Abstract
Norovirus is the leading cause of acute gastroenteritis with most infections caused by GII.4 variants. To understand the evolutionary processes that contribute to the emergence of GII.4 variants, we examined the molecular epidemiology of norovirus-associated acute gastroenteritis in Australia and New Zealand from 893 outbreaks between 2009 and 2012. Throughout the study GII.4 New Orleans 2009 was predominant; however, during 2012 it was replaced by an emergent GII.4 variant, Sydney 2012. An evolutionary analysis of capsid gene sequences was performed to determine the origins and selective pressures driving the emergence of these recently circulating GII.4 variants. This revealed that both New Orleans 2009 and Sydney 2012 share a common ancestor with GII.4 Apeldoorn 2007. Furthermore, pre-epidemic ancestral variants of each virus were identified up to two years before their pandemic emergence. Adaptive changes at known blockade epitopes in the viral capsid were also identified that likely contributed to their emergence.
Keywords: Norovirus, Gastroenteritis, Emergence, Evolution, Sydney 2012
Introduction
Norovirus (NoV) is a clinically important RNA virus that is estimated to cause almost half of all cases of acute gastroenteritis, globally (Patel et al., 2009). NoV is also the only human enteric virus known to cause pandemics of acute gastroenteritis (Siebenga et al., 2009). NoV infects people of all ages; however, the majority of NoV-associated gastroenteritis outbreaks occur within institutional settings, such as aged-care facilities, hospitals and child care centres (Hall et al., 2013; Tu et al., 2008), thereby affecting the most vulnerable in the community including the elderly, immune-compromised and young children. Control strategies are largely preventative, since there is currently a lack of effective treatments or vaccines available for use against NoV infection, and include isolation, disinfection and hygiene measures.
NoV strains can be classified into six major phylogenetic clades (Fields et al., 2013), referred to as genogroups (GI–GVI), of which, GI, GII and GIV include human viruses. Genogroups are further divided into genotypes, with over 40 currently described (Fields et al., 2013). Despite having such a broad genetic diversity, the majority of NoV infections (over 80%), in both a sporadic and outbreak setting, are caused by GII.4 viruses (Siebenga et al., 2009). Over the last two decades, the epidemiological patterns of NoV GII.4 viruses have mirrored that of influenza A virus, where antigenically novel variants emerge every two to three years to replace their predecessors (Koelle et al., 2006). GII.4 variants have also caused the six pandemics of NoV-associated acute gastroenteritis since 1995, all of which were initiated by the emergence of novel GII.4 variants including; US 1995/96 in 1996 (Noel et al., 1999; White et al., 2002), Farmington Hills 2002 in 2002 (Widdowson et al., 2004), Hunter 2004 in 2004 (Bull et al., 2006), Den Haag 2006b in 2007–2009 (Eden et al., 2010; Tu et al., 2008), New Orleans 2009 in 2009–2012 (Tra et al., 2013; Vega et al., 2011) and most recently the Sydney 2012 variant (Eden et al., 2013; van Beek et al., 2013). Importantly, the emergence of these novel NoV GII.4 variants often coincides with dramatic increases in the occurrence of both community-acquired and institutional outbreaks of acute gastroenteritis, as exemplified in late 2012 with the emergence of Sydney 2012 (Bennett et al., 2013; Fonager et al., 2013; van Beek et al., 2013).
The epidemiological success of the GII.4 viruses has been attributed to a number of factors (Bull and White, 2011), including both rapid nucleotide substitution (Bull et al., 2010) and homologous recombination (Eden et al., 2013). Antigenic novelty is generated through variation in the protruding (P2) domain of the viral capsid, which leads to escape from herd immunity (Debbink et al., 2012; Lindesmith et al., 2008). These changes mostly occur at five sites within the P2 domain (referred to as epitopes A–E) that have been shown to be blockade epitopes using a surrogate neutralisation assay (Lindesmith et al., 2012). NoV is also able to greatly extend its genetic repertoire through recombination, which is common both within and between genotypes (Bull et al., 2007). Recombination typically occurs at the ORF1/2 overlap (Bull et al., 2007), although recombination hotspots have also been identified within ORF2 and at the ORF2/3 boundary (Eden et al., 2013). This is important as it facilitates the exchange of non-structural and structural elements of the genome that can also contribute to antigenic variation and immune evasion, potentially leading to the emergence of novel variants (Eden et al., 2013).
To better understand the evolutionary processes that contribute to the emergence of novel NoV GII.4 variants, we first examine the molecular epidemiological trends of NoV-associated acute gastroenteritis in Australia and New Zealand between 2009 and 2012 through the identification of circulating genogroups, genotypes and GII.4 variants. Secondly, we explore the temporal dominance of GII.4 variants and describe the replacement of the pandemic GII.4 virus, New Orleans 2009 by a novel GII.4 variant, Sydney 2012, following its global emergence in 2012. Importantly, we then reveal that distinct pre-epidemic ancestral variants, of both New Orleans 2009 and Sydney 2012, were found to be in circulation up to two-years prior to their pandemic emergence. Finally, we identify adaptive changes in key antigenic blockade epitopes within the capsid P2 domain of the Sydney 2012 virus that likely contributed to its emergence and global dissemination.
Results
Epidemics of NoV-associated acute gastroenteritis occurred in Australia and New Zealand between 2009 and 2012
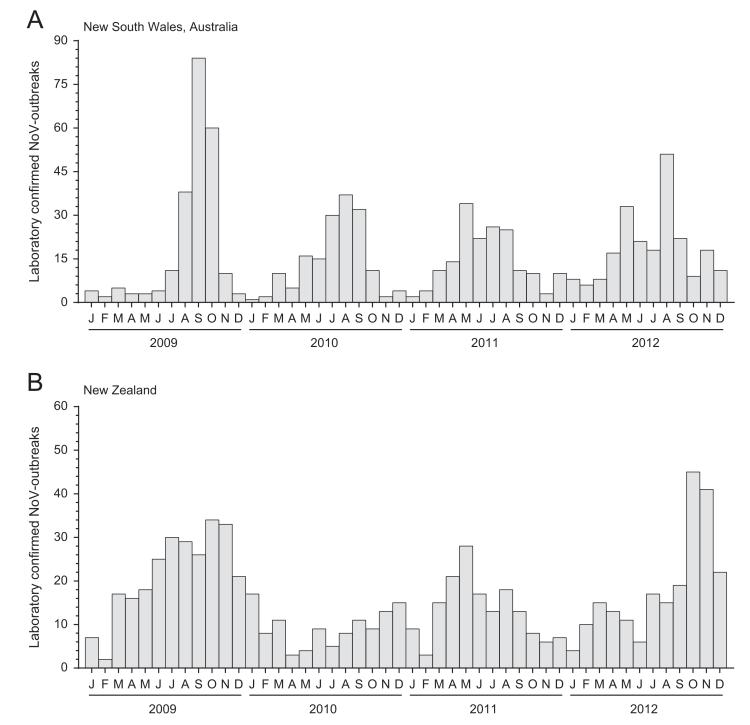
To determine whether there are seasonal patterns of NoV infection or any unusual spikes of NoV incidence in Australia and New Zealand, the number of laboratory confirmed NoV outbreaks was examined each month between January 2009 and December 2012, using data collected by public health authorities (Fig. 1). In NSW, Australia, a distinct peak in NoV activity was observed each year around August during the late-winter period (Fig. 1A); however, the magnitude of the peaks varied. Specifically, a sharp increase in the number of NoV outbreaks was observed between August and October 2009, with 84 outbreaks recorded in September, representing a more than twofold increase compared to the average number of outbreaks (n=41) in the peak winter months for 2010, 2011 and 2012 (Fig. 1A). In contrast, the peaks in NoV activity in New Zealand were generally broader and typically occurred later in the year around October/November, with the exception of 2011, when the peak of NoV outbreaks occurred in May (Fig. 1B).
Fig. 1. Laboratory confirmed number of monthly norovirus outbreaks in Australia and New Zealand between 2009 and 2012.
The number of laboratory confirmed norovirus outbreaks per month is shown between January 2009 and December 2012 for New South Wales, Australia (A) and New Zealand (B). Annual peaks in activity were observed during the late winter period in Australia (A) and the late spring for New Zealand (B), except in 2011 where the peak in activity occurred during autumn.
Circulating NoV genotypes in Australia and New Zealand from 2009 to 2012
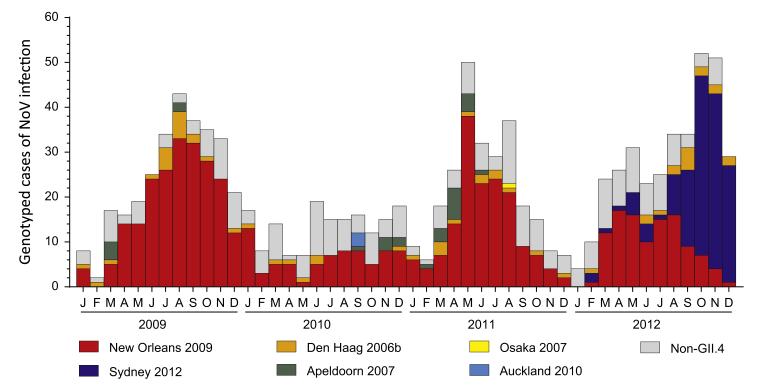
NoV strains from samples collected in this study were genotyped to determine the epidemiology and prevalence of circulating genotypes and GII.4 variants over time in NSW, Australia and New Zealand (Table 1). Additionally, the genotyping results of both countries were combined and plotted by month to examine and identify any shifts in prevalence amongst NoVs in the Oceania region with a focus on circulating GII.4 variants (Fig. 2). Between 2009 and 2012, the most commonly identified genotype was GII.4 (75.2%, n=790/1051) causing 75.0% of all outbreaks investigated (n=670/893 outbreaks) with the GII.4 variant – New Orleans 2009 (Prototype: GU445325|NewOrleans1805|US|2009) – the most predominant strain identified overall (53.0%, n=557/1051) (Table 1 and Fig. 2). However, during late 2012, GII.4 Sydney 2012 (Prototype: JX459908| NSW0514|AU|2012) emerged and replaced New Orleans 2009 as the predominant strain in circulation causing 27.7% and 51.8% of outbreaks during 2012 in Australia and New Zealand, respectively (Table 1 and Fig. 2). Other GII.4 variants detected in this study included Den Haag 2006b (40 outbreaks during 2009–2012), Apeldoorn 2007 (27 outbreaks during 2009–2011) and a single outbreak caused by Osaka 2007 during 2011 (Table 1 and Fig. 2). Furthermore, in New Zealand, a novel GII.4 variant, here referred to as Auckland 2010 (Prototype: KF060124|NLV-10–640|NZ|2010), that likely represents a pre-epidemic version of Sydney 2012, caused three epidemiologically linked outbreaks during 2010.
Table 1. Prevalence of NoV genotypes identified during 2009-2012.
| Year | Genotype | NSW, Australia |
New Zealand |
||||
|---|---|---|---|---|---|---|---|
| No. of outbreaks | % of total outbreaks | No. of unassigned | % of total unassigned | No. of outbreaks | % of total outbreaks | ||
| 2009 | GII.4 New Orleans 2009 | 24 | 82.8 | 3 | 100 | 187 | 72.5 |
| GII.4 Den Haag 2006b | 2 | 6.9 | 0 | 0 | 17 | 6.6 | |
| GII.4 Apeldoorn 2007 | 2 | 6.9 | 0 | 0 | 4 | 1.6 | |
| GII (non-GII.4) | 1 | 3.4 | 0 | 0 | 27 | 10.5 | |
| GI | 0 | 0 | 0 | 0 | 21 | 7.8 | |
| Mixed GI/GII | 0 | 0 | 0 | 0 | 2 | 0.8 | |
| Annual total – 2009 | 29 | 100 | 3 | 100 | 258 | 100 | |
| 2010 | GII.4 New Orleans 2009 | 19 | 79.2 | 11 | 42.3 | 46 | 40.7 |
| GII.4 Den Haag 2006b | 0 | 0 | 4 | 15.4 | 3 | 2.7 | |
| GII.4 Apeldoorn 2007 | 0 | 0 | 0 | 0 | 6 | 5.3 | |
| GII.4 Auckland 2010 | 0 | 0 | 0 | 0 | 3 | 2.7 | |
| GII (non-GII.4) | 3 | 12.5 | 10 | 38.5 | 39 | 34.5 | |
| GIV | 2 | 8.3 | 0 | 0 | 0 | 0 | |
| GI | 0 | 0 | 1 | 3.8 | 16 | 14.2 | |
| Annual total – 2010 | 24 | 100 | 26 | 100 | 113 | 100 | |
| 2011 | GII.4 New Orleans 2009 | 22 | 78.6 | 46 | 66.7 | 91 | 57.6 |
| GII.4 Den Haag 2006b | 3 | 10.7 | 6 | 8.7 | 4 | 2.5 | |
| GII.4 Apeldoorn 2007 | 0 | 0 | 1 | 1.4 | 15 | 9.5 | |
| GII.4 Osaka 2007 | 0 | 0 | 0 | 0 | 1 | 0.6 | |
| GII (non-GII.4) | 2 | 7.1 | 16 | 23.2 | 36 | 22.8 | |
| GI | 1 | 3.6 | 0 | 0 | 10 | 6.3 | |
| Mixed GI/GII | 0 | 0 | 0 | 0 | 1 | 0.6 | |
| Annual total – 2011 | 28 | 100 | 69 | 100 | 158 | 100 | |
| 2012 | GII.4 New Orleans 2009 | 36 | 55.4 | 29 | 48.3 | 43 | 19.7 |
| GII.4 Sydney 2012 | 18 | 27.7 | 14 | 23.3 | 113 | 51.8 | |
| GII.4 Den Haag 2006b | 6 | 9.2 | 6 | 10.0 | 5 | 2.3 | |
| GII (non-GII.4) | 5 | 7.7 | 11 | 18.3 | 45 | 20.6 | |
| GI | 0 | 0 | 0 | 0 | 9 | 4.1 | |
| Mixed GI/GII | 0 | 0 | 0 | 0 | 3 | 1.4 | |
| Annual total – 2012 | 65 | 100 | 60 | 100 | 218 | 100 | |
| Grand total 2009-2012 | 146 | – | 158 | – | 747 | – | |
Fig. 2. Prevalence of circulating norovirus genotypes in the Oceania region.
The genotyping results for Australia and New Zealand were combined then plotted by month between 2009 and 2012 to highlight the cause of each norovirus epidemic as well as the shift in prevalence between New Orleans 2009 and Sydney 2012 that occurred during late 2012. The prevalence of each GII.4 variants is shown with non-GII.4s grouped separately and coloured according to the key provided.
In total, 18 non-GII.4 genotypes were identified, which included GI (n=7), GII (n=10) and GIV (n=1) genotypes, and were collectively responsible for between 16.9% and 43.6% (mean=24.8%) of genotyped cases of NoV infection each year with no seasonal peaks (Fig. 2). Throughout the study, GII.6 was the most commonly identified non-GII.4 genotype (51 outbreaks) (Supplementary Tables 1 and 2). Finally, there were five instances where both NoV GI and GII were identified in the same outbreak (Table 1).
Replacement of New Orleans 2009 by Sydney 2012 as the predominant NoV
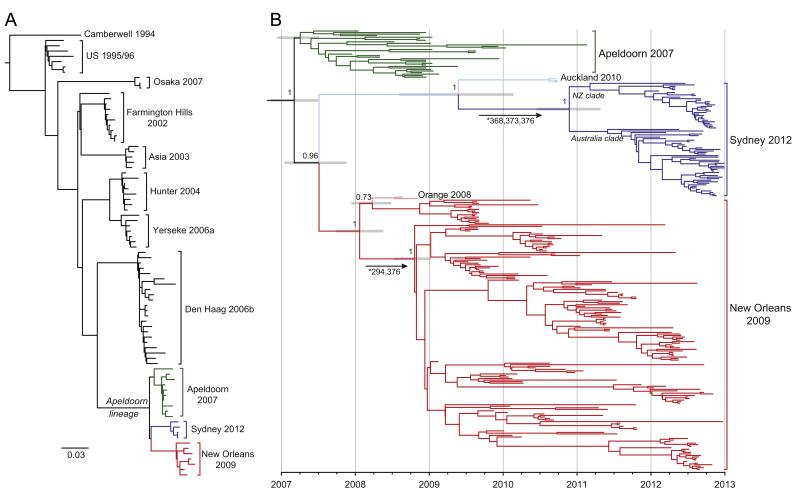
The emergent GII.4 variant, Sydney 2012, steadily replaced New Orleans 2009 as the predominant NoV in circulation in both Australia and New Zealand between September and December 2012 (Fig. 2). Despite both viruses being novel recombinants with distinct ORF1 regions, the ORF2/3 regions of New Orleans 2009 and Sydney 2012 share a common ancestor with Apeldoorn 2007 (Eden et al., 2013). Therefore, to determine the origins and timing of the emergence of Sydney 2012, we performed a Bayesian MCMC analysis (Fig. 3) using complete GII.4 capsid sequences (encoded by ORF2) of Sydney 2012 strains and closely related variants, which included Apeldoorn 2007 and New Orleans 2009, as well as the pre-epidemic variants, Orange 2008 (Prototype: GQ845367|NSW001P|AU|08) and Auckland 2010, here, collectively referred to as the Apeldoorn-lineage (Fig. 3A). As a group, the capsid sequences of the Apeldoorn-lineage viruses have evolved with a mean rate of 7.20 × 10 −3 (95% HPD=6.43 × 10 −3−7.99 × 10 −3) nucleotide substitutions per site per year and coalesce to point early in 2007 (credible range of October 2006–June 2007) (Fig. 3B). The Apeldoorn 2007 variants themselves sit close to the root of the tree and were the first to emerge within this group (Fig. 3B). Furthermore, both New Orleans 2009 and Sydney 2012 capsids are derived from an early Apeldoorn 2007-like virus that was in circulation during 2007 (credible range January–November 2007). Critically, the identification of Auckland 2010 revealed that pre-epidemic forms of Sydney 2012 were in circulation at least since 2010 (Fig. 3B) with the common ancestor of Auckland 2010 and Sydney 2012 temporally located between July 2008 and March 2010. In late 2010, the Sydney 2012 clade then diverged into two distinct sub-clades with the majority of New Zealand strains part of one sub-clade (76.7%, n=23/30) and the majority of Australian strains (NSW and SA) in the other sub-clade (79.5%, n=35/44) (Fig. 3B). The Australia sub-clade also included Sydney 2012 strains from other regions including Hong Kong (Chan and Chan, 2013) and Taiwan (KC243078|Taichung|TW|2012). Finally, the evolutionary analysis revealed that following the initial emergence of the pre-epidemic variant, Orange 2008, the New Orleans 2009 variants have similarly diverged into numerous sub-lineages and within each, a number of distinct temporal clusters were observed that mostly coincided with known epidemic periods (Fig. 3B).
Fig. 3. Temporal evolutionary analysis of the GII.4 Apeldoorn-lineage capsid.
(A) Maximum likelihood phylogeny derived from capsid protruding domain sequences (n=84) showing the major GII.4 variants identified since 1996 (with a Camberwell 1994 outgroup). The Apeldoorn-lineage forms a distinct group of variants that included Apeldoorn 2007, New Orleans 2009 and Sydney 2012, highlighted green, red and blue, respectively. The scale bar indicates the number of nucleotide substitutions per site. (B) Bayesian Maximum Clade Credibility phylogeny of the Apeldoorn-lineage from 286 complete capsid sequences, of which 221 were derived from this study. The posterior probabilities for key nodes are shown for those >0.80. The node bars show the 95% highest probability densities interval for node ages. Clades are coloured as panel A with the pre-epidemic variants Orange 2008 and Auckland 2010 shown in lighter shades of red and blue, respectively. The x-axis is scaled to time (years) with individual tips representing time points of sample collection. Branches marked with an asterisk and arrows indicate that the sites listed are under significant positive selection specifically along these branches and match those shown in Fig. 4.
Antigenic adaptation may have contributed to the emergence of NoV GII.4 Sydney 2012
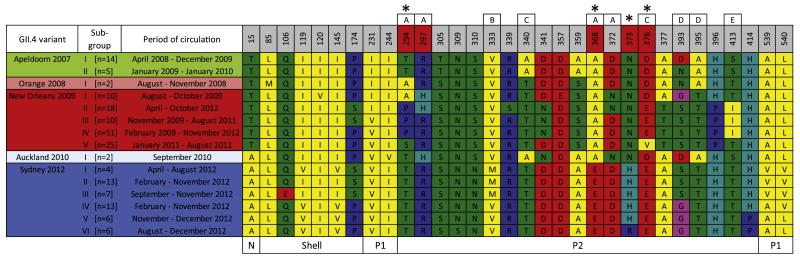
A number of studies have highlighted the role that antigenic variation plays in facilitating the emergence of viruses of the GII.4 lineage (Lindesmith et al., 2008, 2012). We therefore examined the GII.4 capsid sequences for evidence of positive selection, particularly at putative antigenic sites, as well as the presence of clusters with potential antigenic differences (Fig. 4). Overall, 32 variable positions in the GII.4 capsid sequence (total 540 codons) were identified when comparing the GII.4 variants of the Apeldoorn-lineage, that is, Apeldoorn 2007, Orange 2008, New Orleans 2009, Auckland 2010 and Sydney 2012. These variable sites mostly localised (65.6%, n=21/32) to the external surface of the capsid protein (P2 domain). Importantly, four of these sites, amino acid positions 294, 368, 373 and 376 (asterisks, Fig. 4) were found to be under significant positive selection using all three methods employed – iFEL, FUBAR and MEME. The dN/dS values were 3.40, 3.60, 1.61 and 3.30 for positions 294, 368, 373 and 376, respectively (all four positions p<0.027). Furthermore, the MEME analysis revealed that the positive selection at sites 368, 373 and 376 was episodic and occurred transiently along the specific branch of the phylogeny leading to Sydney 2012 and similarly, sites 294 and 376 for the major epidemic lineage of New Orleans 2009 (Fig. 3B). Finally, within each variant additional variation was observed with up to six sub-groups identified that contained distinct amino acid changes (Fig. 4). It is therefore possible that the amino acid changes at these sites, especially those under significant positive selection, may have contributed to the emergence of both Sydney 2012 and New Orleans 2009.
Fig. 4. Antigenic variation in the GII.4 Apeldoorn-lineage.
The GII.4 capsid sequence alignment of variants in the Apeldoorn-lineage used for the temporal evolutionary analysis was examined for antigenic clusters within and between different GII.4 variants as well as for evidence of positive selection. Asterisks indicate sites – 294, 368, 373 and 376 – that are under significant positive selection. Labelled boxes above each position indicate sites within the known blockade epitopes A–E that are important determinants of viral antigenicity. Amino acids have been coloured based on the properties of their side-chains: blue for positive charged – R, H, K; red for negative charged – D, E; Green for polar uncharged – S, T, N, Q; yellow for hydrophobic – A, V, I, L, M, F, Y, W; pink for special cases – C, U, G, P.
Discussion
Since the late 1990s, the molecular epidemiology of NoV-associated acute gastroenteritis has been characterised by the occurrence of annual epidemics of disease and the periodic emergence and pandemic spread of novel variants of the NoV GII.4 lineage (Bull and White, 2011; Eden et al., 2013; Lindesmith et al., 2008; Siebenga et al., 2009). When a new GII.4 variant is identified it often becomes predominant and can be associated with atypical increases in the incidence of acute gastroenteritis. This phenomenon was observed in the current study where an increase in the number of NoV infections occurred in the Oceania region, primarily in New Zealand, between October and November 2012 following the emergence of the novel GII.4 variant, Sydney 2012 (Figs. 1 and 2). Furthermore, during late 2012, similar increases in NoV infections were observed in other regions including parts of Europe (Giammanco et al., 2013; van Beek et al., 2013) and Asia (Chan and Chan, 2013; Rahman et al., 2013; Shen et al., 2013). However, in other regions, such as North America no atypical increase in NoV activity was observed following the emergence of Sydney 2012 (Centres for Disease and Prevention, 2013; Hasing et al., 2013; Leshem et al., 2013). Despite this, the global emergence of Sydney 2012 was characterised by a rapid increase in its prevalence, which in less than three months, saw the switch from New Orleans 2009 to Sydney 2012 as the predominant NoV strain in circulation, highlighting the rapid spread of novel GII.4 variants in the global population.
Antigenic variation is an important factor contributing to the emergence of novel NoVs (Lindesmith et al., 2008, 2012) with most variation between different GII.4 variants localising to five evolving blockade epitopes (A–E) within the capsid P2 domain (Lindesmith et al., 2012). Epitope A is believed to be the most important determinant of antigenic change, following the isolation and mapping of human-derived antibodies to this site using a surrogate neutralization assay (Lindesmith et al., 2012). In this study, four sites within the P2 domain (294, 368, 373, 376) were identified with novel, variant specific mutations that were subject to significant positive selection according to phylogenetic methods employed (Figs. 3 and 4). Two of these sites (294, 368) are positioned within epitope A, one (376) was in epitope C and another site (373) was not associated with any previously defined epitope; however, is structurally adjacent to epitope A. It is therefore possible that the adaptive changes at sites 368, 373 and 376 in the Sydney 2012 capsid and 294 and 376 within the New Orleans 2009 capsid contributed to their emergence through the generation of antigenic variation. This hypothesis is supported by experimental evidence, where a recent study demonstrated, using both monoclonal antibodies and human-derived polyclonal sera, that Sydney 2012 was indeed antigenically distinct to its immediate pandemic precursors, New Orleans 2009 and Den Haag 2006b (Debbink et al., 2013). More specifically, residues 294 and 368 were identified, through mutational studies, as critical determinants of Sydney 2012 antigenicity, of which both sites were found to be under positive selection in this study (Figs. 3 and 4). In addition to those sites under positive selection, a number of other capsid amino acid changes were observed within the major GII.4 variants (Fig. 4) that may have altered the antigenic or receptor binding properties of the virus. For example, variation was observed in the capsid protein sequence of the two major Sydney 2012 phylogenetic clades, the Australia and New Zealand clades from Fig. 3, at positions 333 and 393, which each form part of known blockade epitopes – B and D, respectively. In another example, Sydney 2012 sub-group VI was found to have a unique change at position 373 (which sits adjacent to epitope A) when compared to the other five Sydney 2012 sub-groups (Fig. 4). This sub-group contained only strains identified in the city of Adelaide, South Australia between August and December 2012, which highlighted that these sub-groups found within each major GII.4 variant were mostly defined by geographic and temporal clustering and are more likely explained by founder effects rather than distinct adaptive antigenic changes. Further evidence for this was that the sub-groups mostly formed into discrete genetic clusters when mapped onto the phylogeny (data not shown).
It is also important to consider other factors that may have contributed to the emergence of Sydney 2012. The NoV capsid P2 domain also contains sites involved in the binding of Histo-Blood Group Antigens (HBGAs), which are considered attachment factors and play a role in determining susceptibility to infection (Lindesmith et al., 2008). In these regions would likely be important determinants of transmissibility for a newly emerged virus such as Sydney 2012. The primary HBGA/capsid binding occurs at site I (residues 344–346, 374, 440–444), whilst a secondary stabilising interaction occurs at site II (residues 387–396), which also overlaps with blockade epitope D (Shanker et al., 2011). In this case, none of the residues found to be under positive selection localised specifically to the either HBGA binding site; however, some were structurally adjacent, therefore, a role in HBGA binding variation contributing to the emergence of Sydney 2012 cannot be excluded.
Previous work has also demonstrated the importance of recombination in the emergence and evolution of novel GII.4 variants (Eden et al., 2013). Indeed, despite sharing a common ancestor, Apeldoorn 2007, in the ORF2 region, both New Orleans 2009 and Sydney 2012 are ORF1/2 recombinants with evolutionary distinct ORF1 regions, Yerseke 2006a for New Orleans 2009 and Osaka 2007 for Sydney 2012 (Eden et al., 2013). Currently, little is known about how the non-structural proteins encoded by ORF1 contribute to the emergence of novel NoV strains, however, there may be possible differences in T or B-cell epitopes or even through variation in mutation and replication rates, which may impact both overall viral fitness and immune escape (Bull et al., 2010; Eden et al., 2013).
This study also revealed that for the major pandemic GII.4 variants, New Orleans 2009 and Sydney 2012, early pre-epidemic forms, Orange 2008 and Auckland 2010, respectively, were in circulation up to two years prior to their global epidemic spread. Importantly, both these pre-epidemic variants were only associated with limited outbreaks within a specific region and did not demonstrate a global distribution (Eden et al., 2010; Thornley et al., 2013). One explanation for this initial, limited prevalence was that these pre-epidemic intermediates were found to be more similar to their shared ancestral predecessor, Apeldoorn 2007, at the key antigenic blockade epitopes A–E in the viral capsid (Fig. 4), despite possessing the novel ORF1 recombinant regions associated with their pandemic successors, New Orleans 2009 and Sydney 2012 (Eden et al., 2013). This not only confirms recombination as an important evolutionary mechanism that facilitates the emergence of novel variants and increases diversity in the global NoV population but also suggests that novel GII.4 variants may circulate in the population at low levels before acquiring the necessary P2 domain mutations to facilitate their emergence as a pandemic virus.
The identification of the pre-epidemic variants, Orange 2008 and Auckland 2010, highlights the necessity for thorough surveillance programs that will aid the identification and reporting of novel variants before they cause widespread outbreaks of disease. Furthermore, with improved surveillance and sampling of across all scales of NoV epidemiology, the accuracy of the substitution rate estimations and other useful evolutionary analyses, similar to those presented in this study, will increase and in turn help to better reveal the origins of novel NoV GII.4 variants including the timings and sources of their emergence. The evidence presented by this (Fig. 3) and previous studies (Eden et al., 2013) suggests that the genesis of Sydney 2012 was marked by a recombination event between early Apeldoorn 2007 and Osaka 2007 variants sometime around 2007 and consequently, pre-epidemic variants of Sydney 2012 must have been in circulation since this period. Only one such pre-epidemic variant, Auckland 2010, has been identified to-date, therefore, greater surveillance is likely required to identify emerging GII.4 variants and gain better insight into their origins. The global NoroNet network currently fulfils an important surveillance role and provides a means for regional and national labs to submit information regarding circulating NoVs and provides early warnings about the identification and epidemic potential of novel GII.4 variants such as with Sydney 2012 (van Beek et al., 2013).
In conclusion, this study described the epidemic emergence of a novel GII.4 variant, Sydney 2012 and its replacement of New Orleans 2009 as the predominant NoV strain in circulation, globally. Importantly, adaptive changes in the capsid P2 domain were identified that specifically mapped to key antigenic epitopes. Together, this supports the hypothesis that antigenic variation plays a critical role in the emergence and evolution of viruses of the NoV GII.4 lineage.
Materials and methods
Outbreak data and specimen collection
All stools samples were collected between January 2009 and December 2012 as part of routine surveillance programs; therefore, human ethics was not specifically requested for this study. Furthermore, patient consent was not required as specimens were de-identified and did not include any patient demographic information. In Australia, NoV-infected stools (n=304) were collected through the South Eastern Area Laboratory Services at the Prince of Wales Hospital, New South Wales (NSW). The monthly totals of institutional gastroenteritis outbreaks were provided by the communicable disease branch of the NSW Ministry of Health. In New Zealand, stool samples (n=747) were collected from cases of gastroenteritis, which were referred by New Zealand public health units to the Norovirus Reference Laboratory, Institute of Environmental Science and Research Ltd., for testing as part of New Zealand Ministry of Health national norovirus outbreak surveillance.
Outbreak specimens were defined as those with two or more temporally linked (within a week of collection) NoV-positive specimens collected from the same institution or location; all remaining specimens, which could not be assigned to any particular outbreak, were classified as ‘unassigned’; however, most consisted of individual presentations to hospital emergency departments.
Detection of NoV GI and GII viral RNA
For stool samples collected in Australia, viral RNA was extracted from stool suspensions as previously described (Tu et al., 2008) and reversed transcribed using the SuperScript™ VILO cDNA synthesis kit (Invitrogen, Carlsbad, USA). A real-time RT-PCR targeting the 5′ end of NoV GI and GII capsid genes was employed using iQ SYBR Green Supermix (Bio-Rad, Hercules, US) and the primers COGIF/GISKR (Kageyama et al., 2003; Kojima et al., 2002) and G2F3/G2SKR (Hansman et al., 2004; Kojima et al., 2002) for NoV GI and GII detection, respectively. The reaction conditions were as those described in the iQ SYBR Green Supermix manufacturer’s instructions with an annealing temperature of 55 1C and an extension time of 60 s. PCR products were purified and sequenced as described previously (Eden et al., 2010).
For the New Zealand samples, viral RNA was extracted from stool suspensions then assayed for NoV GI and GII using a duplex real-time RT-PCR as described previously (Greening et al., 2012). At least one NoV-positive sample from each outbreak was then selected for a second genotyping RT-PCR that was performed using SuperScript® III One-Step RT-PCR System with Platinum® Taq (Invitrogen) and primers Mon432/G1SKR and Mon433/G2SKR (Anderson et al., 2003; Kojima et al., 2002) for NoV GI and GII, respectively. PCR products were purified and sequenced as described previously (Greening et al., 2012).
Viral genotyping and sequence analysis
The NoV genotype was determined from partial capsid sequences for Australian samples or both partial polymerase and capsid for New Zealand samples by phylogenetic analysis. The raw sequence reads were first edited with FinchTV v1.4 (Geospiza, Seattle, USA) and then assembled in MEGA5 (Tamura et al., 2011). The relationship of viral sequences was then compared to known references strains using a phylogenetic approach (Kroneman et al., 2011, 2013).
Temporal analysis of NoV GII.4 capsid evolution
Complete capsid gene sequences were obtained from representative GII.4 strains collected in this study along with an additional ten sequences from GII.4 Sydney 2012 strains collected in South Australia (SA) during late 2012 using a sequencing approach described previously (Bull et al., 2012). In total, 221 complete GII.4 capsid sequences were sequenced, representing 146 outbreaks (NSW – 108, New Zealand – 32, SA – 6) and 75 unassigned infections (NSW – 71, SA – 4), and then combined with another 65 GII.4 capsid sequences derived from public databases (GenBank) to produce a final alignment of 286 GII.4 capsid sequences. The evolutionary history and divergence times were then estimated for the combined capsid dataset using the Bayesian Markov Chain Monte Carlo (MCMC) method available in BEAST v1.7.4 (Drummond and Rambaut, 2007). An uncorrelated lognormal relaxed clock model (Drummond et al., 2006) was used with the General Time Reversible (GTR) model of nucleotide substitution and incorporating a proportion of invariant sites (I) and a gamma distribution of among-site rate variation with 4 rate categories (Γ4) (i.e. the GTR+I+Γ4 model), as well as a Gaussian Markov Random Fields (GMRF) Bayesian Skyride tree prior (Minin et al., 2008). For this analysis, two independent chains of 100 million generations were run with 10% burn-in until convergence and then combined. Statistical uncertainty is reflected in values of the 95% Highest Probability Density (HPD). The Maximum Clade Credibility (MCC) tree was then inferred from the posterior distribution of trees with node heights scaled to mean values and posterior probabilities showing the support for individual nodes on the tree.
Analysis of molecular adaptation in the NoV GII.4 capsid gene
The HyPhy web server (Pond and Frost, 2005) (available at http://www.datamonkey.org/) was used to investigate sites within the capsid that were under likely positive selection. Three codon-based methods were used that compare the ratio of non-synonymous to synonymous substitutions per site (dN/dS); (a) a Fixed-Effects Likelihood along interior branches (iFEL) (Pond et al., 2006), (b) a Fast, Unconstrained Bayesian AppRoximation (FUBAR), and (c) Mixed Effects Model of Evolution (MEME), which can consider transient (episodic) selective pressures. The significance threshold was set to p=0.05 for iFEL, posterior probability=0.95 for FUBAR and Bayes factor=100 for MEME.
Nucleotide sequence accession numbers
The GenBank accession numbers for strains sequenced in this study are: JQ613526-JQ613573, KF059996-KF060153 and KF1-77429-KF177451. Furthermore, all sequence alignments used in this study are available from the authors by request.
Supplementary Material
Acknowledgments
This work was supported in part by the National Health and Medical Research Council and the New Zealand Ministry of Health. E.C.H. acknowledges support through an NHMRC Australia Fellowship. M.F.B. is supported by the Wellcome Trust (098511/Z/12/Z).
The authors wish to thank Jennie Musto, Neil Franklin and Dr. Jeremy McAnulty from the NSW Ministry of Health for the provision of data and Dawn Croucher, Malet Rivera-Aban and Laetitia Kaas from ESR for assistance with the New Zealand norovirus analysis.
References
- Anderson AD, Heryford AG, Sarisky JP, Higgins C, Monroe SS, Beard RS, Newport CM, Cashdollar JL, Fout GS, Robbins DE, Seys SA, Musgrave KJ, Medus C, Vinje J, Bresee JS, Mainzer HM, Glass RI. A waterborne outbreak of Norwalk-like virus among snowmobilers-Wyoming, 2001. J. Infect. Dis. 2003;187:303–306. doi: 10.1086/346239. [DOI] [PubMed] [Google Scholar]
- Bennett S, MacLean A, Miller RS, Aitken C, Gunson RN. Increased norovirus activity in Scotland in 2012 is associated with the emergence of a new norovirus GII.4 variant. Eur. Commun. Dis. Bull. 2013:18. [PubMed] [Google Scholar]
- Bull RA, Eden JS, Luciani F, McElroy K, Rawlinson WD, White PA. Contribution of intra- and interhost dynamics to norovirus evolution. J. Virol. 2012;86:3219–3229. doi: 10.1128/JVI.06712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, Eden JS, Rawlinson WD, White PA. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 2010;6:e1000831. doi: 10.1371/journal.ppat.1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, Tanaka MM, White PA. Norovirus recombination. J. Gen. Virol. 2007;88:3347–3359. doi: 10.1099/vir.0.83321-0. [DOI] [PubMed] [Google Scholar]
- Bull RA, Tu ET, McIver CJ, Rawlinson WD, White PA. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 2006;44:327–333. doi: 10.1128/JCM.44.2.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, White PA. Mechanisms of GII.4 norovirus evolution. Trends Microbiol. 2011;19:233–240. doi: 10.1016/j.tim.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Centers for Disease and Prevention Emergence of New Norovirus Strain GII.4 Sydney—United States, 2012. MMWR. Morbidity and Mortality Weekly Report. 2013;62:55. [PMC free article] [PubMed] [Google Scholar]
- Chan MC, Chan PK. Complete genome sequence of a novel recombinant human norovirus genogroup II genotype 4 strain associated with an epidemic during summer of 2012 in Hong Kong. Genome Announc. 2013:1. doi: 10.1128/genomeA.00140-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbink K, Lindesmith LC, Donaldson EF, Baric RS. Norovirus immunity and the great escape. PLoS Pathog. 2012;8:e1002921. doi: 10.1371/journal.ppat.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbink K, Lindesmith LC, Donaldson EF, Costantini V, Beltramello M, Corti D, Swanstrom J, Lanzavecchia A, Vinje J, Baric RS. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J. Infect. Dis. 2013 doi: 10.1093/infdis/jit370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden JS, Bull RA, Tu E, McIver CJ, Lyon MJ, Marshall JA, Smith DW, Musto J, Rawlinson WD, White PA. Norovirus GII.4 variant 2006b caused epidemics of acute gastroenteritis in Australia during 2007 and 2008. J. Clin. Virol. 2010;49:265–271. doi: 10.1016/j.jcv.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Eden JS, Tanaka MM, Boni MF, Rawlinson WD, White PA. Recombination within the pandemic norovirus GII.4 lineage. J. Virol. 2013;87:6270–6282. doi: 10.1128/JVI.03464-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BN, Knipe DM, Howley PM. Fields Virology. 6th ed. Vol. 1. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2013. (online resource (2 v. (xx, 2456, I-2482 p.)) [Google Scholar]
- Fonager J, Hindbaek LS, Fischer TK. Rapid emergence and antigenic diversification of the norovirus 2012 Sydney variant in Denmark, October to December, 2012. Eur. Commun. Dis. Bull. 2013:18. [PubMed] [Google Scholar]
- Giammanco GM, De Grazia S, Tummolo F, Bonura F, Calderaro A, Buonavoglia A, Martella V, Medici MC. Norovirus GII.4/Sydney/2012 in Italy, winter 2012-2013. Emerging Infect. Dis. 2013;19:1348–1349. doi: 10.3201/eid1908.130619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening GE, Hewitt J, Rivera-Aban M, Croucher D. Molecular epidemiology of norovirus gastroenteritis outbreaks in New Zealand from 2002-2009. J. Med. Virol. 2012;84:1449–1458. doi: 10.1002/jmv.23349. [DOI] [PubMed] [Google Scholar]
- Hall AJ, Lopman BA, Payne DC, Patel MM, Gastanaduy PA, Vinje J, Parashar UD. Norovirus disease in the United States. Emerging Infect. Dis. 2013;19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman GS, Katayama K, Maneekarn N, Peerakome S, Khamrin P, Tonusin S, Okitsu S, Nishio O, Takeda N, Ushijima H. Genetic diversity of norovirus and sapovirus in hospitalized infants with sporadic cases of acute gastroenteritis in Chiang Mai, Thailand. J. Clin. Microbiol. 2004;42:1305–1307. doi: 10.1128/JCM.42.3.1305-1307.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasing ME, Lee BE, Preiksaitis JK, Tellier R, Honish L, Senthilselvan A, Pang XL. Emergence of a new norovirus GII.4 variant and changes in the historical biennial pattern of norovirus outbreak activity in Alberta, Canada, from 2008 to 2013. J. Clin. Microbiol. 2013;51:2204–2211. doi: 10.1128/JCM.00663-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle K, Cobey S, Grenfell B, Pascual M. Epochal evolution shapes the phylodynamics of interpandemic in fluenza A (H3N2) in humans. Science. 2006;314:1898–1903. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods. 2002;100:107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- Kroneman A, Vega E, Vennema H, Vinje J, White PA, Hansman G, Green K, Martella V, Katayama K, Koopmans M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013;158:2059–2068. doi: 10.1007/s00705-013-1708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroneman A, Vennema H, Deforche K, v d Avoort H, Penaranda S, Oberste MS, Vinje J, Koopmans M. An automated genotyping tool for entero-viruses and noroviruses. J. Clin. Virol. 2011;51:121–125. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Leshem E, Wikswo M, Barclay L, Brandt E, Storm W, Salehi E, DeSalvo T, Davis T, Saupe A, Dobbins G, Booth HA, Biggs C, Garman K, Woron AM, Parashar AJ, Vinje J, Hall AJ. Effects and clinical significance of GII.4 Sydney norovirus, United States, 2012-2013. Emerging Infect. Dis. 2013:1231–1238. doi: 10.3201/eid1908.130458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith LC, Beltramello M, Donaldson EF, Corti D, Swanstrom J, Debbink K, Lanzavecchia A, Baric RS. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 2012;8:e1002705. doi: 10.1371/journal.ppat.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith LC, Donaldson EF, Lobue AD, Cannon JL, Zheng DP, Vinje J, Baric RS. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 2008;5:e31. doi: 10.1371/journal.pmed.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel JS, Fankhauser RL, Ando T, Monroe SS, Glass RI. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 1999;179:1334–1344. doi: 10.1086/314783. [DOI] [PubMed] [Google Scholar]
- Patel MM, Hall AJ, Vinje J, Parashar UD. Noroviruses: a comprehensive review. J. Clin. Virol. 2009;44:1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, Brown AJ. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput. Biol. 2006;2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Nahar S, Afrad MH, Faruque AS, Azim T. Norovirus variant GII.4/Sydney/2012, Bangladesh. Emerging Infect. Dis. 2013;19:1347–1348. doi: 10.3201/eid1908.130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J. Virol. 2011;85:8635–8645. doi: 10.1128/JVI.00848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Qian F, Li Y, Hu Y, Yuan Z, Zhang J. Novel norovirus GII.4 variant, Shanghai, China, 2012. Emerging Infect. Dis. 2013;19:1337–1339. doi: 10.3201/eid1908.130026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenga JJ, Vennema H, Zheng DP, Vinje J, Lee BE, Pang XL, Ho EC, Lim W, Choudekar A, Broor S, Halperin T, Rasool NB, Hewitt J, Greening GE, Jin M, Duan ZJ, Lucero Y, O’Ryan M, Hoehne M, Schreier E, Ratcliff RM, White PA, Iritani N, Reuter G, Koopmans M. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 2009;200:802–812. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornley CN, Hewitt J, Perumal L, Van Gessel SM, Wong J, David SA, Rapana JP, Li S, Marshall JC, Greening GE. Multiple outbreaks of a novel norovirus GII.4 linked to an infected post-symptomatic food handler. Epidemiol. Infect. 2013;141:1585–1597. doi: 10.1017/S0950268813000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tra, My PV, Lam HM, Thompson CN, Phuc HL, Tuyet PT, Vinh H, Hoang NV, Minh P, Vinh NT, Thuy CT, Nga TT, Hau NT, Chinh NT, Thuong TC, Tuan HM, Campbell JI, Clements AC, Farrar J, Boni MF, Baker S. The dynamics of GII.4 norovirus in Ho Chi Minh City, Vietnam. Infect. Genet. Evol. 2013;18:335–343. doi: 10.1016/j.meegid.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu ET, Bull RA, Greening GE, Hewitt J, Lyon MJ, Marshall JA, McIver CJ, Rawlinson WD, White PA. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin. Infect. Dis. 2008;46:413–420. doi: 10.1086/525259. [DOI] [PubMed] [Google Scholar]
- van Beek J, Ambert-Balay K, Botteldoorn N, Eden JS, Fonager J, Hewitt J, Iritani N, Kroneman A, Vennema H, Vinje J, White PA, Koopmans M, NoroNet Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Eur. Commun. Dis. Bull. 2013;18:8–9. [PubMed] [Google Scholar]
- Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinje J. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerging Infect. Dis. 2011;17:1389–1395. doi: 10.3201/eid1708.101837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PA, Hansman GS, Li A, Dable J, Isaacs M, Ferson M, McIver CJ, Rawlinson WD. Norwalk-like virus 95/96-US strain is a major cause of gastroenteritis outbreaks in Australia. J. Med. Virol. 2002;68:113–118. doi: 10.1002/jmv.10177. [DOI] [PubMed] [Google Scholar]
- Widdowson MA, Cramer EH, Hadley L, Bresee JS, Beard RS, Bulens SN, Charles M, Chege W, Isakbaeva E, Wright JG, Mintz E, Forney D, Massey J, Glass RI, Monroe SS. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 2004;190:27–36. doi: 10.1086/420888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.