Abstract
Bacterial biofilms are complex multicellular communities that are often associated with the emergence of large-scale patterns across the biofilm. How bacteria self-organize to form these structured communities is an area of active research. We have recently determined that the emergence of an intricate network of trails that forms during the twitching motility mediated expansion of Pseudomonas aeruginosa biofilms is attributed to an interconnected furrow system that is forged in the solidified nutrient media by aggregates of cells as they migrate across the media surface. This network acts as a means for self-organization of collective behavior during biofilm expansion as the cells following these vanguard aggregates were preferentially confined within the furrow network resulting in the formation of an intricate network of trails of cells. Here we further explore the process by which the intricate network of trails emerges. We have determined that the formation of the intricate network of furrows is associated with significant remodeling of the sub-stratum underlying the biofilm. The concept of stigmergy has been used to describe a variety of self-organization processes observed in higher organisms and abiotic systems that involve indirect communication via persistent cues in the environment left by individuals that influence the behavior of other individuals of the group at a later point in time. We propose that the concept of stigmergy can also be applied to describe self-organization of bacterial biofilms and can be included in the repertoire of systems used by bacteria to coordinate complex multicellular behaviors.
Keywords: Self-organisation, twitching motility, biofilms, collective behaviour, Pseudomonas aeruginosa
The study of the emergence of large-scale pattern formation in biotic and abiotic systems is of broad scientific interest. Within biological systems pattern formation is a consequence of self-organization and collective motion displayed by the individual organisms belonging to a system or group.1,2 Collective behaviors are observed ubiquitously in nature from higher animals such as flocks of birds, schools of fish, social behaviors of ants and termites and herd migrations through to group behaviors observed in communities of microorganisms such as the active expansion of bacterial biofilms. It has been speculated that the emergence of self-organized pattern formation offers adaptive advantages for the system to respond to the surrounding environment.1,3
A common feature often displayed by these collective phenomena is the formation of trails that lead to the emergence of dramatic patterns of large-scale order.4 This is true for the development of bacterial communities, which are often characterized by extensive spatiotemporal patterns and multicellular structures.1,2,5-8 Understanding the mechanisms that govern the self-organized behaviors that lead to the emergence of these patterns is an area of active research.2,3,8,9
An example of the self-organized emergence of striking patterns in bacterial communities is observed at the edges of actively expanding biofilms of Pseudomonas aeruginosa when cultured at the interface of solidified nutrient media and a coverslip. Under these conditions, the biofilms rapidly expand via type IV pili (tfp)-mediated twitching motility producing an extensive and intricate interconnected network of cells.10,11 We recently set out to investigate how these actively expanding P. aeruginosa biofilm communities self-organize to produce such dramatic large-scale patterns.11 We found that during active biofilm expansion, cells self-organize into highly aligned aggregates (rafts) that plough a network of interconnected furrows which physically confine the following cells, resulting in the emergence of the lattice-like network of trails that is a characteristic feature of these biofilms.11 Here we have further explored the process by which the intricate network of trails is formed in actively expanding interstitial biofilms of P. aeruginosa.
Refinement of the trail network remodels the semi-solid substratum
In our previous study, we utilized tapping mode atomic force microscopy (AFM) to image and analyze the furrows within the semi-solid media once the cells had been removed via washing.11 However, our AFM imaging system was limited to a relatively small scan size and we were unable to accurately correlate AFM scan regions with specific regions of the biofilm visualized by phase-contrast microscopy. To overcome these limitations, in this study we have utilized correlative phase-contrast microscopy and 3D optical profilometry. The latter is a non-contact mode of imaging that permits visualization of a large area and thus enables acquisition of a “birds-eye” overview of the furrow network beneath the expanding biofilm.
In this study we employed low magnification time-lapse phase-contrast microscopy to obtain a large field of view to follow the formation of the intricate trail network. Visualization of time series captured every 2s for 1hr (Movie S1) confirmed our previous observations that the interconnected trail network forms as a consequence of aggregates of cells forging new intersecting furrows. This results in the creation of an extensive, intricate trail network that is stable over time.
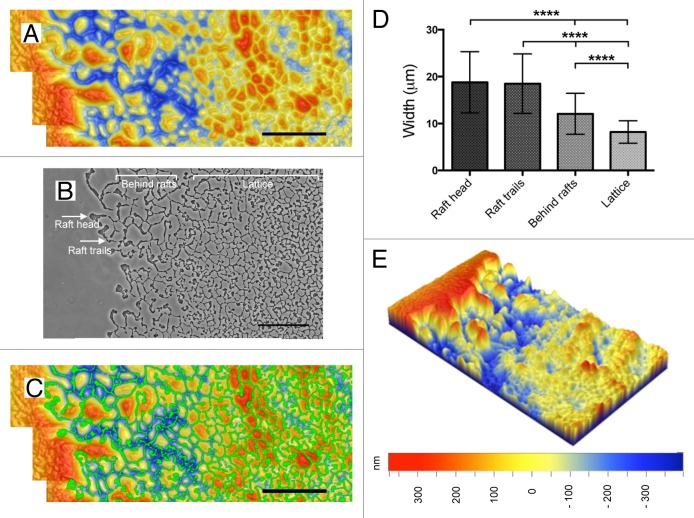
Immediately following cessation of the time-series capture, the cells were washed from the biofilm. We then utilized 3D optical profilometry to visualize the topography of the substrate beneath the biofilm. This technique confirmed our previous observations that the substrate underlying the interstitial biofilm contains an intricate network of furrows (Fig. 1A). However, in this study we were able to correlate the field of view obtained with phase contrast microscopy with the 3D optical profilometer image of the same region. As shown in Figure 1A-C, the network of trails of cells of the interstitial biofilm correlates extremely well with the underlying furrow system such that the phase contrast image of the cellular trail network fits easily within the 3D optical profilometer image of the underlying furrow network (Fig. 1B and C).
Figure 1. Analyses of the interconnected furrow network that guide self-organization of P. aeruginosa interstitial biofilms. (A) High magnification 3D optical profilometery scan of the region where the time-lapse phase-contrast microscopy (Movie S1) was performed. Overlapping scan regions were manually stitched to provide a larger field of view. Scale bar indicates 100 μm. (B) Overlapping phase contrast microscopy images of the region observed in Movie S1 were captured after the time-series was completed and manually stitched to provide a larger field of view that captures the leading edge rafts that had migrated out of frame in Movie S1. Scale bar indicates 100 μm. Relevant morphological features of the interstitial biofilms are labeled. (C) Correlation of the biofilm cell trails to the underlying furrow network. The stitched image depicted in (B) was false colored green and overlaid onto the 3D optical profilometery image depicted in (A). The cellular network lays completely within the furrow network. Scale bar indicates 100 μm. (D) Analysis of furrow widths from the different areas within the interstitial biofilms indicated in (B). Analysis was performed using the high magnification profilometery scans with the number of measurements for each group as follows: raft head n = 21, raft trails n = 30, behind rafts n = 43, and lattice n = 77. Analyses are depicted as mean ± SD P value < 0.0001 determined using a one-way ANOVA test. (E) 3D optical profilometery image (500μm x 280μm) of the underlying semi-solid media where Movie S1 was performed using low magnification to obtain an overview of the furrow network relative to the virgin territories. Height scale is equivalent for (A), (C), and (E).
We have also performed a detailed analysis of furrow widths using data obtained with both 3D optical profilometry and AFM. Both techniques yielded equivalent values and indicate that the widths are narrower in the furrows of the lattice network compared with the furrows at the outermost regions of the biofilm (Fig. 1D). We found that the widths of the furrows beneath the raft head and raft trails were equivalent (mean widths of 18.79 ± 6.52 μm and 18.50 ± 6.33 μm, respectively) whereas the widths of the furrows in the network behind the rafts and in the older, more intricate lattice network became progressively narrower (furrow mean width of 12.07 ± 4.37 μm and 8.20 ± 2.40 μm, respectively; Figure 1D). These observations suggest that sustained cellular traffic through the network refines the wider channels forged by the advancing rafts.
We expected that the impact of sustained cellular traffic throughout the network would result in the furrows becoming progressively deeper in the older lattice regions of the furrow network. However, as noted previously, the furrows are shallower in these regions of the biofilm.11 3D optical profilometry enabled us to gain an understanding of how the various regions of the furrow network are situated relative to each other and the virgin territory. These analyses revealed that the furrows of the raft trails at the leading edge have high walls with their top edges equivalent to the absolute height of the virgin substrate and also that the absolute depth of the base of the furrow is often lower than that of the furrows in the older lattice network located further back in the biofilm (Fig. 1A and E). In contrast, the lattice furrows have low walls and bases that are situated higher than that of the leading edge furrows (Fig. 1A and E). These observations suggest that the formation of the intricate furrow network is associated with significant remodeling of the semi-solid media that occurs when new intersections are forged, resulting in the formation of an intricate interconnected network of narrow furrows with shallow walls. Interestingly, both our AFM and 3D optical profilometry data were obtained several days after removal of the cells from the media. This indicates that the furrow network is a consequence of physical changes in the media such that in the absence of cells the media does not return to its original state.
P. aeruginosa interstitial biofilm expansion is mediated by stigmergic self-organization
Stigmergy is a mechanism of self-organization that was first introduced by the French entomologist Grassé in 1959 to describe the social behaviors of insects such as ants and termites.12 It is a concept used to describe self-organization of group activities via mechanisms that involve indirect communication mediated by alteration of the environment. The underlying principle of stigmergy is that by modifying the local environment, an individual can indirectly influence the actions of another individual at a later time thereby leading to the emergence of apparently coordinated collective behavior, accounting for the formation of complex structures, even by relatively simple “agents” that lack self-awareness or planning ability.13 Stigmergy describes many diverse collective behaviors observed in nature, including the building of nests by termites and wasps, the laying down of pheromone trails that connect ant colonies to food sources,12,14-16 herd migration by animals (for example, the migration of herds of caribou through deep snow), and the emergence of tracks formed by the traffic of pedestrians and hikers.17-19 The potential of the concept of stigmergy has recently been recognized by other branches of science and is now being applied to describe various facets of human society and technology.20-22
With the increasing interest into the concept of stigmergy since it was first proposed in 1959, many theorists have over the years defined various categories of stigmergy, such as sematectonic and marker based,23 passive and active,24,25 and qualitative and quantitative13 to better understand the different forms of communication that influences the group. In their essence, though, these forms of stigmergy essentially differentiate between physical changes to the environment or chemical signals driving the self-organizing behavior of the group. For example, trail following during herd migrations and human traffic are examples of stigmergy driven by physical cues whereas the following of pheromone trails by ants is an example of chemical signal driven stigmergy.
Interestingly, while bacteria display a variety of self-organized collective behaviors that resemble those of animals and insects,1,2 the concept of stigmergy has yet to be applied to any self-organized activity of bacteria. We have shown that P. aeruginosa coordinates the expansion of its interstitial biofilms through the creation of an interconnected network within the semi-solid media. This network is initiated by the advancing aggregates of the cells, with the following cells preferentially confined to these trails.11 Since these “bulldozer” aggregates alter their immediate environment by forging the furrow network, which then influence the movements of cells that traverse the area at a later time, this process of physical self-organization of bacteria into networks of trails is, by definition, a stigmergic phenomenon. To our knowledge this is the first time that physical remodeling of the substrata has been identified as a mechanism for stigmergic self-organization of bacterial communities.
By definition, the tendency of bacterial cells to follow slime trails could also be considered stigmergic phenomena. Examples include the following of slime trails by Myxococcus xanthus and Proteus miriabilis during gliding and swarming motilities,26,27 respectively and the following of Psl exopolysaccharide trails by P. aeruginosa during the early stages of the formation of biofilms under hydrated conditions.28 Therefore, we contend that stigmergy can be included in the repertoire of systems used by bacteria to coordinate complex multicellular behaviors. This new understanding of stigmergy as an important self-organizing principle in bacteria opens the possibility of applying modern molecular genetics to explore stigmergy and to develop testable behavioral models using a model biological system. This will ultimately lead to a greater understanding of stigmergic behaviors in other systems. Furthermore, as the active expansion of bacterial biofilms is important in the spread of infection (e.g., along implanted medical devices) and in biofouling of marine and industrial surfaces, the study of bacterial stigmergy may lead to novel approaches to the development of antimicrobial interventions that impede biofilm expansion.
Materials and Methods
Interstitial biofilm assay
Molten 0.4xLB solidified with 8g/L gellan gum (MP Biomedicals) was poured over sterile glass slides and allowed to set at room temperature. Slides were dried briefly to remove excess surface moisture. Media was inoculated with cells from an overnight plate culture and covered with a sterile glass coverslip and incubated at 37 °C in humid conditions for approximately 6 h prior to imaging.10,11
Imaging techniques
Interstitial biofilm expansion was imaged using an Olympus IX71 wide field inverted microscope (Olympus) with phase contrast optics. Time series were captured at a rate of one frame every 2s for 1 h. During phase contrast microscopy a temperature of 37 °C was maintained using an environmental control chamber (Solent Scientific Ltd).
To analyze the topography of the substratum beneath the biofilm, the coverslip was removed and cells washed away with RO water as detailed in Gloag et al. 2013.11 The topography of the washed interstitial biofilms was imaged using either tapping mode atomic force microscopy (AFM) or 3D optical profilometry. AFM was performed as detailed in Gloag et al. 201311 and analyzed using MFP-3D AFM (Asylum Research) software. 3D optical profilometry was performed using a Contour GT-K1 3D optical surface profiler (Bruker, Germany) fitted with 20x and 50x objectives. Images were analyzed using Vision 4.20 (Veeco, USA) and Image SXM (University of Liverpool) software and graphed in Prism 6 (GraphPad Software Inc.). Analyses are depicted as mean ± SD.
Manual stitching of images and correlation of phase contrast microscopy and profilometry images was performed using Adobe Photoshop CS4 Extended, version 11.0 (Adobe Systems Inc.).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Camazine S, et al. Self-organization in biological systems. Princeton University Press, 2003. [Google Scholar]
- 2.Vicsek T, Zafeiris A. Collective motion. Phys. Rep.- Rev. Sec. Phys. Lett. 2012;517:71–140. [Google Scholar]
- 3.Levine H, Ben-Jacob E. Physical schemata underlying biological pattern formation-examples, issues and strategies. Phys Biol. 2004;1:14–22. doi: 10.1088/1478-3967/1/2/P01. [DOI] [PubMed] [Google Scholar]
- 4.Boissard E, Degond P, Motsch S. Trail formation based on directed pheromone depostition. J Math Biol. 2012 doi: 10.1007/s00285-012-0529-6. [DOI] [PubMed] [Google Scholar]
- 5.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–67. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–90. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Jacob E. From snowflake formation to growth of bacterial colonies II: Cooperative formation of complex colonial patterns. Contemp Phys. 1997;38:205–41. doi: 10.1080/001075197182405. [DOI] [Google Scholar]
- 9.Grammaticos B, Badoual M, Aubert M. An (almost) solvable model for bacterial pattern formation. Physica D. 2007;234:90–7. doi: 10.1016/j.physd.2007.07.002. [DOI] [Google Scholar]
- 10.Semmler AB, Whitchurch CB, Mattick JS. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology. 1999;145:2863–73. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 11.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A. 2013;110:11541–6. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasse P. La reconstruction du nid et les coordinations interindividuelles chez bellicositermes natalensis et cubitermes sp. La theorie de la stigmergie: essai d’interpretation du comportement des termites constructeurs. Insectes Soc. 1959;6:41–81. doi: 10.1007/BF02223791. [DOI] [Google Scholar]
- 13.Theraulaz G, Bonabeau E. A brief history of stigmergy. Artif Life. 1999;5:97–116. doi: 10.1162/106454699568700. [DOI] [PubMed] [Google Scholar]
- 14.Bonabeau E, Theraulaz G, Deneubourg JL, Aron S, Camazine S. Self-organization in social insects. Trends Ecol Evol. 1997;12:188–93. doi: 10.1016/S0169-5347(97)01048-3. [DOI] [PubMed] [Google Scholar]
- 15.Dorigo M, Bonabeau E, Theraulaz G. Ant algorithms and stigmergy. Future Gener Comput Syst. 2000;16:851–71. doi: 10.1016/S0167-739X(00)00042-X. [DOI] [Google Scholar]
- 16.Downing HA, Jeanne RL. Nest construction by the paper wasp, Polistes: a test of stigmergy theory. Anim Behav. 1988;36:1729–39. doi: 10.1016/S0003-3472(88)80112-X. [DOI] [Google Scholar]
- 17.Goldstone RL, Roberts ME. Self‐organized trail systems in groups of humans. Complexity. 2006;11:43–50. doi: 10.1002/cplx.20135. [DOI] [Google Scholar]
- 18.Helbing D, Schweitzer F, Keltsch J, Molnar P. Active walker model for the formation of human and animal trail systems. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1997;56:2527–39. doi: 10.1103/PhysRevE.56.2527. [DOI] [Google Scholar]
- 19.Helbing D, Keltsch J, Molnár P. Modelling the evolution of human trail systems. Nature. 1997;388:47–50. doi: 10.1038/40353. [DOI] [PubMed] [Google Scholar]
- 20.Marsh L, Onof C. Stigmergic epistemology, stigmergic cognition. Cogn Syst Res. 2008;9:136–49. doi: 10.1016/j.cogsys.2007.06.009. [DOI] [Google Scholar]
- 21.Parunak HVD. in Environments for Multi-Agent Systems II 163-186 (Springer, 2006). [Google Scholar]
- 22.Doyle MJ, Marsh L. Stigmergy 3.0: From ants to economies. Cogn Syst Res. 2013;21:1–6. doi: 10.1016/j.cogsys.2012.06.001. [DOI] [Google Scholar]
- 23.Wilson EO. Sociobiology: The new synthesis. 186-188 (Cambridge, MA: Harvard University Press, 1975). [Google Scholar]
- 24.Holland O. Multi-agent systems: Lessons from social insects and collective robotics. In Adaptation, Coevolution and Learning in Multiagent Systems: Papers from the 1996 AAAI Spring Symposium. Menlo Park, CA: AAAI Press. 57-62 (1996). [Google Scholar]
- 25.Holland O, Melhuish C. Stigmergy, self-organization, and sorting in collective robotics. Artif Life. 1999;5:173–202. doi: 10.1162/106454699568737. [DOI] [PubMed] [Google Scholar]
- 26.Burchard RP. Trail following by gliding bacteria. J Bacteriol. 1982;152:495–501. doi: 10.1128/jb.152.1.495-501.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stahl SJ, Stewart KR, Williams FD. Extracellular slime associated with Proteus mirabilis during swarming. J Bacteriol. 1983;154:930–7. doi: 10.1128/jb.154.2.930-937.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luijten E, Parsek MR, Wong GC. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–91. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.