Abstract
The growing epidemic of type 2 diabetes mellitus (T2DM) and obesity is largely attributed to the current lifestyle of over-consumption and physical inactivity. As the primary platform controlling metabolic and energy homeostasis, mitochondria show aberrant changes in T2DM and obese subjects. While the underlying mechanism is under extensive investigation, epigenetic regulation is now emerging to play an important role in mitochondrial biogenesis, function, and dynamics. In line with lifestyle modifications preventing mitochondrial alterations and metabolic disorders, exercise has been shown to change DNA methylation of the promoter of PGC1α to favor gene expression responsible for mitochondrial biogenesis and function. In this article we discuss the epigenetic mechanism of mitochondrial alteration in T2DM and obesity, and the effects of lifestyle on epigenetic regulation. Future studies designed to further explore and integrate the epigenetic mechanisms with lifestyle modification may lead to interdisciplinary interventions and novel preventive options for mitochondrial alteration and metabolic disorders.
Keywords: mitochondrial alteration, epigenetic, type 2 diabetes, obesity, lifestyle
Introduction
Type 2 diabetes mellitus (T2DM) and obesity are growing epidemics due to the current lifestyle of consumption of energy-rich foods and physical inactivity.1-3 Surplus nutrient supply overloads and hyperpolarizes mitochondria, the primary metabolic platform and energy generator in mammalian cells, leading to the accumulation of incompletely oxidized substrates or intermediates (e.g., fatty acids and diacylglycerol) and overproduction of reactive oxygen species (ROS) (Fig. 1).2,4-9 As such, stress-sensitive kinases such as c-Jun N-terminal kinase and protein kinase C are activated and impair insulin action (insulin resistance) by phosphorylating insulin receptor substrates (Fig. 1).2,5,7,8 Accordingly, a decrease in mitochondrial function or mitochondrial DNA (mtDNA) copy number has been correlated with insulin resistance and dysregulated lipid metabolism.7,10-13 A vicious cycle may develop due to negative feedback regulation loops. For instance, insulin resistance can dysregulate electron transport chain and impair mitochondrial function and biogenesis.14,15 In addition, ROS induces further mitochondrial damage and leads to pancreatic β-cell dysfunction and insulin secretory deficiency, thereby deteriorating metabolic disturbance and energy imbalance in various tissues (Fig. 1).16-19 Nevertheless, properly restricted calorie intake or maintaining a physically active lifestyle can significantly improve mitochondrial integrity and function and protect against metabolic syndromes.20-22 The interactions between mitochondria and lifestyle stimuli shown in these studies suggest that epigenetic regulation may underlie mitochondria-mediated metabolism and energy homeostasis. Indeed, DNA methylation of the promoters of genes encoding PGC1α and Tfam (the regulators of mitochondrial biogenesis), and microRNAs that regulate mitochondrial function and dynamics have been identified in obese and diabetic subjects.13,23-28 Recently DNA methyltransferase 1 (DNMT1) was found to translocate to the mitochondria and control the expression of transcripts from the heavy and light strands of mtDNA, which is accompanied with the presence of cytosine 5-hydroxymethylcytosine and 5-methylcytosine in mtDNA.29 In addition, mitochondrial microRNAs have been discovered in purified mitochondria, which may regulate various cellular processes including RNA turnover, apoptosis, cell cycle, and nucleotide metabolism.30,31 Thus, understanding the epigenetic mechanism of mitochondrial alteration in obesity and diabetes may open a new window of prevention or therapeutics for the metabolic diseases. In this article we discuss the latest epigenetic studies and mechanisms of mitochondrial alteration in diabetes and obesity, and review the emerging evidence of lifestyle modifications influencing the epigenetic traits.
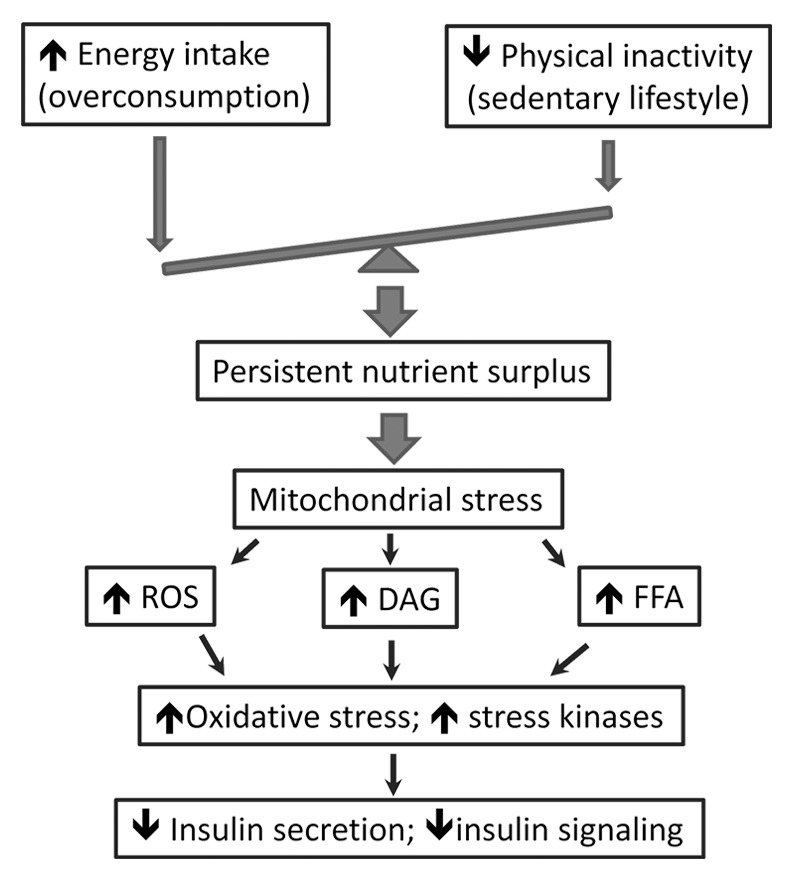
Figure 1. Mitochondrial stress in type 2 diabetes and obesity. The current lifestyle of overconsumption and physical inactivity poses a persistent nutrient surplus, leading to mitochondrial stress and the accumulation of reactive oxygen species (ROS) and metabolite intermediates (FFA and DAG) that can trigger oxidative stress and activation of stress-sensitive kinases. Impaired insulin secretion and sensitivity (i.e., the hallmarks of T2DM) occur as a result of stress-induced β-cell dysfunction and insulin resistance. FFA, free fatty acid; DAG, diacylglycerol.
Mitochondria in Energy Metabolism
Anabolism and catabolism converge on mitochondria, which convert nutrient influx into energy through ATP production and uncoupling protein (UCP)-mediated thermogenesis (Fig. 2A).8,32,33 Mitochondria also convert nutrients into chemical building blocks for macromolecule biosynthesis and energy storage (Fig. 2A).32,33 In response to increased nutrient supply, mitochondrial biogenesis and number is upregulated.34 However, persistent nutrient surplus can override the adaptation and lead to mitochondrial overloading and dysfunction, which also holds true when mitochondrial capacity declines with aging.2,4,6,7 By contrast, calorie restriction has been shown to improve mitochondrial biogenesis and respiration efficiency, which reduces ROS production and promotes metabolic homeostasis.20,35 Mechanistically, mitochondrial biogenesis is regulated by energy sensors such as PKA/CREB, AMPK, and SIRT1 (Fig. 2B).33,36,37 During nutrient or energy depletion (e.g., fasting, exercise, or calorie restriction), the level of cAMP and ratio of AMP/ATP increase, which trigger the signaling cascades of PKA/CREB, AMPK, and SIRT1 that can activate PGC1α, the master regulator of mitochondrial biogenesis.38 As a result, the downstream targets (i.e., NRF1, NRF2 and Tfam) are induced by PGC1α, resulting in upregulation of mitochondrial biogenesis and activity (Fig. 2B).14,33,36
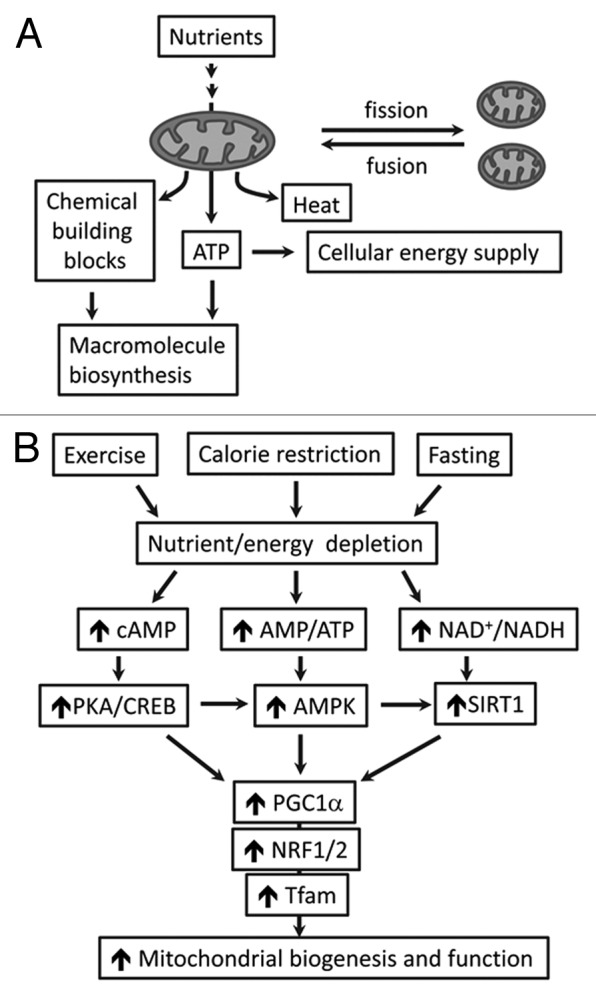
Figure 2. The role of mitochondria in energy metabolism. (A) Mitochondria undergo frequent fusion and fission (the dynamic processes that maintain mitochondrial integrity) and underpin energy homeostasis (ATP generation and thermogenesis) and macromolecule biosynthesis. (B) Energy or nutrient stimuli (e.g., exercise, fasting or calorie restriction) regulate mitochondrial biogenesis and function through pathways involving cAMP-PKA/CREB, AMP/ATP-AMPK, and NAD+/NADH-SIRT1. As the key regulator of mitochondria, PGC1α funnels the signaling to NRF1/2-Tfam and upregulates the genes that are associated with mitochondrial oxidative metabolism. cAMP, cyclic adenosine monophosphate; PKA, protein kinase A; CREB, cAMP response element-binding protein; AMP, 5′-adenosine monophosphate; AMPK, 5′-adenosine monophosphate-activated protein kinase; NAD+, nicotinamide adenine dinucleotide; NADH, reduced form of NAD+; SIRT1, sirtuin (silent mating type information regulation 2 homolog) 1; PGC1α, peroxisome proliferator-activated receptor γ coactivator 1α; NRF1/2, nuclear respiratory factors 1 and 2; Tfam, mitochondrial transcription factor A.
Additional mechanisms that regulate mitochondrial number and integrity include the dynamic processes of mitochondrial fusion and fission (or division) (Fig. 2A).14,39 Although opposing, the fusion and fission processes work in concert to maintain mitochondrial morphology, size, and number.14,39 It has been shown that the fusion and fission can facilitate mixing and exchanging mitochondrial contents, including outer membranes and inner membranes, as well as matrix contents (e.g., mtDNA and metabolites),39 thereby maintaining the integrity of the organelles. Dysregulation of the mitochondrial dynamics leads to inhibition of content mixing/exchange in addition to abnormal morphology, and it is associated with mtDNA instability and reduced respiratory capacity.14,39 It was found that high glucose (i.e., the mimic of hyperglycemia, a hallmark of T2DM) can upregulate mitochondrial fission protein Fis1, resulting in augmented mitochondrial fragmentation and ROS overproduction.6 In addition, dysregulation of mitochondrial fusion protein (mitofusin, Mfn) was observed in insulin-resistant or diabetic subjects, suggesting that alterations in this regulatory pathway may contribute to the pathophysiology of insulin resistance and T2DM.14,40,41
T2DM and obesity are characterized by impaired nutrient and energy control. In line with to such changes, the mitochondria have been found to adopt epigenetic mechanisms, including DNA methlylation, histone modification, and microRNA, to carry the signals of biogenesis,13,23-26,42 functional regulation,43-45 and dynamics.27 Below we discuss the epigenetic mechanisms in detail.
Epigenetic Regulation of Mitochondrial Biogenesis
As the key regulator of mitochondrial biogenesis (Fig. 2B),38 PGC1α is sensitive to environmental factors such as aerobic capacity, age, birth weight, and sex.46 In addition, alcohol consumption has been correlated with specific single-nucleotide polymorphism in PPARGC1A (the PGC1α-encoding gene) in African Americans and Caucasians.47 The activity and expression of PGC1α can be regulated by histone deacetylases (HDACs), the enzymes that increase PGC1α activity by deacetylating PGC1α protein (e.g., SIRT1)14 or control PPARGC1A expression through deacetylating chromatin structure protein histones (e.g., HDAC3).42 In insulin-resistant and diabetic obese mice, SIRT1 is less active, and PGC1α exists primarily in an acetylated form; however, activation of SITR1 by small molecules prevents hyperacetylation of hepatic PGC1α protein and enhances mitochondrial biogenesis and function.14 In a study of obese diabetic mice, treatment with HDAC3 inhibitors promoted mitochondrial oxidative metabolism in skeletal muscle and adipose tissue, which increased energy expenditure and improved glucose tolerance and insulin sensitivity.42 These effects were associated with elevated expression and activity of PGC1α and the downstream target Tfam, thereby increasing mitochondrial biogenesis and energy expenditure. It is known that HDAC3 is recruited onto transcription factor MEF2 (myocyte enhancer factor 2) to form a repressive complex with HDAC4, HDAC5, and nuclear receptor corepressor (NCoR), which represses the transcriptional activation of PGC1α by MEF2;42,48,49 however, treatment with HDAC3 inhibitors reduces the recruitment of HDAC3 onto MEF2, thereby removing the transcriptional repression and increasing PGC1α expression.42
Epigenetic studies have also suggested that the expression of PGC1α may be controlled at the level of DNA methylation of PPARGC1A promoter, at both cytosine-guanine dinucleotide (CpG) sites and non-CpG sites.13,23-25 The increase in DNA methylation of PPARGC1A promoter appears to be a shared mechanism by multiple tissues in patients with T2DM or nonalcoholic fatty liver diseases (NAFLD), which reduces PPARGC1A expression and mitochondrial content, resulting in impaired insulin secretion from pancreatic islets28 and reduced insulin sensitivity in the liver (Fig. 3).13 Whole-genome promoter methylation analysis of skeletal muscle biopsies reveals cytosine hypermethylation of PPARGC1A in T2DM patients compared with normal glucose-tolerant subjects, and the highest portion of non-CpG methylation levels are negatively correlated with PPARGC1A expression and mitochondrial content (Fig. 3).23 In monozygotic twins, higher DNA methylation at CpG sites of promoters of PPARGC1A was present in skeletal muscle from type 2 diabetic vs. non-diabetic twins (13.9 ± 6.2% vs. 9.0 ± 4.5%, P = 0.03), which may account for the monozygotic twins discordant for T2DM.25 It has also been shown that both prenatal (i.e., low birth weight) and postnatal (i.e., 5-day high-fat overfeeding) environmental factors can change PPARGC1A methylation.50 Importantly, altered PPARGC1A methylation induced by the short-term high-fat overfeeding in normal birth weight subjects was significantly reversed by switching back to normal diet.50 In addition, a positive correlation exists between maternal pregestational body mass index (BMI) and PPARGC1A promoter methylation in the umbilical cord of newborns (r = 0.41, P = 0.0007).24 The strong associations of high-fat overfeeding, birth weight, and maternal pregestational BMI with PPARGC1A methylation suggest that interventions through lifestyle modification could be an effective strategy to prevent the aberrant epigenetic traits and mitochondrial alteration.
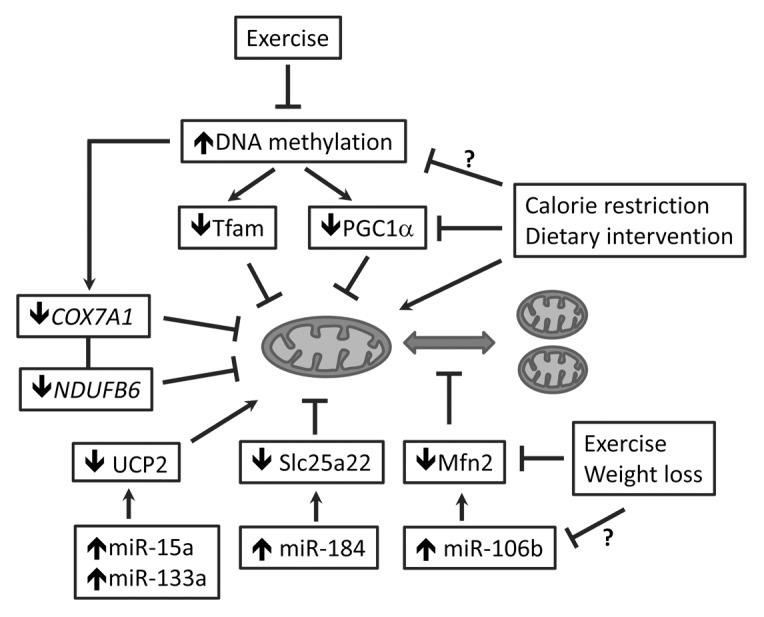
Figure 3. The epigenetic mechanisms of mitochondrial regulation in T2DM and obesity. Increased DNA methylation represses expression of PGC1α and Tfam, the key regulators of mitochondrial biogenesis. Epigenetic regulation of mitochondrial function includes: (1) DNA hypermethylation that represses the genes of mitochondrial oxidative metabolism (e.g., COX7A1 and NDUFB6); and (2) microRNAs (miR15a, miR133a, and miR-184) that repress mitochondrial substrate carrier (Slc25a22) or uncoupling protein (UCP2). microRNA (miR-106b) was also identified to regulate mitochondrial dynamic protein (Mfn2). Behavioral intervention such as exercise has been shown to change the epigenetic signature (DNA methylation) and improve mitochondrial biogenesis and function through PGC1α and Tfam. The mitochondria-promoting effects have also been reported for dietary intervention, calorie restriction, and weight loss; whether and how an epigenetic mechanism is involved in the mitochondrial regulation deserves further investigation. PGC1α, peroxisome proliferator-activated receptor γ coactivator 1α; Tfam, mitochondrial transcription factor A; UCP2, uncoupling protein 2; Mfn2, mitofusin 2;sirt COX7A1, cytochrome c oxidase subunit VIIa polypeptide 1; NDUFB6, NADH dehydrogenase (ubiquinone) 1 β subcomplex, 6.
As a downstream target of PGC1α, Tfam senses the signal of mitochondrial biogenesis and regulates mtDNA replication.14,51 The promoter of human Tfam has 67 CpG dinucleotides, and methylation of Tfam promoter at NRF1-binding site reduces the promoter activity by 90% (Fig. 3).52 According to a recent study of DNA methylation of Tfam promoter in peripheral leukocytes (white blood cells) from high-school students, the ratio of the promoter methylated DNA/unmethylated DNA was inversely correlated with fasting plasma insulin (r = −0.26, P < 0.004), homeostasis model assessment index (r = −0.27, P < 0.002), and obesity (r = −0.27, P < 0.002).26 However, the mechanistic link between insulin resistance and increased methylation of Tfam promoter remains elusive.26 Given that DNA hypermethylation represses Tfam promoter activity, the increased DNA methylation of Tfam promoter suggests a tendency of impaired mitochondrial biogenesis and function in the peripheral leukocytes. Indeed, leukocytes from T2DM patients were found to have compromised mitochondria and elevated ROS and pro-inflammatory cytokines, both of which have been implicated in insulin resistance and T2DM pathophysiology.53 Thus, future studies designed to establish the epigenetic signature of Tfam promoter in peripheral blood and its role in T2DM pathophysiology might lead to new biomarkers for diagnosis and treatment of this disease.
Epigenetic Regulation of Mitochondrial Function
Mitochondrial function underpins insulin secretion from pancreatic β-cell to maintaining systemic glycemic and nutrient homeostasis.18 In response to elevated nutrient influx, ATP production through mitochondrial-coupled respiration (oxidative phosphorylation) is stimulated, which closes the ATP-sensitive K+-channels on the plasma membrane of pancreatic β-cell and depolarizes the cell. As a result, voltage-gated Ca2+ channels are open, and Ca2+ influx raises cytosolic Ca2+ concentration to trigger insulin secretion.18 Uncoupling oxidative phosphorylation from mitochondrial respiration may reduce ATP production and lead to β-cell dysfunction and insulin secretory deficiency in T2DM.19,54 By contrast, suppression or silencing of mitochondrial uncoupling protein UCP2 in β-cell promotes insulin secretion, suggesting UCP2 can be a target to treat insulin deficiency.55,56 To this end, microRNA (e.g., miR-15a) was recently identified and found to promote insulin synthesis and secretion by inhibiting UCP2 (Fig. 3).43 The expression of miR-15a increased significantly in mouse islets after treatment with a high concentration of glucose (33 mM) for 1 hour, accompanied with upregulated expression and biosynthesis of insulin. However, persistent high glucose (33 mM for 3 days) reduced miR-15a level and insulin synthesis in mouse islets, suggesting dysregulation of miR-15a may account for hyperglycemia-induced compromise of insulin secretion in T2DM.43 Mechanistic study indicates that miR-15a directly inhibited UCP-2 gene expression and reduced UCP2 3′-UTR luciferase reporter activity, in line with lower endogenous UCP-2 protein levels.43 Another microRNA that regulates insulin secretion is miR-184, whose expression in pancreatic islets is negatively correlated with insulin secretion.57 microRNA-184 may regulate insulin secretion through repression of Slc25a22, a mitochondrial glutamate carrier that controls cytosolic glutamate and the activities of insulin granules and secretory vesicles during insulin exocytosis (Fig. 3).58 Transfection of MIN6 islet β-cell line with miR-184 reduces 70% of 3′-UTR luciferase reporter signal, indicative of an interaction of miR-184 with Slc25a22 3′-UTR. Ectopic expression of miR-184 in MIN6 cells suppresses Slc25a22 expression and reduces insulin secretion.58
microRNA regulation of UCP2 appears to play a different role in skeletal muscle. It was shown that the muscle-specific microRNA, miR-133a, suppresses UCP2 expression in skeletal muscle and promotes muscle development.44 During myogenic differentiation, MyoD can upregulate miR-133a, which, in turn, silences UCP2, the brake of muscle development.44 In addition, epigenetic regulation of genes encoding mitochondrial proteins affects insulin responsiveness and glucose uptake in skeletal muscle. The elderly or individuals with insulin resistance and T2DM show reduced mitochondrial oxidative capacity in skeletal muscle, concomitant with downregulation of an array of genes responsible for oxidative phosphorylation, including COX7A1 (the subunit of cytochrome c oxidase, or complex IV in the respiratory chain) and NDUFB6 (subunit in complex I in the respiratory chain).7,45,59 Bisulphite sequencing analysis showed that DNA methylation of the COX7A1 promoter was significantly higher in muscle from elderly than young twins (19.9 ± 8.3% vs 1.8 ± 2.7%; P = 0.035), which was associated with a lower expression of COX7A1 in elderly than in young twins (1.00 ± 0.05 vs 1.68 ± 0.06; P = 0.0005).45 An aging-induced epigenetic mechanism also occurs to NDUFB6 regulation: the skeletal muscles from elderly subjects have higher DNA methylation of the NDUFB6 promoter but lower NDUFB6 expression than from young twins.60 Regression analysis of mitochondrial function and oxidative capacity with the expression of COX7A1 and NDUFB6 revealed positive correlations of these 2 gene levels with total body aerobic capacity and insulin-stimulated glucose uptake.45,60 These findings suggest that epigenetic (DNA methylation) regulation of mitochondrial function underlies age-dependent susceptibility to insulin resistance and metabolic programming.
Epigenetic Regulation of Mitochondrial Dynamics
Dysregulation of mitochondrial dynamics alters mitochondrial morphology, metabolism, and intracellular signaling, and it has been implicated in various human diseases, including T2DM and obesity.14,39,40 The skeletal muscles in non-diabetic obese subjects and lean or obese T2DM patients have exhibited significantly lower expression of Mfn2, the regulator of mitochondrial dynamics and network, when compared with control groups.40 The same study suggests that Mfn2 expression is directly proportional to insulin sensitivity, but inversely proportional to the body mass index; however, body weight loss through bilio-pancreatic diversion significantly increases Mfn2 expression in skeletal muscle.40 Using gene-chips and mRNA sequencing, Gallagher et al. identified a highly expressed microRNA, miR-106b, in the skeletal muscle of diabetes patients.61 This microRNA is also highly expressed in mice with insulin resistance induced by high-fat diet.62 Luciferase activity assay combined with mutational analysis revealed that the 3′-UTR of Mfn2 possesses miR-106b binding sites, and that miR-106b targets the 3′-UTR and silences Mfn2 expression.27 Gain of miR-106b function impairs mitochondrial function, whereas loss of miR-106b function promotes mitochondrial function and insulin sensitivity (Fig. 3).27 However, there was no in vivo evidence reported, and it would be of importance for further in vivo studies to validate the effect of silencing miR-106b on mitochondrial dynamics and function, as well as its effects on insulin sensitivity.
Conclusion and Perspectives
Epigenetic regulation of mitochondria affects the biogenesis, function, and dynamics of the organelles. DNA hypermethylation suppresses the expression of PGC1α and Tfam, thereby reducing mitochondrial biogenesis and oxidative metabolism; microRNAs interact with the 3′-UTR of target genes (e.g., UCP2 and Mfn2) and silence the gene expression, which changes mitochondrial respiration and morphology (Fig. 3). In obese and T2DM individuals, the dysregulation of DNA methylation and microRNA has been increasingly discovered, and it is strongly associated with aberrant mitochondrial traits and loss of metabolic homeostasis. In addition, epigenetic dysregulation of mitochondria has been reported in diabetic complications, such as diabetic cardiovascular diseases,63-65 diabetic retinopathy,66 NAFLD,12,13 and cardiorenal dysfunction.67
Epigenetic signatures feature the interactions between environmental factors and genes in human beings. Social and lifestyle factors (such as dietary pattern, physical activity, micro-nutrition status, socioeconomic status, and BMI) have been correlated with human global DNA methylation,68-73 suggesting that lifestyle modification and behavioral intervention may reverse or prevent epigenetic dysregulation of mitochondrial function and metabolism. Indeed, exercise intervention was found to reduce DNA methylation of the promoters of metabolism-regulating genes in both human and rodent skeletal muscles, including PPARGC1A (Fig. 3).71,74 The exercise-induced DNA hypomethylation led to a dose-dependent upregulation of PGC1α, consistent with an enhanced mitochondrial biogenesis and metabolism.23,71,75 Physical activity also modulates methylation of hepatic mitochondrial NADH dehydrogenase 6 (MT-ND6) and its transcriptional activity, which ameliorates the histological severity of NAFLD.12 By contrast, physical inactivity induced by 9-day bed rest increases PPARGC1A promoter methylation and significantly reduces the expression of PPARGC1A and other genes that are associated with mitochondrial function, accompanied with development of insulin resistance; however, these changes can be partly reversed by a 4-week retraining, underscoring the promising potential of physical activity in preventing aberrant epigenetic traits and mitochondrial alteration.76 In addition, weight loss, calorie restriction, and dietary intervention have also been shown to improve mitochondrial biogenesis, network, and function, albeit an epigenetic mechanism remains to be defined (Fig. 3).20-22,40,41,77 Given that lifestyle interventions effectively delay and prevent the onset of diabetes and obesity,78-82 it will be of interest to examine whether and how epigenetic mechanisms of mitochondrial regulation contribute to the efficacy of interventions. Future studies exploring these epigenetic signatures might lead to novel molecular diagnostic options and interdisciplinary interventions for aberrant mitochondrial traits and metabolic disorders.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
This work was supported by the start-up funds from HNFE, CALS, and Fralin Life Science Institute at Virginia Tech (Z.C.) and NIH grant 5R18DK091811-02 (F.A.A. and Z.C.). Publication of this article was supported by Virginia Tech's Open Access Subvention Fund.
Glossary
Abbreviations:
- AMPK
5′ adenosine monophosphate-activated protein kinase
- BMI
body mass index
- COX7A1
cytochrome c oxidase subunit VIIa polypeptide 1
- CpG
cytosine-guanine dinucleotide
- CREB
cAMP response element-binding protein
- DNMT1
DNA methyltransferase 1
- HDAC
histone deacetylase
- MEF2
myocyte enhancer factor 2
- Mfn
mitofusin
- mtDNA
mitochondrial DNA
- MT-ND6
mitochondrial NADH dehydrogenase 6
- NAFLD
nonalcoholic fatty liver disease
- NCoR
nuclear receptor corepressor
- NDUFB6
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6
- NRF
nuclear respiratory factor
- OXPHOS
oxidative phosphorylation
- PGC1α
peroxisome proliferator-activated receptor γ coactivator 1α
- PKA
protein kinase A
- PPARGC1A
the gene encoding PGC1α
- ROS
reactive oxygen species
- SIRT1
sirtuin (silent mating type information regulation 2 homolog) 1
- T2DM
type 2 diabetes mellitus
- Tfam
mitochondrial transcription factor A
- UCP
uncoupling protein
- 3′UTR
3′-untranslated region
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/28189
References
- 1.Hamilton MT, Hamilton DG, Zderic TW. Role of low energy expenditure and sitting in obesity, metabolic syndrome, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56:2655–67. doi: 10.2337/db07-0882. [DOI] [PubMed] [Google Scholar]
- 2.Fisher-Wellman KH, Neufer PD. Linking mitochondrial bioenergetics to insulin resistance via redox biology. Trends Endocrinol Metab. 2012;23:142–53. doi: 10.1016/j.tem.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muoio DM, Newgard CB. Obesity-related derangements in metabolic regulation. Annu Rev Biochem. 2006;75:367–401. doi: 10.1146/annurev.biochem.75.103004.142512. [DOI] [PubMed] [Google Scholar]
- 4.Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 2012;15:595–605. doi: 10.1016/j.cmet.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu T, Robotham JL, Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103:2653–8. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–7. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Z, Tseng Y, White MF. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol Metab. 2010;21:589–98. doi: 10.1016/j.tem.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Z, Ristow M. Mitochondria and metabolic homeostasis. Antioxid Redox Signal. 2013;19:240–2. doi: 10.1089/ars.2013.5255. [DOI] [PubMed] [Google Scholar]
- 10.Gianotti TF, Sookoian S, Dieuzeide G, García SI, Gemma C, González CD, Pirola CJ. A decreased mitochondrial DNA content is related to insulin resistance in adolescents. Obesity (Silver Spring) 2008;16:1591–5. doi: 10.1038/oby.2008.253. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Z, White MF. Foxo1 in hepatic lipid metabolism. Cell Cycle. 2010;9:219–20. doi: 10.4161/cc.9.2.10567. [DOI] [PubMed] [Google Scholar]
- 12.Pirola CJ, Gianotti TF, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castaño GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–63. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 13.Sookoian S, Rosselli MS, Gemma C, Burgueño AL, Fernández Gianotti T, Castaño GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 14.Cheng Z, Guo S, Copps K, Dong X, Kollipara R, Rodgers JT, Depinho RA, Puigserver P, White MF. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med. 2009;15:1307–11. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sleigh A, Raymond-Barker P, Thackray K, Porter D, Hatunic M, Vottero A, Burren C, Mitchell C, McIntyre M, Brage S, et al. Mitochondrial dysfunction in patients with primary congenital insulin resistance. J Clin Invest. 2011;121:2457–61. doi: 10.1172/JCI46405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonnard C, Durand A, Peyrol S, Chanseaume E, Chauvin MA, Morio B, Vidal H, Rieusset J. Mitochondrial dysfunction results from oxidative stress in the skeletal muscle of diet-induced insulin-resistant mice. J Clin Invest. 2008;118:789–800. doi: 10.1172/JCI32601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadagurski M, Cheng Z, Rozzo A, Palazzolo I, Kelley GR, Dong X, Krainc D, White MF. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. J Clin Invest. 2011;121:4070–81. doi: 10.1172/JCI46305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maechler P. Mitochondrial function and insulin secretion. Mol Cell Endocrinol. 2013;379:12–8. doi: 10.1016/j.mce.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 19.Maechler P, Wollheim CB. Mitochondrial function in normal and diabetic beta-cells. Nature. 2001;414:807–12. doi: 10.1038/414807a. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Montalvo A, de Cabo R. Mitochondrial metabolic reprogramming induced by calorie restriction. Antioxid Redox Signal. 2013;19:310–20. doi: 10.1089/ars.2012.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szendroedi J, Phielix E, Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat Rev Endocrinol. 2012;8:92–103. doi: 10.1038/nrendo.2011.138. [DOI] [PubMed] [Google Scholar]
- 22.Toledo FG, Goodpaster BH. The role of weight loss and exercise in correcting skeletal muscle mitochondrial abnormalities in obesity, diabetes and aging. Mol Cell Endocrinol. 2013;379:30–4. doi: 10.1016/j.mce.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–98. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Gemma C, Sookoian S, Alvariñas J, García SI, Quintana L, Kanevsky D, González CD, Pirola CJ. Maternal pregestational BMI is associated with methylation of the PPARGC1A promoter in newborns. Obesity (Silver Spring) 2009;17:1032–9. doi: 10.1038/oby.2008.605. [DOI] [PubMed] [Google Scholar]
- 25.Ribel-Madsen R, Fraga MF, Jacobsen S, Bork-Jensen J, Lara E, Calvanese V, Fernandez AF, Friedrichsen M, Vind BF, Højlund K, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One. 2012;7:e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gemma C, Sookoian S, Dieuzeide G, García SI, Gianotti TF, González CD, Pirola CJ. Methylation of TFAM gene promoter in peripheral white blood cells is associated with insulin resistance in adolescents. Mol Genet Metab. 2010;100:83–7. doi: 10.1016/j.ymgme.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Yang L, Gao YF, Fan ZM, Cai XY, Liu MY, Guo XR, Gao CL, Xia ZK. microRNA-106b induces mitochondrial dysfunction and insulin resistance in C2C12 myotubes by targeting mitofusin-2. Mol Cell Endocrinol. 2013;381:230–40. doi: 10.1016/j.mce.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Ling C, Del Guerra S, Lupi R, Rönn T, Granhall C, Luthman H, Masiello P, Marchetti P, Groop L, Del Prato S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–22. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–5. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, Singh R. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One. 2012;7:e44873. doi: 10.1371/journal.pone.0044873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandiera S, Matégot R, Girard M, Demongeot J, Henrion-Caude A. MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic Biol Med. 2013;64:12–9. doi: 10.1016/j.freeradbiomed.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 32.De Pauw A, Tejerina S, Raes M, Keijer J, Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol. 2009;175:927–39. doi: 10.2353/ajpath.2009.081155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–59. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner N, Bruce CR, Beale SM, Hoehn KL, So T, Rolph MS, Cooney GJ. Excess lipid availability increases mitochondrial fatty acid oxidative capacity in muscle: evidence against a role for reduced fatty acid oxidation in lipid-induced insulin resistance in rodents. Diabetes. 2007;56:2085–92. doi: 10.2337/db07-0093. [DOI] [PubMed] [Google Scholar]
- 35.López-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–73. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–33. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng Z, Schmelz EM, Liu D, Hulver MW. Targeting mitochondrial alterations to prevent type 2 diabetes – Evidence from studies of dietary redox active compounds. Mol Nutr Food Res. 2014 doi: 10.1002/mnfr.201300747. [DOI] [PubMed] [Google Scholar]
- 38.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–35. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 39.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–87. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 40.Bach D, Naon D, Pich S, Soriano FX, Vega N, Rieusset J, Laville M, Guillet C, Boirie Y, Wallberg-Henriksson H, et al. Expression of Mfn2, the Charcot-Marie-Tooth neuropathy type 2A gene, in human skeletal muscle: effects of type 2 diabetes, obesity, weight loss, and the regulatory role of tumor necrosis factor alpha and interleukin-6. Diabetes. 2005;54:2685–93. doi: 10.2337/diabetes.54.9.2685. [DOI] [PubMed] [Google Scholar]
- 41.Hernández-Alvarez MI, Thabit H, Burns N, Shah S, Brema I, Hatunic M, Finucane F, Liesa M, Chiellini C, Naon D, et al. Subjects with early-onset type 2 diabetes show defective activation of the skeletal muscle PGC-1alpha/Mitofusin-2 regulatory pathway in response to physical activity. Diabetes Care. 2010;33:645–51. doi: 10.2337/dc09-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galmozzi A, Mitro N, Ferrari A, Gers E, Gilardi F, Godio C, Cermenati G, Gualerzi A, Donetti E, Rotili D, et al. Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes. 2013;62:732–42. doi: 10.2337/db12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun LL, Jiang BG, Li WT, Zou JJ, Shi YQ, Liu ZM. microRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res Clin Pract. 2011;91:94–100. doi: 10.1016/j.diabres.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Wang K, Chen J, Guo J, Yin Y, Cai X, Guo X, Wang G, Yang R, Zhu L, et al. In vitro evidence suggests that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is an indispensable step in myogenic differentiation. J Biol Chem. 2009;284:5362–9. doi: 10.1074/jbc.M807523200. [DOI] [PubMed] [Google Scholar]
- 45.Rönn T, Poulsen P, Hansson O, Holmkvist J, Almgren P, Nilsson P, Tuomi T, Isomaa B, Groop L, Vaag A, et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia. 2008;51:1159–68. doi: 10.1007/s00125-008-1018-8. [DOI] [PubMed] [Google Scholar]
- 46.Ling C, Poulsen P, Carlsson E, Ridderstråle M, Almgren P, Wojtaszewski J, Beck-Nielsen H, Groop L, Vaag A. Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J Clin Invest. 2004;114:1518–26. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards TL, Velez Edwards DR, Villegas R, Cohen SS, Buchowski MS, Fowke JH, Schlundt D, Long J, Cai Q, Zheng W, et al. HTR1B, ADIPOR1, PPARGC1A, and CYP19A1 and obesity in a cohort of Caucasians and African Americans: an evaluation of gene-environment interactions and candidate genes. Am J Epidemiol. 2012;175:11–21. doi: 10.1093/aje/kwr272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–6. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grégoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, Wong J, Seto E, Yang XJ. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol. 2007;27:1280–95. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brøns C, Jacobsen S, Nilsson E, Rönn T, Jensen CB, Storgaard H, Poulsen P, Groop L, Ling C, Astrup A, et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J Clin Endocrinol Metab. 2010;95:3048–56. doi: 10.1210/jc.2009-2413. [DOI] [PubMed] [Google Scholar]
- 51.Leigh-Brown S, Enriquez JA, Odom DT. Nuclear transcription factors in mammalian mitochondria. Genome Biol. 2010;11:215. doi: 10.1186/gb-2010-11-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi YS, Kim S, Pak YK. Mitochondrial transcription factor A (mtTFA) and diabetes. Diabetes Res Clin Pract. 2001;54(Suppl 2):S3–9. doi: 10.1016/S0168-8227(01)00330-8. [DOI] [PubMed] [Google Scholar]
- 53.Hernandez-Mijares A, Rocha M, Apostolova N, Borras C, Jover A, Bañuls C, Sola E, Victor VM. Mitochondrial complex I impairment in leukocytes from type 2 diabetic patients. Free Radic Biol Med. 2011;50:1215–21. doi: 10.1016/j.freeradbiomed.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 54.Chan CB, De Leo D, Joseph JW, McQuaid TS, Ha XF, Xu F, Tsushima RG, Pennefather PS, Salapatek AM, Wheeler MB. Increased uncoupling protein-2 levels in beta-cells are associated with impaired glucose-stimulated insulin secretion: mechanism of action. Diabetes. 2001;50:1302–10. doi: 10.2337/diabetes.50.6.1302. [DOI] [PubMed] [Google Scholar]
- 55.Chan CB, Saleh MC, Koshkin V, Wheeler MB. Uncoupling protein 2 and islet function. Diabetes. 2004;53(Suppl 1):S136–42. doi: 10.2337/diabetes.53.2007.S136. [DOI] [PubMed] [Google Scholar]
- 56.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolmeson C, Esguerra JL, Salehi A, Speidel D, Eliasson L, Cilio CM. Differences in islet-enriched miRNAs in healthy and glucose intolerant human subjects. Biochem Biophys Res Commun. 2011;404:16–22. doi: 10.1016/j.bbrc.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 58.Morita S, Horii T, Kimura M, Hatada I. MiR-184 regulates insulin secretion through repression of Slc25a22. PeerJ. 2013;1:e162. doi: 10.7717/peerj.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev. 2010;31:364–95. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ling C, Poulsen P, Simonsson S, Rönn T, Holmkvist J, Almgren P, Hagert P, Nilsson E, Mabey AG, Nilsson P, et al. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest. 2007;117:3427–35. doi: 10.1172/JCI30938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gallagher IJ, Scheele C, Keller P, Nielsen AR, Remenyi J, Fischer CP, Roder K, Babraj J, Wahlestedt C, Hutvagner G, et al. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010;2:9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen GQ, Lian WJ, Wang GM, Wang S, Yang YQ, Zhao ZW. Altered microRNA expression in skeletal muscle results from high-fat diet-induced insulin resistance in mice. Mol Med Rep. 2012;5:1362–8. doi: 10.3892/mmr.2012.824. [DOI] [PubMed] [Google Scholar]
- 63.Baseler WA, Thapa D, Jagannathan R, Dabkowski ER, Croston TL, Hollander JM. miR-141 as a regulator of the mitochondrial phosphate carrier (Slc25a3) in the type 1 diabetic heart. Am J Physiol Cell Physiol. 2012;303:C1244–51. doi: 10.1152/ajpcell.00137.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magenta A, Greco S, Gaetano C, Martelli F. Oxidative stress and microRNAs in vascular diseases. Int J Mol Sci. 2013;14:17319–46. doi: 10.3390/ijms140917319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yildirim SS, Akman D, Catalucci D, Turan B. Relationship between downregulation of miRNAs and increase of oxidative stress in the development of diabetic cardiac dysfunction: junctin as a target protein of miR-1. Cell Biochem Biophys. 2013;67:1397–408. doi: 10.1007/s12013-013-9672-y. [DOI] [PubMed] [Google Scholar]
- 66.Kowluru RA. Mitochondria damage in the pathogenesis of diabetic retinopathy and in the metabolic memory associated with its continued progression. Curr Med Chem. 2013;20:3226–33. doi: 10.2174/09298673113209990029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aroor AR, Mandavia C, Ren J, Sowers JR, Pulakat L. Mitochondria and Oxidative Stress in the Cardiorenal Metabolic Syndrome. Cardiorenal Med. 2012;2:87–109. doi: 10.1159/000335675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanyam MA, Diez-Roux AV, Pilsner JR, Villamor E, Donohue KM, Liu Y, Jenny NS. Social factors and leukocyte DNA methylation of repetitive sequences: the multi-ethnic study of atherosclerosis. PLoS One. 2013;8:e54018. doi: 10.1371/journal.pone.0054018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, Vishwanatha JK, Morabia A, Santella RM. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics. 2011;6:293–9. doi: 10.4161/epi.6.3.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, Kaur M, Vishwanatha JK, Santella RM, Cardarelli R. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr. 2011;141:1165–71. doi: 10.3945/jn.110.134536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrès R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–11. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Zhang FF, Santella RM, Wolff M, Kappil MA, Markowitz SB, Morabia A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics. 2012;7:606–14. doi: 10.4161/epi.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perng W, Rozek LS, Mora-Plazas M, Duchin O, Marin C, Forero Y, Baylin A, Villamor E. Micronutrient status and global DNA methylation in school-age children. Epigenetics. 2012;7:1133–41. doi: 10.4161/epi.21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nitert MD, Dayeh T, Volkov P, Elgzyri T, Hall E, Nilsson E, Yang BT, Lang S, Parikh H, Wessman Y, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61:3322–32. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iversen N, Krustrup P, Rasmussen HN, Rasmussen UF, Saltin B, Pilegaard H. Mitochondrial biogenesis and angiogenesis in skeletal muscle of the elderly. Exp Gerontol. 2011;46:670–8. doi: 10.1016/j.exger.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Alibegovic AC, Sonne MP, Højbjerre L, Bork-Jensen J, Jacobsen S, Nilsson E, Faerch K, Hiscock N, Mortensen B, Friedrichsen M, et al. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am J Physiol Endocrinol Metab. 2010;299:E752–63. doi: 10.1152/ajpendo.00590.2009. [DOI] [PubMed] [Google Scholar]
- 77.Abete I, Parra D, Martinez JA. Legume-, fish-, or high-protein-based hypocaloric diets: effects on weight loss and mitochondrial oxidation in obese men. J Med Food. 2009;12:100–8. doi: 10.1089/jmf.2007.0700. [DOI] [PubMed] [Google Scholar]
- 78.Gregg EW, Chen H, Wagenknecht LE, Clark JM, Delahanty LM, Bantle J, Pownall HJ, Johnson KC, Safford MM, Kitabchi AE, et al. Look AHEAD Research Group Association of an intensive lifestyle intervention with remission of type 2 diabetes. JAMA. 2012;308:2489–96. doi: 10.1001/jama.2012.67929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perreault L, Pan Q, Mather KJ, Watson KE, Hamman RF, Kahn SE, Diabetes Prevention Program Research Group Effect of regression from prediabetes to normal glucose regulation on long-term reduction in diabetes risk: results from the Diabetes Prevention Program Outcomes Study. Lancet. 2012;379:2243–51. doi: 10.1016/S0140-6736(12)60525-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith-Ray RL, Almeida FA, Bajaj J, Foland S, Gilson M, Heikkinen S, Seagle H, Estabrooks PA. Translating efficacious behavioral principles for diabetes prevention into practice. Health Promot Pract. 2009;10:58–66. doi: 10.1177/1524839906293397. [DOI] [PubMed] [Google Scholar]
- 81.Almeida FA, Shetterly S, Smith-Ray RL, Estabrooks PA. Reach and effectiveness of a weight loss intervention in patients with prediabetes in Colorado. Prev Chronic Dis. 2010;7:A103. [PMC free article] [PubMed] [Google Scholar]
- 82.Estabrooks PA, Glasgow RE, Xu S, Dzewaltowski DA, Lee RE, Thomas D, Almeida FA, Thayer AN, Smith-Ray RL. Building a multiple modality, theory-based physical activity intervention: The development of CardiACTION! Psychol Sport Exerc. 2011;12:46–53. doi: 10.1016/j.psychsport.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]


