-responsive phosphorylation sites have also been identified across the different SCAR complex members.15
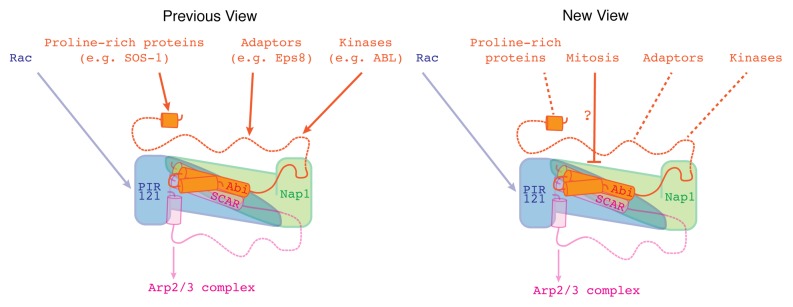
Figure 1. The role of Abi within the SCAR complex. The SCAR complex is composed of PIR121 (blue), Nap1 (green), HSPC300 (hidden in this image), SCAR (magenta), and Abi (orange). The SCAR complex promotes activation of the Arp2/3 complex (magenta arrow). Abi has long been considered a key regulator of the SCAR complex. In combination with the Rac/PIR121 interaction (blue arrow), Abi was thought to activate actin polymerization by coupling SCAR to various signaling and adaptor proteins (orange arrows) via its C-terminal SH3 domain (metazoans only) and polyproline tail (dashed ribbons). However, having deleted the majority of Dictyostelium Abi, it is now evident that Abi is not required for SCAR complex activation. Instead, Abi likely acts to tune the activity of the SCAR by integrating both positive and negative signals (dashed lines). One possible negative input could be acting through the N-terminal first α-helix to suppress SCAR activity during mitosis. However, the regulators that bind this domain have yet to be identified.
In vivo, SCAR is entirely dependent on its regulatory complex for stability, and in the absence of any one complex member, SCAR is rapidly degraded.3,16 Herein lies the problem that has confounded the study of the individual SCAR complex members: the inability to separate the function of the individual complex members from their requirement for complex stability. Numerous studies have endeavored to replace different SCAR complex members with mutant or truncated proteins in order to determine their function.17,18 However, all too often, the effect on complex stability has been ignored.
Most of Abi is dispensable
Previously, our lab has characterized the individual SCAR complex member gene disruptants in Dictyostelium discoideum.16,19-21 These mutants typically exhibit a scrA null phenotype due to the degradation of SCAR. We reasoned that if SCAR complex stability could be restored in the different SCAR complex member nulls, we could finally address the relative role of these proteins to the activity of the complex as a whole. In particular we sought to identify functional domains required for normal SCAR complex recruitment and activity. To achieve this, we set about generating a deletion series of the Dictyostelium PIR121, Nap1, and Abi with the objective of identifying minimal fragments of these proteins that could stabilize the complex. This would enable us to assign specific functions to the absent domains. Incremental truncation of both PIR121 and Nap1 failed to yield any fragments that could stabilize SCAR. Although not available at the time, the SCAR complex crystal structure subsequently revealed the highly convoluted conformation of both PIR121 and Nap1 within the complex, which undoubtedly underlies the failure of this approach in theses cases.
In contrast, in our recent paper we demonstrated that we could successfully delete 239 of the 332 amino acids comprising Abi and retain both SCAR and SCAR complex stability in the Dictyostelium abiA null.1 Surprisingly, we found that none of this sequence was necessary for robust recruitment of the SCAR complex to the tips of pseudopodia. The suppressed rate of pseudopod formation in the abiA null was also restored, implying that the majority of Abi sequence is not required for SCAR complex activation either. Although the N-terminus of Abi was specifically involved in regulating the SCAR complex during cytokinesis, apparently negatively, we could find no phenotype associated with a loss of the C-terminal polyproline tail.
What is the true role of Abi within the SCAR complex?
As summarized in Figure 1, it was concluded that SCAR complex localization and activity do not depend on any signals that are transduced through Abi.1 Instead, we suggest that Abi serves to modulate the activity of SCAR, particularly during events such as cytokinesis. Despite low detailed amino acid identity, a C-terminal polyproline tail is a consistent feature of almost all Abi homologs, which implies that the polyproline domain does have a universal role in the regulation of the SCAR complex. We propose it acts as a non-essential signal integrator that tunes the activity of the SCAR complex after it has been activated by alternate pathways.
Such a role is consistent with the evolutionary recent acquisition of a C-terminal SH3 domain found in metazoan Abi homologs. Even though this domain has been implicated in the regulation of the mammalian SCAR complex, it is not required for SCAR complex recruitment during actin-based protrusion.14,22
The attainment of multicellularity in metazoa has been accompanied with a huge increase in regulatory complexity, and cells within a tissue have very different requirements of the SCAR complex in comparison to free-living amoeba. For example, unlike amoebae that are constantly on the move, metazoan cells within a tissue presumably suppress SCAR complex activity until it is required during carefully choreographed events. We propose that Abi is the obvious candidate for the application of these additional layers of control.
Future directions and conclusions
Although deletion series analysis allowed us to explore the role of Abi within the SCAR complex, this has proved too crude a method to similarly investigate the function of the other complex members such as PIR121 and Nap1. We believe a subtler approach will be required to mutate these complex members while preserving complex stability. For instance, our lab has recently successfully used phosphomimetic mutation to study the activation of SCAR.23 Few phosphosites have been identified within PIR121 and Nap1. PIR121 and Nap1 are also both so large that systematic alanine scanning mutagenesis is a daunting prospect. However, given the fragility of the SCAR complex, it may prove the only available option.
The intricacy of the SCAR complex continues to block investigation from every angle, and as a result we have barely scratched the surface of how these proteins collectively function. Despite the difficulty, we believe that genetic approaches could still offer the key to unlocking the secrets of the SCAR complex.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Davidson AJ, Ura S, Thomason PA, Kalna G, Insall RH. Abi is required for modulation and stability of the SCAR/WAVE complex, but not localization or activity. Eukaryot Cell. 2013;12:1509–16. doi: 10.1128/EC.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veltman DM, King JS, Machesky LM, Insall RH. SCAR knockouts in Dictyostelium: WASP assumes SCAR’s position and upstream regulators in pseudopods. J Cell Biol. 2012;198:501–8. doi: 10.1083/jcb.201205058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–75. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Hahne P, Sechi A, Benesch S, Small JV. Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 2001;492:215–20. doi: 10.1016/S0014-5793(01)02239-6. [DOI] [PubMed] [Google Scholar]
- 5.Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–56. doi: 10.1016/S0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 6.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–44. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–3. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 8.Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16:561–3. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollitt AY, Insall RH. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J Cell Sci. 2009;122:2575–8. doi: 10.1242/jcs.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Insall RH. Understanding eukaryotic chemotaxis: a pseudopod-centred view. Nat Rev Mol Cell Biol. 2010;11:453–8. doi: 10.1038/nrm2905. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–8. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson AJ, Insall RH. Actin-based motility: WAVE regulatory complex structure reopens old SCARs. Curr Biol. 2011;21:R66–8. doi: 10.1016/j.cub.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura N, Iwamatsu A, Kaibuchi K. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273:291–5. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- 14.Innocenti M, Frittoli E, Ponzanelli I, Falck JR, Brachmann SM, Di Fiore PP, Scita G. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J Cell Biol. 2003;160:17–23. doi: 10.1083/jcb.200206079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–24. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibarra N, Blagg SL, Vazquez F, Insall RH. Nap1 regulates Dictyostelium cell motility and adhesion through SCAR-dependent and -independent pathways. Curr Biol. 2006;16:717–22. doi: 10.1016/j.cub.2006.02.068. [DOI] [PubMed] [Google Scholar]
- 17.Leng Y, Zhang J, Badour K, Arpaia E, Freeman S, Cheung P, Siu M, Siminovitch K. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc Natl Acad Sci U S A. 2005;102:1098–103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ring C, Ginsberg MH, Haling J, Pendergast AM. Abl-interactor-1 (Abi1) has a role in cardiovascular and placental development and is a binding partner of the alpha4 integrin. Proc Natl Acad Sci U S A. 2011;108:149–54. doi: 10.1073/pnas.1012316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blagg SL, Stewart M, Sambles C, Insall RH. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr Biol. 2003;13:1480–7. doi: 10.1016/S0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- 20.Pollitt AY, Insall RH. Abi mutants in Dictyostelium reveal specific roles for the SCAR/WAVE complex in cytokinesis. Curr Biol. 2008;18:203–10. doi: 10.1016/j.cub.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 21.Pollitt AY, Insall RH. Loss of Dictyostelium HSPC300 causes a scar-like phenotype and loss of SCAR protein. BMC Cell Biol. 2009;10:13. doi: 10.1186/1471-2121-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stradal T, Courtney KD, Rottner K, Hahne P, Small JV, Pendergast AM. The Abl interactor proteins localize to sites of actin polymerization at the tips of lamellipodia and filopodia. Curr Biol. 2001;11:891–5. doi: 10.1016/S0960-9822(01)00239-1. [DOI] [PubMed] [Google Scholar]
- 23.Ura S, Pollitt AY, Veltman DM, Morrice NA, Machesky LM, Insall RH. Pseudopod growth and evolution during cell movement is controlled through SCAR/WAVE dephosphorylation. Curr Biol. 2012;22:553–61. doi: 10.1016/j.cub.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]


