Abstract
The humanized monoclonal antibody H27K15 specifically targets human CD115, a type III tyrosine kinase receptor involved in multiple cancers and inflammatory diseases. Binding of H27K15 to hCD115 expressing cells inhibits the functional effect of colony-stimulating factor-1 (CSF-1), in a non-competitive manner. Both homology modeling and docking programs were used here to model the human CD115 extracellular domains, the H27K15 variable region and their interaction. The resulting predicted H27K15 epitope includes mainly the D1 domain in the N-terminal extracellular region of CD115 and some residues of the D2 domain. Sequence alignment with the non-binding murine CD115, enzyme-linked immunosorbent assay, nuclear magnetic resonance spectroscopy and affinity measurements by quartz crystal microbalance revealed critical residues of this epitope that are essential for H27K15 binding. A combination of computational simulations and biochemical experiments led to the design of a chimeric CD115 carrying the human epitope of H27K15 in a murine CD115 backbone that is able to bind both H27K15 as well as the murine ligands CSF-1 and IL-34. These results provide new possibilities to minutely study the functional effects of H27K15 in a transgenic mouse that would express this chimeric molecule.
Keywords: CD115, CSF-1, IL-34, epitope mapping, homology modeling, protein-protein docking, quartz crystal microbalance, NMR
Introduction
CD115, also known as macrophage colony-stimulating factor receptor (M-CSF-R) or CSF1-R, is a member of the type III tyrosine kinase receptor family.1 CD115 has two identified physiological ligands: colony-stimulating factor 1 (CSF-1) and interleukin-34 (IL-34). CSF-1 is a growth factor that regulates the survival, growth and differentiation of myeloid lineage cells including macrophages, dendritic cells and osteoclasts.2-4 The functions of the recently discovered cytokine IL-34 are less well known, but included some biological effects that are shared with CSF-1 and others that are distinctly different from CSF-1 activities.5,6 Overexpression of CD115 has been reported in a wide variety of human tumors7-9 and inflammatory diseases;3,10,11 therefore, this molecule has been identified as a potential target for a monoclonal antibody (mAb)-based therapy. The CD115 extracellular region is composed of five consecutive Ig-like C2-type domains, D1 to D5, with D5 being the closest domain to plasma membrane (Uniprot P07333). The binding sites for both CSF-1 and IL-34 have recently been reported12,13 as located at the D2 and D3 domains interface.
H27K15 is a humanized mAb developed by Transgene that specifically binds the human CD115 (hCD115),14 but not mouse CD115 (mCD115). The H27K15 mAb has been previously shown to inhibit the activity of the CSF-114 in a non-competitive manner. The goal of this work was to identify the epitope of H27K15 and to characterize the hCD115 residues involved in the hCD115/H27K15 interaction for the design of a chimeric h/mCD115 able to bind H27K15 and the murine ligands.
To achieve this purpose, we resorted to computational methods (Accelrys Discovery Studio version 3.1) such as homology modeling using Modeler15 and rigid-body docking algorithms ZDOCK16 + RDOCK17 to simulate the antibody-receptor interaction.18,19
The predicted interaction model was then validated by measuring the binding of H27K15 to various CD115 mutants and chimeras using enzyme-linked immunosorbent assays (ELISAs) and quartz crystal microbalance (QCM) affinity measurements. Interaction with an essential epitope was confirmed by nuclear magnetic resonance (NMR) spectroscopy using synthetic peptides mixed with H27K15 at different concentrations.
By combining computational modeling analyses and binding experiments, we identified the residues of the epitope in D1 and D2 domains and, for some of them, characterized their contribution to ligand binding of H27K15 mAb. The resulting information was then used to design a chimeric human-murine CD115 containing critical human residues in a murine CD115 backbone enabling the binding of both H27K15 and the murine ligands CSF-1 and IL-34 without affecting significantly their respective affinities.
Results
Modeling of the H27K15 Fv antibody
Three-dimensional models of the H27K15 variable fragment (Fv) were generated by homology modeling of the VH and VL chains, and VH/VL interface as described in the methods section.
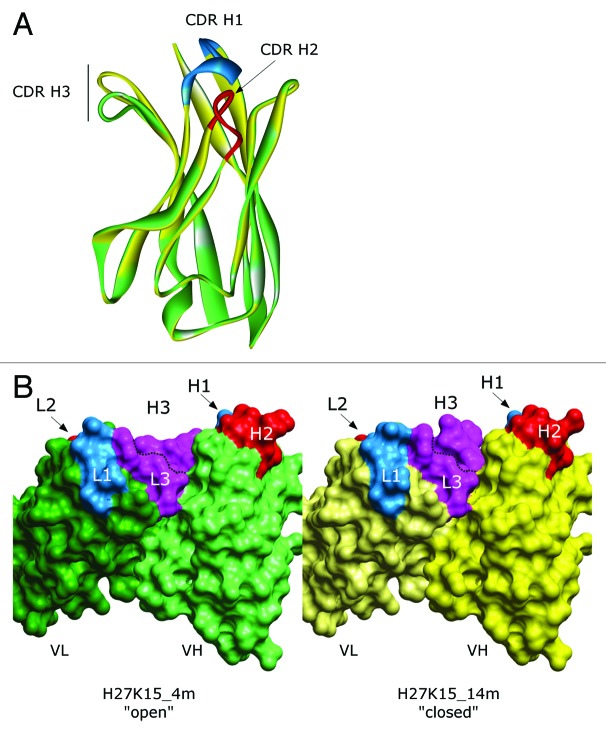
The modeling of the complementarity-determining regions (CDR)20 loops led to two major models, each with a different CDR H3 loop conformation (Fig. 1A). The mobility of this loop confers an open or a closed configuration of the paratope (Fig. 1B). Discrete optimized protein energy (DOPE) scores21 of both conformations are in the same range (-23,026 for the “open” form named “4m” and -22,854 for the “closed” form named “14m”), suggesting that they both may exist. Thus, the epitope predictions based on protein-protein docking were performed with both forms.
Figure 1. Homology models of the H27K15 mAb variable region. (A) Ribbon diagram of VH conformations. Green: model named “4m” with opened CDR H3 conformation; yellow, model with closed CDR H3 conformation named “14m”; CDR H1 and CDR H2 are shown in blue and red, respectively. (B) Solvent accessible surface of H27K15 4m (left panel) and 14m (right panel) Fv fragments; light chains (dark green, 4m; light yellow, 14m) and heavy chains (light green, 4m; dark yellow, 14m) are indicated together with CDR1 (blue), CDR2 (red) and CDR3 (purple).
Epitope prediction based on computational modeling and docking
Interaction of H27K15 Fv models on hCD115D1-D3 structure was first analyzed by protein-protein docking analyses on hCD115 model built from the mCD115D1-D3 crystal structure (3EJJ), the template with the highest sequence homology at that time (56% sequence identity with hCD115). As shown in Figure S1A, 54,000 poses were processed all around the CD115D1-D3 except the region flanking the C-terminal part of the D3 domain, artificially accessible via the missing D4 and D5 domains of the protein. The docking results, as illustrated in Figure S1B, were processed as follows: The top 200 poses from ZDOCK16 were re-ranked by ZRANK22 and clustered. All clustered poses were then evaluated with RDOCK17 and the cluster with the highest density of poses was considered. The best pose was kept for each conformation based on the lowest RDOCK energy, -17.3 kcal.mol−1 and -28.8 kcal.mol−1 for the 4m and the 14m conformations, respectively. The hCD115 residues shown in Figure 2 (highlighted in gray) contain atoms within 5 Å of any atom of the 4m and the 14m antibody conformations. The binding area on hCD115 models (1,600 Å2) for each conformation is shown in surface representation in Figure S2. The imprint of the H27K15 4m conformation on the hCD115 model is mainly located on a side of the D1 domain, whereas the one of the H27K15 14m conformation slightly slid to a small part of the D2 domain (positions P113, V114 and E116).
Figure 2. Residues of the hCD115 model and crystal in interaction with the best docked pose of either 4m or 14m H27K15 mAb conformation. Residues with atoms distance below 5 Å from any VH and VL atom are filled in gray. *The theoretical epitope (filled in black) is defined as the combination of the interacting residues found in each pair of model+crystal.
The hCD115D1-D3/IL-34 co-crystal published afterwards (PDB 4DKD13) was used to validate both the modeling of hCD115 and the use of this model in protein-protein docking analyses. Indeed, despite an unresolved loop (G-D-P-L) in the D1 domain of the crystal (Fig. S3A), the main chains root-mean-square deviation (RMSD) between the two structures was only of 1.4 Å for D1-D2 domains (Fig. S3B). Moreover, docking results with the H27K15 models on the hCD115 crystal structure led to the identification of nearly the same interacting residues (Fig. 2).
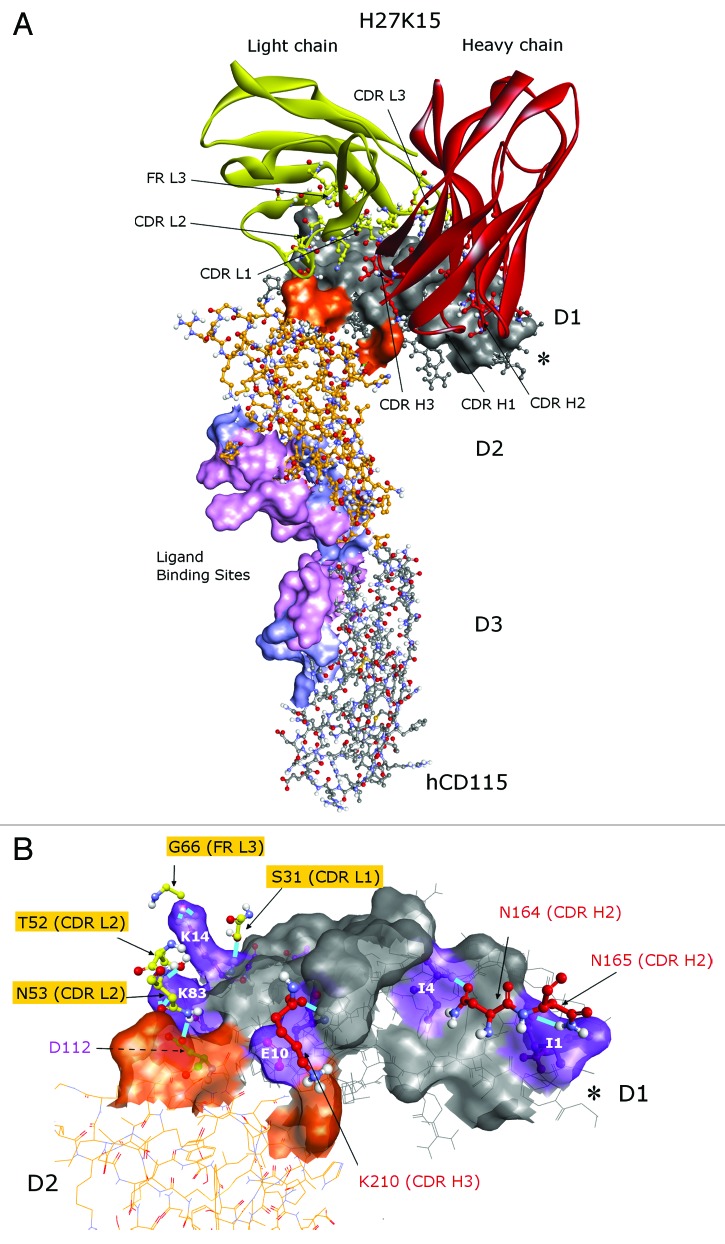
A theoretical epitope was thus defined as follows: any residue of both model and crystal hCD115 that was predicted to interact with H27K15 4m or 14m models. It corresponds to the 28 residues highlighted in black in Figure 2 located mainly in the D1 domain (residues 1 to 24 and 75 to 83) and in the D2 domain (113, 114 and 140). Notably, with 26 out of the 28 residues, the interface between the H27K15 4m and the hCD115 crystal was the closest to the theoretical epitope. This complex was therefore selected as the interaction model between H27K15 and hCD115 for the rest of this work. The interaction surface of 1,762 Å2 (Fig. 3A) covers the D1 domain (in gray) and a small portion of the D2 domain (orange). This region does not overlap the known ligands binding sites, both located at the D2-D3 junction of the CD115, as deduced from the co-crystals.13 Residues of the 6 CDRs of H27K15 are involved in the interaction, as well as some residues of the framework region 3 of the light chain (FR L3). Interaction types were monitored, including hydrogen, polar and van der Waals bonds. Hydrogen bonds involved residues from CDR H2 (N164:I4 and N165:I1), CDR H3 (K210:E10), CDR L1 (S31:K14), CDR L2 (T52:K83, N53:K83, N53:D112) and FR L3 (G66:K14) residues (Fig. 3B). Residues from the CDR H1 and CDR L3 were involved in less energetic polar bonds (Fig. S4A) and van der Waals interactions (Fig. S4B) with hCD115 residues, therefore completing the entire interaction surface.
Figure 3. Predicted epitope of the H27K15 mAb. Best docked pose of the H27K15 4m model on the hCD115 crystal. (A) Red, heavy chain; yellow, light chain of H27K15 Fv; gray, D1 domain; orange, D2 domain of the hCD115. Surface corresponds to the interface area between H27K15 and hCD115. Residues specifically interacting with IL-34 (light blue) and interacting with both IL-34 and CSF-1 (light pink) are also indicated as ligand binding sites surface. (B) hCD115 residues forming hydrogen bonds are indicated in purple. *indicates the position of the G-D-P-L unresolved loop. Views were generated using Discovery Studio v3.5 (Accelrys).
Experimental validation of the epitope prediction by ELISA
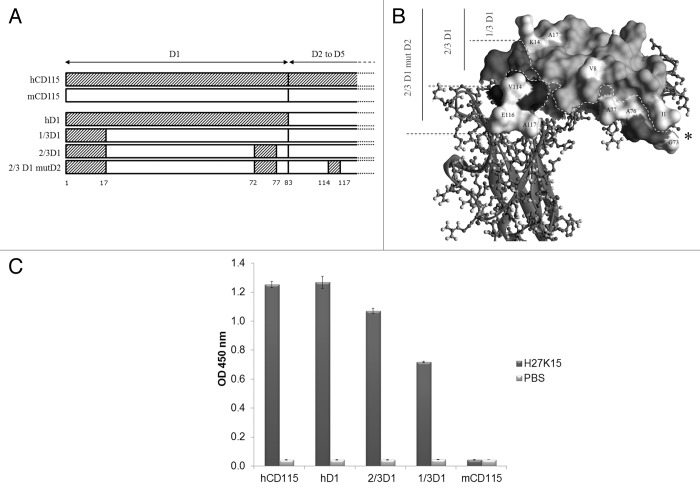
To validate the epitope and to characterize in more detail the contribution of each part of D1 regions on the binding of H27K15, ELISA experiments were performed using human-mouse CD115 chimeric constructs. Those chimeras correspond to mCD115D1-D5 bearing either the whole hCD115 D1 domain (named hD1) or specific regions of the human D1 domain, namely 1/3D1 and 2/3D1 (see Fig. 4A and B for details). Chimera 1/3D1 contains the 17 first residues of hCD115 and displays a humanized surface of 1,467 Å2. The 2/3D1 construct corresponds to the 1/3D1 plus 5 human specific residues within the last 14 residues of D1 domain and displays a humanized surface area of 2,068 Å2.
Figure 4. ELISA experiments. Schematic (A) and structural (B) views of the constructs are shown. Human specific residues are indicated in white; * indicates the position of the G-D-P-L unresolved loop. Calculated surface of humanized areas are 1 467 Å2, 2 068 Å2 and 2 407 Å2 for 1/3D1, 2/3D1, and 2/3D1mutD2, respectively. (C) Double sandwich ELISA experiment with H27K15 mAb. The purified chimeric proteins were coated on plate and incubated in the presence of either H27K15 (dark gray bars) or PBS (light gray bars). Bound H27K15 was revealed with an anti-Fc human-HRP antibody followed by TMB incubation.
H27K15 binds hCD115, but not the murine mCD115 (Fig. 4C). H27K15, however, is able to bind all the chimeric receptors tested, including the construct with the minimal modification (i.e., 1/3D1) that bears the 17 first residues of the hCD115. Therefore, those 17 residues define an essential epitope required for a detectable binding of H27K15 by ELISA (Fig. 4C).
Epitope fine characterization
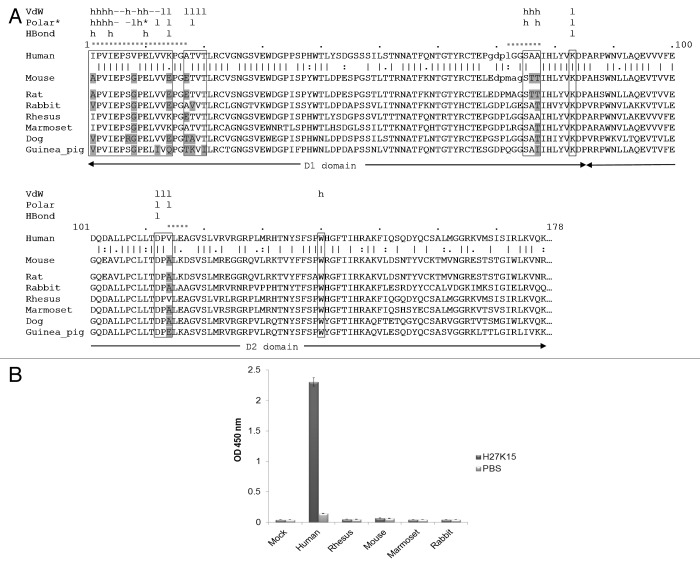
Because D3 domain does not appear to be involved in the binding to H27K15, sequences from the D1-D2 regions of hCD115 and mCD115 were aligned (Fig. 5A). Annotations from the docking results and the interaction monitoring were added to the sequence to identify the critical residues responsible for the non-recognition of mCD115 by the H27K15 mAb. The three hCD115 regions involved in the chimeric constructs mentioned previously are indicated by a dashed line above the sequence. At the binding interface (boxes Fig. 5A), few residues are different between the human and the mouse CD115 (gray). More precisely, only 4 positions (I1A, V8G, K14E and A17E), within the 20 residues of the essential epitope, are different between the human and the mouse CD115 (gray).
Figure 5. Cross species experiments. (A) Sequence alignment of the D1-D2 domains of CD115 from different species. Predicted interaction sites of the epitope are highlighted in boxes. Residues of the epitope that differ from the human sequence are indicated in gray. The unresolved residues of the human and mouse crystal structures loops are represented in lower case. Interaction types are shown above the sequence, VdW (van der Waals), * indicates an electrostatic polar interaction. The mAb chains involved in interactions are also mentioned: h, heavy chain; l, light chain; -, both chains. Dashed segments indicate the human fragments used for the chimeric CD115 constructs. (B) Double sandwich ELISA experiment with H27K15 mAb. The transiently expressed CD115D1-D5 constructs from various species fused with an 8 histidine tag were captured with a coated anti-penta histidine antibody, then incubated in the presence of either biotinylated H27K15 (dark gray bars) or PBS (light gray bars). Bound H27K15 was revealed by streptavidin-HRP followed by TMB incubation.
In addition to the murine one, CD115D1-D2 sequences from other animal species were aligned (Fig. 5A). The residues of the epitope that differed from the human sequence are highlighted in gray. Rat and mouse share 100% sequence identity for all residues of the epitope, including the 4 positions cited above. Rabbit CD115 displays 4 differences with the human molecule in the essential epitope, among which two residues (I1V and T18V) differ from the mouse. As expected, rhesus macaque and marmoset CD115 are much closer in homology to the human CD115. Marmoset CD115 differs from hCD115 by only one residue (V8G) within the essential epitope, while rhesus macaque CD115 displays two differences in this area (V8G and A17E). Beside these two residues in the essential epitope, all the residues of macaque CD115 are identical to the human ones in the epitope. Dog and guinea pig sequences exhibited more differences with the human residues within the essential epitope than the other species, and so, were not further considered for experimental binding studies with H27K15. Notably, the V8G substitution was present in all of these CD115s.
To enable selection of an animal model for the in vivo evaluation of H27K15 activity, the binding of H27K15 to the CD115 of three species was further investigated. CD115D1-D5 from rhesus macaque, rabbit and marmoset were thus expressed in HEK 293 cells and purified (Geneart GmbH). The binding of H27K15 to those three CD115 was evaluated by ELISA. As shown in Figure 5B, none of them bound H27K15. Marmoset CD115 only displayed the V8G substitution in the essential epitope, highlighting the potential critical role of this residue. The effect of I1A, V8G, K14E and A17E residues in the essential epitope on the binding of the H27K15 was further individually evaluated in hCD115, first by computational analyses and second by binding experiments on hCD115 mutants.
The effect of each of the 4 different residues on the binding of the antibody was calculated by computational mutagenesis, (Table 1). With weighted mutation energy of 0.38 kcal.mol−1, the I1A mutation was considered to have a neutral effect on the binding, while the V8G mutation was considered to have a destabilizing effect (2.27 kcal.mol−1). The two remaining mutations K14E and A17E were considered as stabilizing the interaction, with weighted mutation energies of -0.95 kcal.mol−1 and -1.83 kcal.mol−1, respectively. Therefore, three mutants (I1A, V8G and A17E) covering negative, neutral and positive effects on interactions, were selected for both in silico protein-protein docking and in vitro binding measurements. Because the A17E mutant had a greater stabilizing effect (-1.83 kcal.mol−1), this substitution was preferred to the K14E mutant.
Table 1. Computational parameters.
| Constructs | Weighted mutation energy | Effect of mutation a | RDOCK scores b |
|---|---|---|---|
| kcal.mol−1 | kcal.mol−1 (% hCD115) | ||
| hCD115 (WT) | NA | NA | -15.0 (100.0) |
| hCD115 I1A | 0.38 | neutral | -12.4 (82.7) |
| hCD115 V8G | 2.27 | destabilizing | -9.5 (63.3) |
| hCD115 A17E | -1.83 | stabilizing | -11.1 (74.0) |
| hCD115 K14E | -0.95 | stabilizing | Not done |
| mCD115 (WT) | NA | NA | Not done |
a Defined by the program with the following rule: stabilizing < -0.5 < neutral < 0.5 < destabilizing. bDocking scores with hCD115 from 4DKD co-crystal (used as reference) and models from computational mutagenesis. NA: Not Applicable
Protein-protein docking between H27K15 Fv and the models computed by the “Build mutant” algorithm revealed that the three mutations were lowering the binding. With RDOCK scores of -12.4 and -9.5 kcal.mol−1, I1A and V8G were thus predicted as the less and the most destabilizing mutations, respectively (Table 1).
Experimental validation
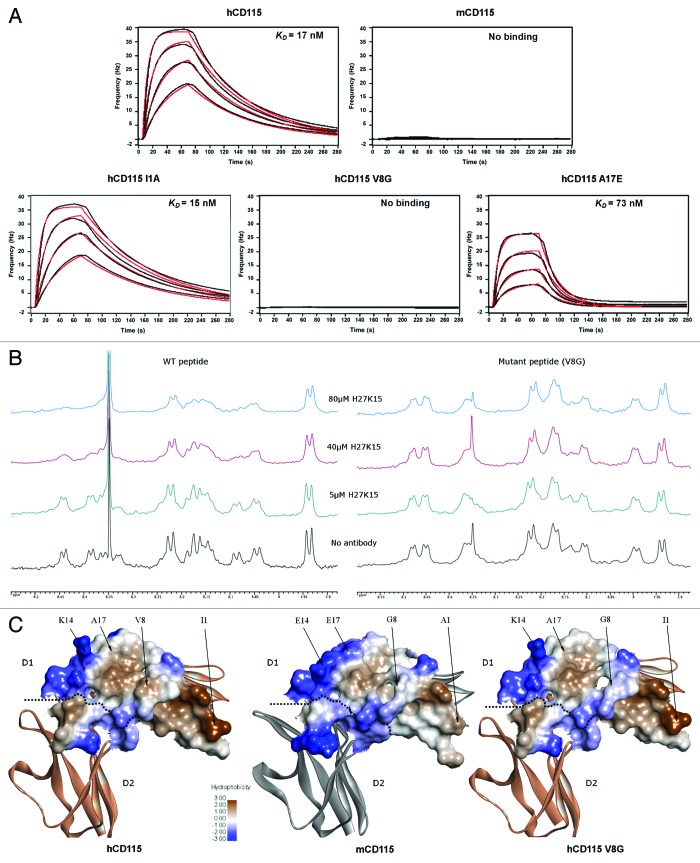
To validate the computational results, H27K15 binding measurements were performed on human CD115D1-D5 constructs with the single mutations I1A, V8G, or A17E. The affinities of H27K15 for these three mutants were measured using QCM technology (Fig. 6A; Table 2). The measured interaction kinetic constants showed that the hCD115 with A17E mutation led to a 4-fold decreased affinity for H27K15, reaching an equilibrium dissociation constant (KD) of 73 nM. I1A substitution had no measurable effect on the binding of H27K15 to hCD115, with KD values of 15 and 17 nM, respectively. The V8G mutation, however, completely annihilated the binding of H27K15 to hCD115. Therefore, affinity measurements confirmed the computational ranking of the mutations effects obtained by RDOCK scores and emphasized the effect of the V8G change on the binding of the H27K15 to the hCD115.
Figure 6. In vitro binding experiments of the H27K15 mAb on various CD115 constructs. (A) Quartz Crystal Microbalance affinity profiles of the immobilized H27K15 mAb in association with hCD115D1-D5, mCD115D1-D5 and the hCD115D1-D5 I1A, V8G and A17E single-point mutants. Raw data (black); model fitting (red). Kinetics parameters are reported in Table 2. (B) HN region of the 1D 1H NMR spectra of 250 µM peptide with the addition of increasing amounts of H27K15 antibody. Left panel, spectra of the WT (hCD115) 23-mer peptide. Right panel, spectra of the V8G mutant 23-mer peptide. (C) hydrophobicity of the interaction surface of the hCD115 and mCD115 crystal structures and the hCD115 V8G variant model based on the hCD115 crystal structure. Positions of the 4 residues of interest are indicated. Hydrophobicity plot and visualization were generated with Discovery Studio v3.5.
Table 2. Kinetics parameters of the interactions of CD115 constructs with mAb H27K15 measured by QCM.
| Constructs | H27K15a | ||
|---|---|---|---|
| ka (105.M−1.s−1) | kd* (10−2.s−1) | KD (nM) | |
| hCD115 (WT) | 7.0 ± 0.06 | 1.2 ± 0.01 | 17 ± 0.1 |
| hCD115 I1A | 6.7 ± 0.08 | 1.0 ± 0.01 | 15 ± 0.2 |
| hCD115 V8G | NB | NB | NB |
| hCD115 A17E | 5.3 ± 0.20 | 3.8 ± 0.01 | 73 ± 2.0 |
| mCD115 (WT) | NB | NB | NB |
a Binding of H27K15 to CD115 constructs. Deduced from curves presented in Figure 6A. *Error considered as 0.01 when error from the fitting model was found below 0.01. NB: No detectable binding.
The critical involvement of the valine at position 8 in the binding of the H27K15 was confirmed by NMR experiments performed using two peptides corresponding to the 23 first residues of hCD115 with or without (WT) the V8G substitution. The H27K15 antibody interaction with the WT (hCD115) 23-mer peptide was monitored using 1D 1H NMR in solution state. The titration of the WT peptide with increasing amounts of the antibody led to a progressive line-broadening of a set of correlation peaks in the HN region of the NMR spectrum, suggesting that an interaction occurs between the peptide and H27K15 (Fig. 6B). The specificity of the interaction was then assessed by performing the titration using the V8G mutant 23-mer peptide. In this latter case, no alteration of the HN correlations was observed, meaning that no antibody-bound form of the peptide was detected in the experimental conditions. These results demonstrated that H27K15 is able to interact specifically with the 23 first residues of hCD115 independently of the rest of the molecule.
To better understand the role of the valine 8 in the binding, we compared the surface hydrophobicity23 of the H27K15 epitope of hCD115, mCD115 and the V8G hCD115 model (Fig. 6C). On the hCD115 surface (left panel), the valine 8 is surrounded by two main hydrophobic patches (in brown), therefore participating in a relatively broad hydrophobic surface all along the epitope. On the mCD115 receptor (middle panel), the hydrophobic patches at the H27K15/receptor surface are dramatically reduced and limited to the region around the A1 residue. The hCD115 V8G model shows a disruption of the hydrophobic surface (right panel) that could explain the striking effect of this substitution on the H27K15 binding.
Since the CD115 of the above-mentioned species all contained the deleterious V8G mutation, it is highly probable that none of them would bind H27K15. Thus, none of these species would be useful as animal models for in vivo evaluation of the H27K15 activity.
Design and selection of the chimeric receptor
Without a relevant species for the determination of H27K15 activity or toxicity, it would be necessary to generate a chimeric CD115D1-D5 protein bearing the optimal area of the H27K15 epitope. The three CD115D1-D5 chimeras previously described were completed with a construction of 2/3D1 mutD2, identical to 2/3D1 plus 3 human substitutions in D2 domain (A114V, K116E and D117A). These extra substitutions were added to have all the human residues from the epitope. K116E and D117A were also added to limit local interferences of mouse-specific residues surrounding the epitope (Fig. 4A and B). The resulting humanized surface area of 2,407 Å2 thus includes the entire 1,762 Å2 predicted epitope area.
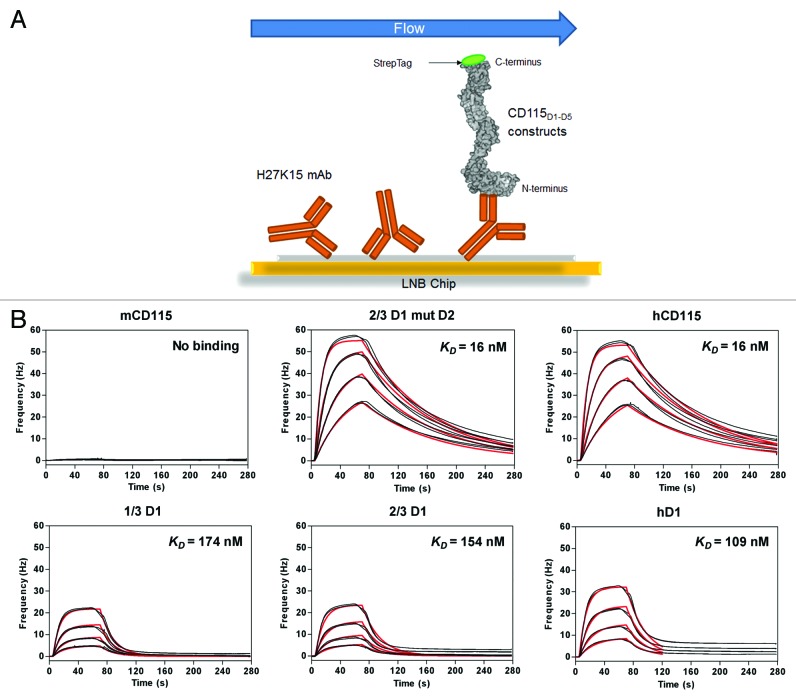
The affinities of the H27K15 for these chimeric receptors were measured using QCM (Fig. 7; Table 3). The H27K15 mAb was immobilized on the chip by amine coupling, prior to the addition of the CD115 constructs in solution (Fig. 7A). The binding affinities of the H27K15 for the chimeric receptors 1/3D1, 2/3D1, and hD1 were 10.5, 10, and 7-fold lower, respectively, than the one of the fully human CD115D1-D5, with KD values ranging from 16 to 168 nM. The additional mutations of three residues in D2 of 2/3D1 (namely 2/3D1 mutD2) yielded a chimeric CD115 with an affinity for H27K15 identical (KD = 16 nM) to the one obtained with the fully human CD115D1-D5 (Fig. 7B). This result showed that the D2 domain plays an important role in the binding affinity of H27K15, while residues within 72–77 of both species are acceptable given binding affinities are indistinguishable for 1/3D1 and 2/3D1.
Figure 7. Selection of the chimeric CD115 target. Experimental design of the chip. (A) Quartz Crystal Microbalance association and dissociation profiles of the chimeric CD115D1-D5 constructs with the immobilized H27K15 mAb (B) Raw data (black); model fitting (red). Kinetics parameters deduced from these curves are reported in Table 3.
Table 3. Kinetics parameters of the interactions of chimeric constructs with mAb measured by QCM.
| Constructs | H27K15a | ||
|---|---|---|---|
| ka (105.M−1.s−1) | kd* (10−2.s−1) | KD (nM) | |
| mCD115 (WT) | NB | NB | NB |
| 2/3D1 mutD2 | 6.2 ± 0.03 | 1.0 ± 0.01 | 16 ± 0.1 |
| hCD115 (WT) | 5.8 ± 0.05 | 0.9 ± 0.01 | 16 ± 0.1 |
| 1/3D1 | 3.2 ± 0.30 | 5.4 ± 0.01 | 168 ± 20.0 |
| 2/3D1 | 2.8 ± 0.90 | 4.5 ± 0.01 | 158 ± 50.0 |
| hD1 | 3.4 ± 1.00 | 3.8 ± 0.01 | 109 ± 0.9 |
a Binding of H27K15 to CD115 constructs. Deduced from curves presented in Figure 7B. *Error considered as 0.01 when error from the fitting model was found below 0.01. NB: No detectable binding
Validation of the chimeric receptor
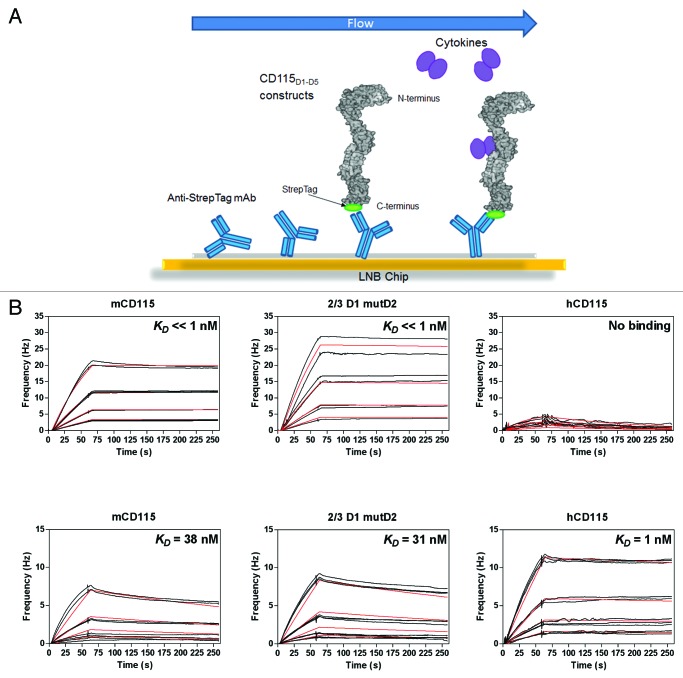
To validate the further use of the 2/3D1 mutD2 receptor in preclinical studies, affinities of the two ligands mCSF-1 and mIL-34 were also measured using QCM (Fig. 8; Table4). The design differs from the previous experiment, since the CD115 constructs were immobilized on the chip using an anti-Tag mAb prior to the addition of the cytokines in solution (Fig. 8A). Both chimeric receptor and the fully murine CD115D1-D5 were able to bind the mCSF-1 with KD values below the nanomolar range (Fig. 8B). As no ligand dissociation was observed for any constructs, it was not possible to calculate an accurate KD (Fig. S5A).
Figure 8. Validation of the chimeric 2/3D1 mutD2 CD115. Experimental design of the chip (A) Quartz Crystal Microbalance association and dissociation profiles of the immobilized CD115D1-D5 constructs with the mouse ligands mCSF-1 (B) or mIL-34 (C). Raw data (black); model fitting (red). Kinetics parameters deduced from these curves are reported in Table 4.
Table 4. Kinetics parameters of the interactions of chimeric constructs with mCSF-1 or with mIL-34 measured by QCM.
| Constructs | mCSF-1a | mIL-34b | |||||
|---|---|---|---|---|---|---|---|
| ka (106.M−1.s−1) | kd (10−4.s−1) | KD (nM) | ka (104.M−1.s−1) | kd* (10−4.s−1) | KD (nM) | ||
| mCD115 (WT) | 2.1 ± 0.01 | < 1 | < < 1 | 5.0 ± 1.00 | 19.0 ± 0.03 | 38 ± 8.0 | |
| 2/3D1 mutD2 | 1.6 ± 0.01 | < 1 | < < 1 | 5.1 ± 0.60 | 16.0 ± 0.01 | 31 ± 3.0 | |
| hCD115 (WT) | NB | NB | NB | 29.0 ± 0.20 | 3.1 ± 0.09 | 1 ± 0.0 | |
| 1/3D1 | 1.0 ± 0.02 | < 1 | < < 1 | 2.7 ± 2.00 | 11.0 ± 0.02 | 41 ± 3.0 | |
| 2/3D1 | 1.4 ± 0.01 | < 1 | < < 1 | 2.5 ± 2.00 | 9.6 ± 0.20 | 38 ± 3.0 | |
| hD1 | 1.6 ± 0.01 | < 1 | < < 1 | 0.8 ± 0.04 | 2.9 ± 0.10 | 37 ± 2.0 | |
a Binding of mCSF-1 to CD115 constructs. Deduced from curves presented in Figure 8B and Figure S5A. bBinding of mIL-34 to CD115 constructs. Deduced from curves presented in Figure 8C and Figure S5B. *Error considered as 0.01 when error from the fitting model was found below 0.01. NB: No detectable binding
The chimeric receptor 2/3D1 mutD2 was also able to interact with mIL-34 with a binding affinity of 31 nM (Fig. 8C), which is similar to the affinity measured for the fully murine CD115D1-D5 (38 nM). Compared with its murine counterpart, the hCD115 exhibits a 30-fold increased affinity for mIL-34 in these conditions. Since all other chimeric constructs, including hD1, interact with mIL-34 with similar affinities (~40 nM), this gain might be due to the presence of human D2-D5 domains (Fig. S5B).
These observations regarding the 2/3D1 mutD2 construct demonstrate that the epitope of H27K15 can be restored in the murine receptor, without impairing the affinity of its natural ligands, which validates the in silico prediction. Even though affinity will not guarantee functionality, it seems worthwhile that 2/3D1 mutD2 could be used in a transgenic mouse for future in vivo studies of H27K15.
Discussion
At the early stage of this work, we used models for both mAb and hCD115 for protein-protein docking because no hCD115 structure was available. In the field of antibody modeling, it is well established that the conservation of the framework and some CDRs allows a high success rate for antibody modeling.24 Modeling the target remains less obvious because it depends on the availability of crystal templates; however, the increasing power and performance of modeling algorithms allowed the use of models for studying protein interactions.25 The recent publications of the structures of hCD115 in combination with either CSF-1 or IL-3413,26 has confirmed both the predicted hCD115 model (D1 and D2 main-chain RMSD 1.4Å between modeled and crystal structure of hCD115) and the theoretical epitope residues (first 24 residues of the N-terminus D1 domain, 9 residues prior to the D1-D2 junction and 3 residues in the D2 domain). The low resolution electron microscopy structure of hCD11526 showed a different orientation of the D1 vs. D2 domains compared with the one published by Ma et al.,13 suggesting a degree of flexibility between these domains. Remarkably, D1-D2 conformation from both mCD115 and hCD115 crystals were relatively well superimposed, unlike the electron microscopy observation. This might be explained by the fact that the observed flexibility could be constrained by the crystallography technique.
In the absence of available H27K15 and H27K15/CD115 crystal structures, the protein-protein docking analyses required experimental validations. We achieved this by measuring interaction by ELISA and binding affinities measurements by QCM with a panel of chimeric and mutated recombinant CD115D1-D5. These series of experiments confirmed mainly the D1 domain and certain residues in D2 as the H27K15 epitope. Moreover, the use of chimeric constructs led to the identification of the first 17 residues of hCD115 as part of an essential epitope (i.e., necessary to detect binding of H27K15). NMR spectroscopy on a 23-mer synthetic peptide demonstrated that this essential epitope is able to interact with H27K15 independently of the rest of the CD115 molecule. The results were quite surprising because of the relative unfolded state of the 23-mer peptide; however, it has been described that a peptide can adopt a folded conformation driven by the microenvironment of the antigen-binding site of antibodies.27
In mouse CD115, which does not bind H27K15, four positions are different among the 17 first residues compared with hCD115. Three of the four mutations were introduced to hCD115D1-D5 as single substitutions and their effect on H27K15 binding was evaluated by QCM. The single mutation V8G alone was shown to completely abolish detectable binding of H27K15. This result was confirmed by NMR spectroscopy with a mutated V8G 23-mer peptide. Moreover, valine at position 8 in the hCD115 sequence is located in the center of a hydrophobic area that may be critical for H27K15/CD115 interaction. CD115 from several animal species bear this single V8G substitution in their CD115, and hence could not be used as models for in vivo H27K15 studies. Therefore, to broadly explore the mechanism of action, a transgenic mouse model should be generated. Several human-mouse CD115 chimeras were constructed to determine the one with the optimal binding (i.e., with an affinity similar to the wild type CD115) for both H27K15 and the murine ligands. A mouse CD115 with a fully human essential epitope (namely 1/3D1) has a 10-fold reduced affinity for H27K15 compared with the fully human CD115. It demonstrates that another part of CD115 than the essential epitope participates to the binding of H27K15. Affinities similar to hCD115 and to mCD115 for H27K15 and mCSF-1, respectively, were obtained with the construct 2/3D1 mutD2. This chimeric CD115 with 8 human substitutions in D1 and 3 in D2 was built to cover the entire in silico predicted epitope.
Murine CSF-1 binding affinity for mCD115 was measured below the nanomolar range due to an undetermined dissociation rate constant (kd). Nevertheless, the affinity remains somehow different from what was measured previously.12 This difference might be due to the technology used (microcalorimetry vs. QCM), the nature and sources of the proteins (e.g., mCSF-1 expressed in insect cells (4 to 150) vs. HEK 293 (36 to 444)) or the design of the experiment (in solution vs. immobilized molecules). Despite these discrepancies, the affinities of mCSF-1 measured here were only used to select the construct with an affinity to mCSF-1 close to the one of wild type mCD115. Results with the mIL-34 were helpful to confirm the selection of the 2/3D1 mutD2 construct for further use, since its affinity to the mIL-34 was similar to the wild type mCD115 as well. Moreover, we demonstrated here that in contrast to mCSF-1, mIL-34 is able to bind human CD115, which had not previously been shown to our knowledge.
None of the residues involved in the H27K15 binding have been shown to be implicated in the CSF-1 or IL-34 binding sites. This observation is in agreement with the fact that H27K15 is a non-competitive partial inhibitor of the CSF-1.14 The H27K15 epitope is not part of the described homodimerization domains of the CD115 (i.e., D4-D5) that occur upon ligand binding. Therefore, there is no clear molecular mechanism of inhibition of CD115 activity that could be deduced from the epitope of H27K15. However, the fact that the Fc of H27K15 plays a major role in inhibition of the CD115 pathway indicates that a trimolecular interaction, CD115/H27K15/Fc receptor, is probably involved at the surface of myeloid lineage cells.14
The generation of a transgenic mouse carrying the 2/3D1 mutD2 CD115 construct could be helpful to gain additional information about the in vivo behavior of H27K15 and also to conduct efficacy or toxicity studies of this antibody.
Methods
Antibody production
The heavy and light chains of humanized anti-CD115 H27K15 mAb were cloned in plasmids pcDNA™3.3 and pOptiVEC™, respectively. Polyclonal pools of stable H27K15 transfected CHO-DG44 were generated (OptiCHO™ Antibody Express Kit, Invitrogen). The mAb H27K15 was purified from the culture supernatants on Protein A Sepharose 4FF (GE Healthcare) followed by gel filtration (HiLoad 26/60 Superdex 200, GE Healthcare). mAb concentration was calculated using the absorbance at 280 nm measured with a spectrophotometer (Specord, Analytik Jena AG) and the mass extinction coefficient, determined using the ProtParam software28 on the ExPASy server (www.expasy.org). Purified mAbs were sterilized by filtration on a 0.2 µm membrane (Nalgene), aliquoted and stored at -80 °C pending use.
Protein and peptide production
Murine CSF-1 (mCSF-1), D1-D5 of mCD115, hCD115 and every associated chimeric construct described for QCM experiments as well as ELISA, were transiently transfected in HEK 293 cells and purified by GeneArt GmbH. All CD115 produced and purified by Geneart had a StrepTag at their C-terminus part that allows their immobilization on QCM chips. For NMR experiments, peptides from the 23 most N-terminal residues of hCD115, including or not the murine V8G mutation (IPVIEPS[V/G]PELVVKPGATVTLRC), were provided by Thinkpeptides (United Kingdom). For ELISA experiments with different species of CD115, 8 histidine tagged CD115D1-D5 constructs were cloned and transiently expressed in CHO-K1 cells.
Homology modeling
Homology modeling of the H27K15 mAb variable fragment (Fv) and human CD115 were performed using Discovery Studio 3.1 (Accelrys Inc). The anti-dansyl immunoglobulin IgG2a crystal29 (PDB 1DLF), the humanized anti-lysozyme Fv fragment crystal30 (PDB 1BVK) and the anti-sars scFv antibody crystal31 (PDB 2GHW) were used as templates for modeling the heavy chain (VH), light chain (VL) and the VH/VL interface of H27K15 Fv, respectively. The sequence homologies (identity and similarity) between each template and H27K15 Fv were of 79% and 88% for VH, 87% and 94% for VL and 65% and 81% for the entire Fv. The “Model Antibody Framework” protocol was first used to generate 100 models of the H27K15 framework region. Then, the “Model Antibody Loops” protocol was used to generate 25 CDR-optimized models from the framework model with the best scoring function. The crystal template used for hCD115 modeling was the murine CD115 crystal12 (PDB 3EJJ, 56% sequence identity with hCD115) because the human CD115 crystal was not available at the initiation of this work. Finally, the human CD115 crystal13 (PDB 4DKD) was used for validation of the models.
Computational mutagenesis
The effect of hCD115 single-point mutations on the binding of the antibody model was calculated with the CHARMm-based “Calculate Mutation Energy (Binding)” protocol. The total free energy difference between the wild-type and mutated structures was calculated as a weighted sum of the van der Waals (0.45), Electrostatic (0.45) and Entropy (0.80) terms. The effect of each mutation on the binding was considered with regard to the following rule: “stabilizing” < -0.5 kcal.mol−1 < ”neutral” < +0.5 kcal.mol−1 < “destabilizing.” The selected single-point mutants were then modeled using the “Build Mutant” protocol32 of Discovery Studio 3.1 (Accelrys Inc) for protein-protein docking analyses.
Docking
Prior to docking, models were typed using the CHARMm Polar H forcefield and were minimized with the adopted basis Newton-Raphson method (ABNR).33,34 ZDOCK16 was used with a 6° angular step size generating 54 000 poses of which the top 2000 were reranked by ZRANK22 and the top 200 were clustered. These poses were then processed with RDOCK17 and only clusters with the highest density of poses were further considered. All docking calculations were made with Discovery Studio 3.1 (Accelrys Inc).
Sequence alignment
CD115 protein sequences from human (P07333), mouse (P09581) and rat (Q00495) were retrieved from Swiss-Prot database; rhesus macaque (F7BTH9), dog (E2RJV2) and Guinea pig (H0VVM7) CD115s were retrieved from TrEMBL database; marmoset and rabbit CD115s were cloned and sequenced internally; Swiss-Prot and TrEMBL are part of the UniProt knowledgebase35 (www.uniprot.org). Global and multiple alignments were performed using the Gap and PileUp programs, respectively, from GCG Wiskonsin Package (Accelrys Inc).
ELISA binding experiments
For ELISA with CD115 of different species, microtiter plates (96-well Nunc Maxisorp) were coated in 100 µl with 500 ng of mouse anti-penta-histidine IgG (Qiagen) overnight at 4 °C and then saturated during 1 h at room temperature (RT) with blocking buffer PBS, 3% BSA. Interaction analysis with H27K15 mAb were performed in the presence of diluted CHO-K1 supernatants transiently expressing soluble 8 histidine tagged human, mouse, rhesus, marmoset and rabbit CD115 proteins (100 µl per well) incubated during 2 h at RT. Plates were rinsed five times in washing buffer containing 50 mM Tris pH 8, 0.14 M NaCl, 0.05% Tween 20 (Sigma-Aldrich), then 100 ng of biotinylated H27K15 mAb or PBS (negative control) were added for 2 h at RT. The washing step was repeated and the binding was detected using streptavidin-HRP complex (Southern Biotech) diluted 5 000-fold in BSA 1%, PBS for 1 h at RT. The TMB (3,3',5,5'-tetramethylbenzidine, Sigma) was added as a substrate (100 µl per well). The absorbance was measured directly at 620 nm or at 450 nm if the reaction was stopped by 100 µl of 2 M H2SO4.
For ELISA with chimeric CD115, the experimental conditions were as described above except purified CD115 constructs were directly coated on plate at 1µg.ml−1 and binding of unlabeled H27K15 was detected with an anti-Fc human HRP (Bethyl).
Affinity measurements
Quartz Crystal Microbalance technology (Attana 200) was used to measure affinities of the different CD115 constructs for H27K15, mCSF-1 or mIL-34. The affinities of CD115 for mCSF-1 and mIL-34 were performed as following: a monoclonal anti-StrepTag (StrepMAB-Immo, IBA) was directly immobilized on chips using amine coupling kit (3501–3001, Attana). CD115-StrepTag proteins were captured on chips via binding to the anti-StrepTag antibody at quantities yielding a signal of about 40Hz. Analyses were performed at a flow rate of 30 µl min−1 and at a temperature of 25 °C in HBST buffer (10 mM HEPES, 150 mM NaCl, 0.005% Tween 20, pH 7.4, Attana). The sensor chip surface was regenerated after each cycle by injecting 10 mM glycine-HCl pH 3. Analyte injections were realized for 70 s before a post-injection phase of 180 s. Blank signal obtained with buffer injections was used as reference and subtracted from each analyte signal. Murine IL-34 was injected at 12, 6, 3, and 1.5 nM (using a molecular weight of 52 kDa) and mCSF-1 was injected at 6, 3, 1.5, and 0.75 nM (using a molecular weight of 94 kDa). Data analyses were performed using Evaluation Software (Attana) and simple model 1:1 was used for data fitting.
For the H27K15/CD115 affinity measurements, the experimental conditions were the same as the ones described above except for the following points: H27K15 was immobilized directly on chips using amine coupling kit at quantities yielding a signal of about 100 Hz; CD115 constructs were injected during 60 s at 172, 86, 43, and 21.5 nM (using a molecular weight of 58 kDa); and the temperature of the experiments was 37 °C.
NMR experiments
NMR experiments were recorded at 295 K on a Bruker Avance III spectrometer equipped with a TCI cryoprobe. H27K15 antibody was purified in PBS buffer pH 7.4, and wild-type and mutant 23-mer peptides were solubilized in PBS buffer. All experiments were performed using unlabeled samples with the addition of 10% D2O v/v (lock signal), using an excitation sculpting scheme for water suppression and a 30 ms spin-lock pulse to filter the antibody signal from the peptide signal.
Supplementary Material
Disclosure of Potential Conflicts of Interest
Most authors are employees of Transgene, the company that patented H27K15.
Acknowledgments
We thank Dr Hugues-Olivier Bertrand from Accelrys for his expertise and his helpful comments about the manuscript. We also gratefully thank Dr Myew-Ling Toh, for critical reading of the manuscript.
Glossary
Abbreviations:
- CDR
complementarity-determining region
- CSF-1
colony-stimulating factor-1
- DOPE
discrete optimized protein energy
- ELISA
enzyme-linked immunosorbent assay
- Fv
variable fragment
- Fc
constant fragment
- GCG
Genetics Computer Group
- HBST
Hepes buffered Steinberg’s solution
- HRP
horseradish peroxidase
- IL-34
interleukin-34
- LNB
low non-specific binding
- PBS
phosphate buffered saline
- PDB
protein data bank
- QCM
quartz crystal microbalance
- mAb
monoclonal antibody
- NMR
nuclear magnetic resonance
- RMSD
root mean square deviation
- TMB
3,3',5,5'-tetramethylbenzidine
- VH
heavy chain variable region
- VL
light chain variable region
- WT
wild-type
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/27736
References
- 1.Sherr CJ. Colony-stimulating factor-1 receptor. Blood. 1990;75:1–12. [PubMed] [Google Scholar]
- 2.Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 3.Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Curr Opin Immunol. 2006;18:39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8:533–44. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 5.Chihara T, Suzu S, Hassan R, Chutiwitoonchai N, Hiyoshi M, Motoyoshi K, Kimura F, Okada S. IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 2010;17:1917–27. doi: 10.1038/cdd.2010.60. [DOI] [PubMed] [Google Scholar]
- 6.Nandi S, Cioce M, Yeung YG, Nieves E, Tesfa L, Lin H, Hsu AW, Halenbeck R, Cheng HY, Gokhan S, et al. Receptor-type protein-tyrosine phosphatase ζ is a functional receptor for interleukin-34. J Biol Chem. 2013;288:21972–86. doi: 10.1074/jbc.M112.442731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers SK. Role of CSF-1 in progression of epithelial ovarian cancer. Future Oncol. 2009;5:1429–40. doi: 10.2217/fon.09.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ide H, Seligson DB, Memarzadeh S, Xin L, Horvath S, Dubey P, Flick MB, Kacinski BM, Palotie A, Witte ON. Expression of colony-stimulating factor 1 receptor during prostate development and prostate cancer progression. Proc Natl Acad Sci U S A. 2002;99:14404–9. doi: 10.1073/pnas.222537099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith HO, Anderson PS, Kuo DY, Goldberg GL, DeVictoria CL, Boocock CA, Jones JG, Runowicz CD, Stanley ER, Pollard JW. The role of colony-stimulating factor 1 and its receptor in the etiopathogenesis of endometrial adenocarcinoma. Clin Cancer Res. 1995;1:313–25. [PubMed] [Google Scholar]
- 10.Kitaura H, Zhou P, Kim HJ, Novack DV, Ross FP, Teitelbaum SL. M-CSF mediates TNF-induced inflammatory osteolysis. J Clin Invest. 2005;115:3418–27. doi: 10.1172/JCI26132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel S, Player MR. Colony-stimulating factor-1 receptor inhibitors for the treatment of cancer and inflammatory disease. Curr Top Med Chem. 2009;9:599–610. doi: 10.2174/156802609789007327. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci U S A. 2008;105:18267–72. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma X, Lin WY, Chen Y, Stawicki S, Mukhyala K, Wu Y, Martin F, Bazan JF, Starovasnik MA. Structural basis for the dual recognition of helical cytokines IL-34 and CSF-1 by CSF-1R. Structure. 2012;20:676–87. doi: 10.1016/j.str.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Haegel H, Thioudellet C, Hallet R, Geist M, Menguy T, Le Pogam F, Marchand JB, Toh ML, Duong V, Calcei A, et al. A unique anti-CD115 monoclonal antibody which inhibits osteolysis and skews human monocyte differentiation from M2-polarized macrophages toward dendritic cells. MAbs. 2013;5:736–47. doi: 10.4161/mabs.25743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 16.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–7. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 18.Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 19.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2006; Chapter 5:Unit 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987;196:901–17. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 21.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–24. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce B, Weng Z. ZRANK: reranking protein docking predictions with an optimized energy function. Proteins. 2007;67:1078–86. doi: 10.1002/prot.21373. [DOI] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Morea V, Lesk AM, Tramontano A. Antibody modeling: implications for engineering and design. Methods. 2000;20:267–79. doi: 10.1006/meth.1999.0921. [DOI] [PubMed] [Google Scholar]
- 25.Schneider C. Can we use homology models in protein-protein docking?: http://accelrys.com/resource-center/case-studies/pdf/zdock_homology_models.pdf, 2005.
- 26.Elegheert J, Desfosses A, Shkumatov AV, Wu X, Bracke N, Verstraete K, Van Craenenbroeck K, Brooks BR, Svergun DI, Vergauwen B, et al. Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. Structure. 2011;19:1762–72. doi: 10.1016/j.str.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keskin O. Binding induced conformational changes of proteins correlate with their intrinsic fluctuations: a case study of antibodies. BMC Struct Biol. 2007;7:31. doi: 10.1186/1472-6807-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al., eds. Protein Identification and Analysis Tools on the ExPASy Server: Humana Press, 2005. [Google Scholar]
- 29.Nakasako M, Takahashi H, Shimba N, Shimada I, Arata Y. The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody. J Mol Biol. 1999;291:117–34. doi: 10.1006/jmbi.1999.2931. [DOI] [PubMed] [Google Scholar]
- 30.Holmes MA, Buss TN, Foote J. Conformational correction mechanisms aiding antigen recognition by a humanized antibody. J Exp Med. 1998;187:479–85. doi: 10.1084/jem.187.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang WC, Lin Y, Santelli E, Sui J, Jaroszewski L, Stec B, Farzan M, Marasco WA, Liddington RC. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J Biol Chem. 2006;281:34610–6. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feyfant E, Sali A, Fiser A. Modeling mutations in protein structures. Protein Sci. 2007;16:2030–41. doi: 10.1110/ps.072855507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. doi: 10.1002/jcc.540040211. [DOI] [Google Scholar]
- 34.Momany FA, Rone R. Validation of the general purpose QUANTA ®3.2/CHARMm® force field. J Comput Chem. 1992;13:888–900. doi: 10.1002/jcc.540130714. [DOI] [Google Scholar]
- 35.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011; 2011:bar009. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.