Abstract
The structure of protein-protein complexes can be constructed by using the known structure of other protein complexes as a template. The complex structure templates are generally detected either by homology-based sequence alignments or, given the structure of monomer components, by structure-based comparisons. Critical improvements have been made in recent years by utilizing interface recognition and by recombining monomer and complex template libraries. Encouraging progress has also been witnessed in genome-wide applications of template-based modeling, with modeling accuracy comparable to high-throughput experimental data. Nevertheless, bottlenecks exist due to the incompleteness of the proteinprotein complex structure library and the lack of methods for distant homologous template identification and full-length complex structure refinement.
Introduction
Proteins are important molecules involved in virtually all cellular functions, including structural support, signal transduction, bodily movement, and defense against pathogens. Most functions are mediated by interactions between proteins. To perform all their various biological functions, the protein-protein interactions must be extremely diverse in the threedimensional structure: individual protein chains may form homo- or hetero-oligomeric, obligate or non-obligate, and transient or permanent complexes. These interactions form an intricate and dynamic network, the interactome, in living cells. Due to the important role in cellular processes, vast efforts have been devoted to uncovering the interactome, primarily by high-throughput experimental techniques [1–2]. However, these methods can at best tell which proteins interact, but are unable to reveal the structural details of such interactions; the latter is essential to understanding the molecular basis of cellular functions and for designing new therapies to regulate these interactions. Therefore, a major long-term goal of modern structural biology is to create a detailed ‘atlas’ of protein-protein interactions [3], containing not only the full interactome but, more challengingly, the atomic-level 3D structures of all protein complexes.
The most accurate structures of protein complexes are provided by X-ray crystallography and NMR spectroscopy; however, these techniques are labor-intensive and time-consuming. There has been a large gap between the number of known interactions and the number of interactions with known structures. Despite significant efforts in traditional structural biology and the structural genomics projects that aim at high-throughput complex structure determination [4], the latest statistics show that only ~6% of the known protein interactions in the human interactome have an associated experimental complex structure [5]. This number is quite low considering that we have a complete or partial experimental structure for ~30% of human proteins. Moreover, while the estimated size of the human interactome ranges from ~130,000 [6] to ~650,000 [7], interactome databases currently contain only ~41,000 binary interactions between human proteins, and many of them may be in error because of the inherent limitations of high-throughput experimental interaction discovery methods such as the yeast two-hybrid method [8]. Therefore, the development of efficient computational methods for discovering new interactions and in particular for large-scale, high-resolution structural modeling of protein-protein interactions is of paramount importance.
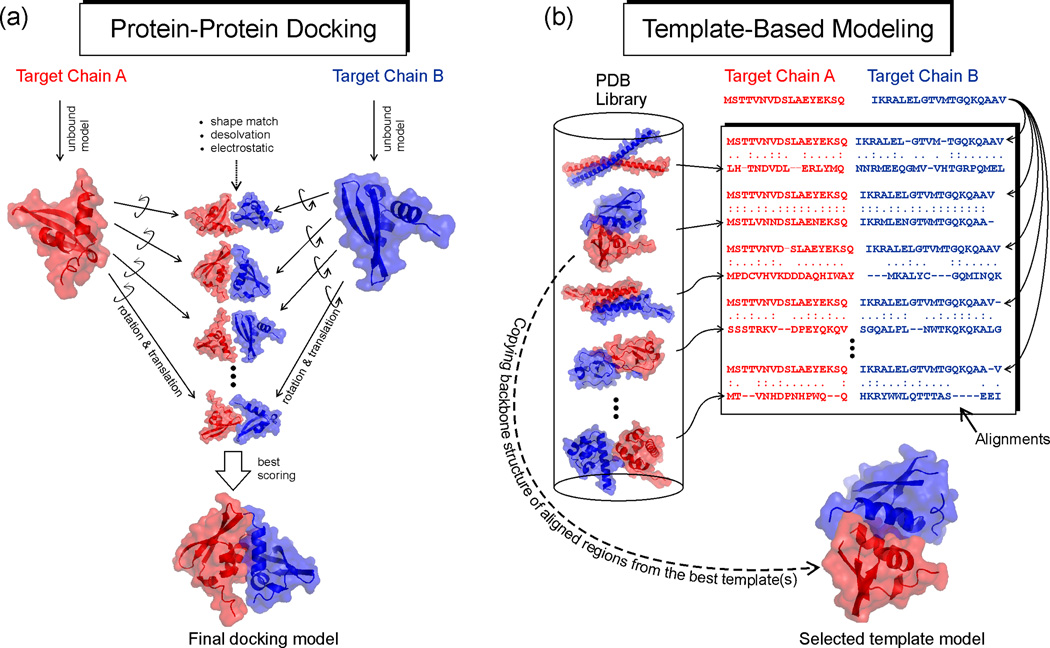
There are two distinct methods for the computational modeling of protein-protein complex structures (Figure 1). In protein-protein docking, complex models are constructed by assembling known structures of the interacting components, which are solved or predicted in the unbound form, through an exhaustive search and selection of various binding orientations (Figure 1a). The docking searches are often based on the shape and solvation matches of the surfaces of the component proteins, and work well for the protein complexes with an interface having obvious shape complementarity and with a large (>1400 Å2) and predominantly hydrophobic interfacial area [9]. But one challenge for rigid-body protein docking is that the accuracy decreases rapidly when the protein chains undergo large conformational changes upon binding [10–11]. Additionally, docking can only be performed when monomer structures of the interacting components are provided; but the experimental structures are in fact unavailable for a major portion of protein domains (–although structural models of the monomer proteins can be generated by computational structure prediction, the rigid-body docking accuracy is sensitive to the errors in the monomer models). The recent progresses in rigid-body protein docking are reviewed in [11–12].
Figure 1.
Two principal protocols for protein complex structure prediction. Red and blue represent sequences and structures of two individual chains. (a) Rigid-body protein-protein docking constructs protein complex structures by assembling known structures of monomer components which are usually solved (or modeled) in their unbound states. The final model is selected from those with the best shape complementarity, desolvation free energy and electrostatic matches between interfaces of the component structures [9–12]. (b) Template-based modeling (TBM) identifies complex structure templates by aligning the amino acid sequences of the target chains with the solved complex structures in the PDB library (shown on the left). The alignment can be generated based on sequence, sequence profile, or a combination of the sequence and structure feature information. The best template of the highest alignment score is selected; and the structure framework in the aligned regions is copied from the template protein which serves as a basis for constructing the structure model of the target [18•,21••,24–25]. Note that (b) only shows a typical protocol of homology-based template detection. There are variants of TBM which detect complex templates by query and template structure comparisons (see Figure 2) [19•–20••,22•–23,30].
The second method is template-based modeling (or TBM), which constructs protein complex structure of unknown targets by copying and refining the structural framework of other related protein-protein complexes whose structure has been experimentally solved (Figure 1b). The method of TBM has long been used to predict the tertiary structure of single-chain proteins, based on the principle that homologous proteins of similar sequences usually take the similar structure [13]; the method was later extended to model tertiary structure for distant homology proteins with the invention of the technique of threading [14], which aims to recognize the template structures without evolutionary relation to the target through incorporating structure information into sequence alignments. The general steps of TBM include finding one or more appropriate template(s); aligning the target sequence with the templates using sequence alignment, profile-based alignment, or threading; building an initial model for the target by copying the structural fragments from the aligned regions of the template(s); replacing the side chains to match the sequence of the target; constructing missing loops and termini; and, finally, refining the model to obtain a full-length atomic structure. Many variations and advanced methods have been developed for TBM [15–16], and it has been highly successful for protein tertiary structure modeling [17]. The TBM of protein-protein complexes is an extension of TBM techniques of single-chain proteins, where an essential step is to match the sequences of both chains with the solved complex structure library to identify appropriate template frameworks. The term TBM is often used interchangeably with ‘homology modeling’ in complex structure prediction although there have been substantial efforts and progress in detecting templates beyond homologous (or evolutionary) relationships [18•–23].
Compared to rigid-body docking, one advantage of TBM of protein-protein complexes lies in that the models are in principle constructed from amino acid sequences, and the structures of the monomer components are not pre-required. In addition, TBM methods construct the interaction models based on complex templates, which are in the bound form (in contrast to the unbound structures used in rigid-body docking) and are expected to be structurally similar to the target in all respects; therefore the TBM methods are usually not sensitive to the type of complex (large or small interface area, permanent or transient interaction) and to the extent of conformational change upon binding. Template-based complex structure prediction methods have significantly advanced in the past few years, and have been applied to whole proteomes with impressive results. In this review, we focus on the introduction and categorization of the most successful TBM methods (listed in Table 1), the identification of the key elements responsible for their success, the integration of TBM with other methods, and how TBM has helped to construct 3D interactomes. As these methods are often used to predict not only the 3D structures of the complexes but also whether two proteins interact, we include structurebased protein interaction prediction methods at the end of our discussion.
Table 1.
List of methods for template-based protein complex structure prediction.
| Methods [Ref.]a | Method typeb |
Interaction predictionc |
Structure predictiond |
Large- scale appe |
Web site |
|---|---|---|---|---|---|
| Interactome3D [65•] | DT | − | refined | [65•] | http://interactome3d.irbbarcelona.org/ |
| InterPreTS [44,48] | DT | + | crude | [30] | http://www.russelllab.org/cgi-bin/tools/interprets.pl |
| ABCLM [30] | DT & TBD | + | unrefined | [30] | |
| SPRING [21••] | MOM | − | crude | [21••] | http://zhanglab.ccmb.med.umich.edu/spring/ |
| COTH [18•] | DT | − | crude | [18•] | http://zhanglab.ccmb.med.umich.edu/COTH/ |
| TACOS | DT & FSS | − | refined | - | http://zhanglab.ccmb.med.umich.edu/TACOS/ |
| Multiprospector [24] | DT | + | crude | [69] | |
| M-TASSER [28] | DT & FSS | − | refined | - | |
| PrePPI [20••] | TBD | + | crude | [20••] | http://bhapp.c2b2.columbia.edu/PrePPI/ |
| Coev2Net [47•] | DT | + | - | [47•] | http://groups.csail.mit.edu/cb/coev2net/ |
| Struct2Net [26,50] | DT | + | crude | [26] | http://groups.csail.mit.edu/cb/struct2net/webserver/ |
| iWrap [27] | DT | + | crude | [27] | http://groups.csail.mit.edu/cb/iwrap/ |
| PRISM [23,46] | TBD | + | unrefined | [23,70] | http://prism.ccbb.ku.edu.tr/ |
| SKV [19•] | TBD | − | unrefined | - | |
| HOMBACOP [25] | DT | + | refined | [72•] | |
| KA [71] | DT & TBD | + | crude | [72•] | |
| HOMCOS [45,49] | DT | − | refined | [49] | http://strcomp.protein.osaka-u.ac.jp/homcos/ |
| THSWP [38] | DT | + | crude | [38] |
The methods without an explicit name are represented by an acronym formed from the authors’ initials.
Type of methods, categorized into dimeric threading (DT), monomer threading and oligomer mapping (MOM), template-based docking (TBD), and full-length complex structure simulation (FSS), following the categorizations in Figure 2 and Table 2.
“+”means that the method provides information about the existence of protein-protein interaction, whereby “-” means that the method does not conduct interaction prediction.
“crude” indicates that the method only provides a raw alignment of query and template proteins with gaps/insertions; “unrefined” means that the monomer chains are continuous but no further refinement was carried out; “refined” refers to the methods with some type of structure optimizations.
The literature that applied the developed methods to a large-score protein complex structure modeling.
Three general pipelines of template-based structure modeling
A standard procedure of conventional template-based complex modeling, starting from the sequences of the complex components, consists of four steps which are essentially identical to those used in TBM of single-chain proteins: First, finding known structures (templates) related to the sequences to be modeled; second, aligning the target sequences to the template structure by sequence- or profile-based methods or threading; third, constructing structural frameworks by copying the aligned regions of template structures; fourth, constructing the unaligned loop regions and adding side-chain atoms. The first two steps are actually conducted in a single procedure of multi-chain threading because the correct selection of templates requires accurate alignments. Similarly, the last two steps are performed simultaneously since the atoms of the core and loop regions interact closely. While most current TBM algorithms focus on the first two steps, i.e. template identification [18•,21••,24–27], there are only a few methods developed for full-length complex structure construction and refinement, which are in general more complicated and time-consuming [28]. In addition, there are other forms of TBM which detect complex frameworks through monomer-based structure comparisons [19•–20••,22•–23,29–30].
The quality of the TBM models essentially depends on the accuracy of the template identifications. There have been roughly three general strategies developed for complex template identification and structure combination, as detailed below (see also Figure 2 and Table 2 for summary and comparisons).
Figure 2.
Flowcharts for the three representative template-based complex structure prediction strategies. (a) Dimeric threading method. The black lines outline a threading procedure, similar to MULTIPROSPECTOR [24], which identifies complex templates from a dimer template library by dimeric query-to-template alignments. Blue lines indicate additional steps that improve upon the base method by utilizing a monomer template library and structural superposition, similar to COTH [18•]. Parts in magenta indicate stages where interface evaluation is used to increase alignment accuracy, ranking, and specificity. (b) Monomer threading and oligomer mapping. The protocol was used in SPRING [21••] where a combined template library containing both monomer and oligomer proteins is used. Monomeric threading is first used to identify a list of templates for each monomer chain where some templates will be parts of oligomers. The complex models are constructed by mapping the top templates of each monomer onto the framework excised from the associated oligomers, and ranked by monomer threading and interface matching scores. (c) Template-based docking. In this protocol, full-length models or experimental structures of the monomer proteins are matched against the dimer template library based on either global fold or interface structure comparisons. Dimer templates are selected from the complexes which have both components structurally similar to monomer structure of the target chains. A similar protocol is used in PrePPI [20••], PRISM [23] and the approach by Vakser et al [19•,22•].
Table 2.
A summary of the main features of different approaches to TBM of protein-protein complexes.
| Approach | Template libraries required |
Monomer template search method |
Complex template search method |
Complex structure construction |
Advantages (limitations) |
|---|---|---|---|---|---|
| Dimeric threading (Fig. 2a without blue parts) | Dimer template library | None | Dimeric threading | Dimer structure copied from template proteins | Alignment considering interfacial interactions dimer library is limited) |
| Extended dimeric threading (Fig. 2a with blue parts) | Dimer template library plus separate monomer template library | Monomeric threading | Dimeric threading | Superposition of monomer templates onto dimeric template | Improved models for individual subunits |
| Monomer threading and oligomer mapping (Fig. 2b) | Combined library of monomer and oligomer structures | Monomeric threading | Framework mapped from monomeric threading | Superposition of monomer templates onto oligomer subunits | A single template library covering different binding modes |
| Template-based docking (Fig. 2c) | Library of complexes or interfaces | Typically starts from monomer structures/models | Monomer to complex structural alignments | Superposition of monomer models onto the complex interface templates | Potential to detect non-homologous templates |
| Full-length complex structure simulation | None | None | None | Reassemble template structures by Monte Carlo simulations | Construction of full-length model and potential of structure refinement |
Dimeric threading
The first and probably the earliest strategy is the dimeric threading method [24], which is an extension of the single-chain threading (or fold-recognition) approach widely-used in tertiary protein structure prediction [14,16]. To detect homologous complex templates, the query sequences of both target chains are matched with the sequences of protein complexes whose structure has already been solved in the Protein Data Bank (PDB), generally through sequence profile-to-profile alignment assisted by secondary structure matching [31–32]. The final template models are selected by a combination of the threading alignment score and an interface evaluation score, the latter calculated by residue-based statistical potentials [33]. In a recent extension of the strategy [18•], the monomer structures identified from the tertiary template library by single-chain threading [31] are superimposed on the complex threading frameworks, which has been shown to improve the modeling accuracy and the coverage of threading alignments (Figure 2a). This strategy can generate high-resolution models when close homologous templates are identified in the libraries.
Monomer threading and oligomer mapping
The second strategy is based on monomer threading and oligomer mapping [21••]. The sequence of one monomer chain (e.g. Chain A) is first threaded through the PDB tertiary structure library to identify the closest homologous template. If the monomer template constitutes a part of a higher-order oligomer, each of the binding partners associated with the oligomer will be mapped to an appropriate threading template of another chain (Chain B) by a pre-calculated look-up table. The complex models are then constructed by superimposing the top monomer threading templates of both chains on the interacting framework excised from the higher-order oligomer structures, which are evaluated by a sum of the monomer threading alignment score and the interface matching score (Figure 2b). The essential difference between this strategy and the dimeric threading method is that this strategy does not include dimeric threading and, therefore, a non-redundant dimeric complex template library is not required. Instead, the monomer-based threading is run over all oligomer structures of the PDB library, and it has therefore the ability to detect complex frameworks associated with different binding modes of the same structure pair, which are often omitted in dimer-based threading on a reduced complex library [21••].
Template-based docking
The third strategy is through structural alignment based recognition and superimposition, also referred to as template-based docking [19•–20••,22•–23,30]. In this pipeline, the full-length monomer models are first taken from the PDB when available, or constructed by homology modeling. The templates of chain interactions are then identified from the solved complex library by requiring that the component chains of the complexes are structurally similar to the monomer models of the target sequences. The structural similarity between the target monomer models and the complex templates is assessed by standard structure alignment programs [34–36] which match either the global fold or the interface fragments. Finally, the complex models are constructed by superimposing the monomer models on the selected frameworks and evaluated by complex scoring functions that measure the structural similarities between the monomer models and the complex template components, and the fit of the interface shapes (Figure 2c). Since the initial target-template associations are established by purely structural alignments, this strategy has the potential to detect distant or non-homologous templates. Template-based docking has been recently reviewed in this journal [37].
Key elements of successful template-based structure prediction
A straightforward approach to template-based protein complex structure modeling is to simply match the monomer query sequences against the sequences of the subunits in a complex template library, and then copy the aligned monomer structures if both chains hit the same complex template [24–25,30]. Although this approach is successful to some extent [38], several improvements have been introduced that brought significant progress in the accuracy and coverage of complex template identification, especially when the homology between the target and the template is hard to detect or nonexistent (see e.g. [21••,39•] for benchmark comparisons of the straightforward approaches with more advanced ones). In this section, we present three key ideas that seem essential for the improved performance of state-of-the-art methods.
Interface evaluation
Even if a clearly homologous complex template exists for a given query protein pair, this does not necessarily mean that the query proteins actually interact or that they interact structurally in the same way. It has been shown that there is a “twilight zone” of sequence similarity (~25% sequence identity) below which it is almost impossible to tell whether domains will interact similarly [40]. Often, interfaces are not topologically conserved between protein families within a superfamily [41]. Therefore, some way of evaluating or scoring interfacial residue interactions should be an essential part of the template recognition of protein complexes [5,18•,21••,33,42–43].
One common approach to evaluating the putative interfaces is by using knowledge-based statistical interfacial potentials, usually residue-based [33], derived from known complex structures in the PDB. An interfacial potential can be used at several stages of the TBM procedure, including assisting in recognizing whether the two proteins interact as in InterPreTS [44]; improving the accuracy and ranking of template alignments as in MULTIPROSPECTOR [24], HOMCOS [45], Struct2Net [26], iWrap [27] and SPRING [21••]; and serving as an energy function term to guide the full-chain structural refinement as in M-TASSER [28] and TACOS (Mukherjee and Zhang, ‘Assemble protein complex structure by template identification and atomic-level structural refinement’, submitted) which, in particular, helps to eliminate the steric clashes and to optimize the interface contacts.
Various other approaches to evaluate interface interactions have also been introduced. In the COTH method, the interfacial residues are predicted from sequences by a neural network, which are then used to constrain the target-template alignments [18•]. In PRISM, which needs monomer structures as input, a combination of structural and evolutionary scores is used to measure the interface similarity between the query and template structures [23,46]. In HOMBACOP, a profile-profile alignment is used between the query and template sequences, with the profiles containing added information from experimental data about the interfacial residues [25]. PrePPI uses a combination of scores associated with the size of the interface and the overlap between predicted interfacial residues of the query proteins and the interface of the template to evaluate putative complex models [20••]. A new, promising approach, Coev2Net, uses a model for the coevolution of the interfacial residues to evaluate putative interfaces for interaction prediction [47•].
Combination of tertiary and quaternary template libraries
The existence of experimental template structures is a precondition of successful TBM prediction. Therefore, it is essential to assess and extend the coverage of the complex template libraries. Most TBM methods, including InterPreTS [44,48], MULTIPROSPECTOR [24], HOMBACOP [25], HOMCOS [45,49], Struct2Net [26,50] and Coev2Net [47•], rely on a complex template library to predict the chain orientations (quaternary structure) and the backbone framework of the targets. However, the coverage of quaternary structure space by the PDB is considerably lower than that of tertiary (monomer) structure space; a recent estimate shows that while there are structural data (experimental structure or a useable homologous template) for ~60–80% of the monomer chains of complex proteins, such coverage of complex structures is <30% [5]. When redundant complex structures (i.e. protein having a sequence identity >70% to other proteins) are filtered out, the library of protein structure complexes in the PDB is nearly six times smaller than the library of monomer structures [18•]. These findings imply that by restricting the template library to complexes alone, correct models can only be constructed for a small portion of protein complexes by TBM.
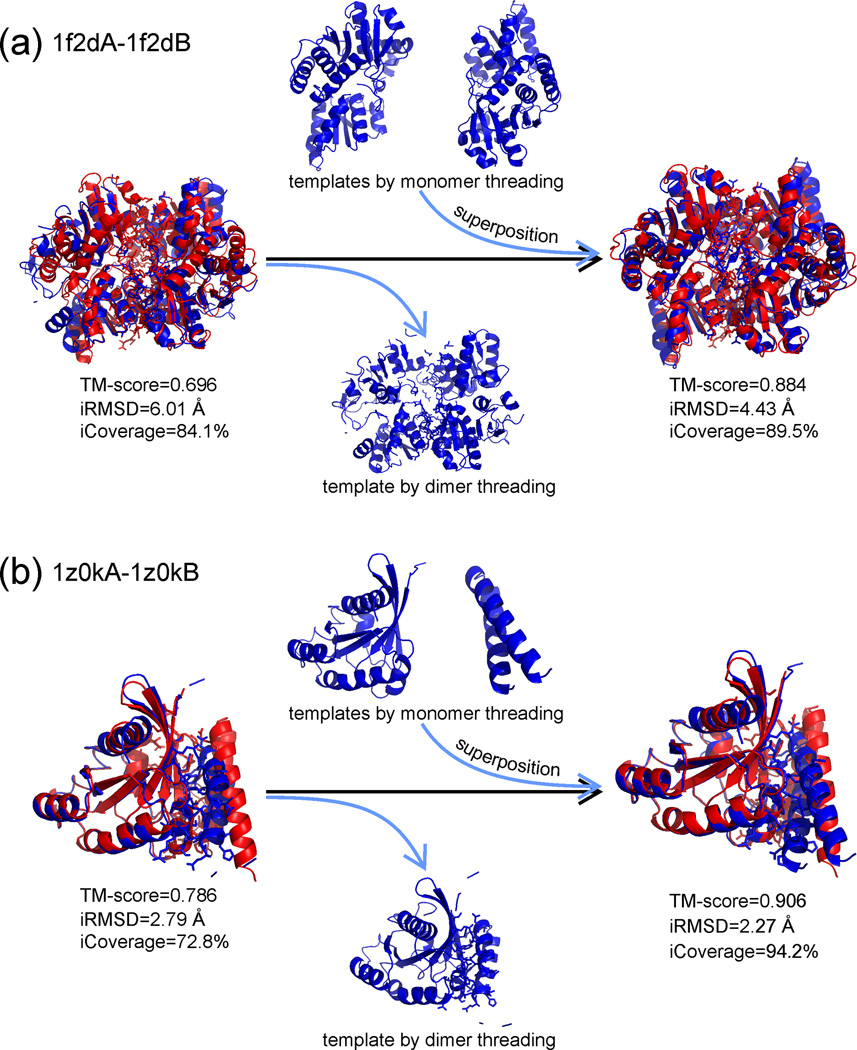
M-TASSER [28] and COTH [18•] are two approaches proposed to extend the quaternary template library by recombining the tertiary structure templates. In COTH, the sequences of both query chains are simultaneously threaded through the tertiary and quaternary structure libraries, and the monomer templates from the tertiary threading are then combined to create a quaternary framework by structurally superposing them onto a quaternary template. It was shown that this recombination of monomer templates on the framework of a complex template can significantly increase the accuracy and coverage of the interface contacts and the overall fold of the complex models (see Figure 3 for two examples from 1f2d and 1z0k) [18•].
Figure 3.
Tertiary structure models from monomer threading were used to improve the model accuracy of dimeric threading models by structural superposition in COTH [18•]. Red cartoons represent experimental structures and blue ones are predicted models from monomer and dimeric threading, with sticks highlighting the interface residues. (a) A homodimer example from the 1-aminocyclopropane-1-carboxylate deaminase (PDB ID: 1f2d), which has the TM-score increased from 0.696 to 0.884 after the structural superposition of the monomer threading models on the dimer threading framework. The interface RMSD (iRMSD) is reduced from 6.01 Å to 4.43 Å with the alignment coverage of interface residues (iCoverage) increasing from 84.1% to 89.5%. (b) A heterodimer example from GTP-Bound Rab4Q67L GTPase (PDB ID: 1zok), where TM-score, iRMSD and iCoverage are improved, after the structure superposition, from 0.786, 2.79 Å, 72.8% to 0.906, 2.27 Å and 94.2%, respectively.
The idea of quaternary template library extension was reinforced in SPRING [21••] which uses monomer threading and oligomer-based mapping to explore the different binding modes of all oligomer structures in the PDB. The complex models are then constructed by structurally aligning the top threading templates of individual chains onto the frameworks selected from the oligomer structures (see also Figure 2b). The template-based docking methods, as illustrated in Figure 2c, also superimpose the full-length monomer structures on the complex templates, and therefore share a similar foundation with COTH and SPRING; but they do not directly use tertiary template libraries to extend the quaternary structural space of the complex template library [19•–20••,22•–23,30].
Utilizing interface templates
The number of protein interaction types, or “quaternary folds”, in nature was estimated to be ~10,000 by Aloy and Russell [51], while a more recent estimate by Garma et al. lowered this number to ~4000 [52]. One difference between these estimations is that Aloy and Russell clustered the interaction types based on sequence identity while Garma et al. used the quaternary structure similarity of the complexes. Despite the difference, the authors of both estimates agreed that the current PDB only covers a small fraction of interaction types, and, extrapolating the current trends of structural biology, decades will pass before a full coverage of the quaternary structure space can be reached. This finding seems to put a strong limit to what TBM algorithms may achieve.
Some novel observations have, however, spurred more optimism. Kundrotas et al. [22•] found that the protein structural alignment program TM-align [34] can identify structural analogs of the monomer components of nearly all target complexes, with a TM-score >0.4, from the set of known complex structures in the PDB; the authors suggested that the current PDB can provide docking templates for almost all protein interactions once the monomer structure is known. However, the success rate of the template-based docking approach using the identified complex analogs is relatively low (~23%) when there are no close homologous templates with a sequence identity >40% with the target for at least one of the chains, indicating that the gain from the structural templates in addition to the sequence-based methods is yet modest, especially for the targets with non-homologous templates (typically having a sequence identity <25%), which the conventional homology-based methods have difficulty with.
Rather than considering the structures of entire complexes, one can focus solely on the protein-protein interfaces, and examine how much they are covered by the current PDB [53••–54•]. It turns out that the protein “interface space” is limited, and even chains with different folds often have similar interfaces. Calculations show that this interface space is degenerate and in fact close to complete, implying that templates of interfaces are probably available in the current PDB to model nearly all protein complexes in the interface regions.
Supported by these findings, several template-based methods introduced techniques exploiting the observed degeneracy of the interface space. One way to achieve this is to continue using complex templates onto which monomer templates are superimposed but to restrict the structural alignment to the interfacial region [19•]. In the PrePPI algorithm [20••], this is performed by using the structural alignment program Ska [35] which allows structural alignments to be considered significant even if only three secondary structure elements are well aligned. SPRING [21••] uses TM-align [34] to align monomer models with complex templates while the alignment is restricted to the interfacial residues.
An alternative solution is to construct a dedicated interface template library for complex structural modeling. In PRISM [23,46], for instance, a non-redundant interface library is used in association with the structural alignment program MultiProt [36] which is capable of aligning segments in a sequence order independent fashion. In ISEARCH [55], a library of domain-domain interfaces (DDI) was used to scan the surfaces of unbound protein structures for interaction sites similar to a known interface, and to guide the construction of complex models. Similarly, the interaction prediction method iWrap [27] uses a dedicated DDI library, SCOPPI [56], along with associated profiles as constructed by the multiple interface alignment algorithm CMAPi [57], to detect novel protein-protein interactions by threading the query to the interface library.
Vakser and coworkers [19•,29] systematically examined the template-based docking methods, by comparing the results obtained from the structural superposition applied to full monomer structures versus interface regions only, using the structural alignment program TM-align for both cases [34]. The authors found that the interface-based alignment generates more accurate structural models, especially when the template is remotely similar to the target and when one component protein can bind different partners at the same site (e.g. enzyme-inhibitor complexes). It was shown that the best modeling results are obtained when the interface region is defined as atom pairs within 12 Å across the interface [58].
Integration of template-based with non-template-based techniques
When a new algorithm is developed, it is important to test its performance when used on its own. Often, however, different algorithms are complementary to each other, and work better in combination than any of them do by themselves. Therefore, for practical purposes, an integrative approach is often favorable. One notable example is the PrePPI algorithm which combines structural modeling with non-structural features such as protein essentiality, coexpression, functional similarity and phylogenetic profiles using a Bayesian network to predict novel protein-protein interactions [20••]. The accuracy of this approach was found to be comparable to experimental high-throughput methods, with a largely complementary coverage. In a similar spirit, the SPRING algorithm [21••] was extended to utilize high-throughput experimental information to help predict whether two query proteins interact (Guerler, Warner, Zhang, ‘Genome-wide prediction and structural modeling of protein-protein interactions in Escherichia coli’, submitted). Recently, an integrative approach with an even wider scope has been proposed, aiming to combine experimental data from several sources (e.g. electron microscopy images [59]) with structures obtained from comparative modeling and protein-protein docking in order to determine the structure of macromolecular complexes at a resolution that is made possible by the available data [60]. This integrative approach has been successfully applied for the structure determination of several very large complexes [61–62].
The combination of template-based modeling with traditional template-free protein-protein docking is particularly appealing. As illustrated in Figure 1a, protein docking is designed to find the relative orientation of the component chains of a complex from their unbound forms, generally based on the shape complementarity and physico-chemical interactions of the interface atoms. Despite the impressive advances made in the past few years, protein-protein docking is still prone to yield false positive predictions, and tends to fail in particular when there is a large conformational change upon binding, as witnessed by the CAPRI blind prediction experiments [11]. Here, template-based prediction clearly has an advantage in modeling the binding-induced conformational changes, provided that an adequate complex template representing the bound conformation is available. In the absence of an interaction template, however, template-free protein-protein docking seems currently be the only choice.
During the testing of SPRING, a TBM method, comparisons were made with the latest version of the template-free docking algorithm ZDOCK [63] on the docking benchmark 3.0 [64], and it was found that SPRING only outperforms ZDOCK if it is provided with complex templates with a relatively high sequence identity to the query proteins. However, the best modeling result could be achieved when the outputs from ZDOCK and SPRING were combined [21••]. A similar observation was also noted in the benchmarking of the COTH method [18•]. Vreven et al. [39•] recently compared two TBM methods (namely, COTH based on multi-chain threading [18•] and PRISM based on interface structure alignment [23]) with ZDOCK [63]. It was shown that the template-based approaches are better at handling complexes that involve binding-induced conformational changes, and threading-based and docking methods are better for modeling of enzyme–inhibitor complex. While similar overall performance was achieved by the three approaches, correct predictions were generally not shared by the various approaches, suggesting again that the best results can be achieved by combining the different methods. The recent emergence of template-based docking [19•–20••,22•–23,30] represents one way to integrate TBM and docking approaches.
Full-length model construction and complex structure refinement
Most of the current TBM methods identify or construct complex templates for the query proteins but do not provide a complete, refined model of the predicted complex containing full-length structure of both chains, since the sequence and threading alignments often contain gaps with missing residues or loops [18•,21••,24–25]. Even methods using full-length monomer models often do not perform any further refinement to optimize the complex structure [19•–20••,22•–23,30]. The missing regions often include important functional sites which have varying structures among proteins from the same families, and are essential for understanding and annotating the functional differences between the different molecules. However, only a few efforts have been devoted to the important problem of full-length complex structure construction and refinement.
In several methods such as HOMCOS [45,49] and Interactome3D [65•], and HOMBACOP [25], the monomer-based comparative modeling programs MODELLER [66] and NEST [67] are used to build complete complex models. Extensive assembly and refinement for protein-protein complex structures are conducted in both M-TASSER [28] and TACOS, which perform Monte Carlo simulations to reassemble the threading fragments using a reduced protein model. The TACOS algorithm, in particular, is an extension of the highly successful I-TASSER program [68] for single chain structure prediction, and uses binding site prediction and long-range inter-chain contact and distance restraints from multiple templates to optimize the relative orientation of the component structures. Benchmark tests of these algorithms demonstrated marked improvements over the initial structures derived from the threading templates, i.e. the final full-length models were closer to the experimental structures than the templates.
Genome-scale protein complex structure predictions
Each cellular process involves a large variety of protein-protein interactions constituting a complex network of pathways. A comprehensive understanding and annotation of such networks requires the availability of structures for all involved proteins and interactions. The prediction methods that do not perform extensive refinement and structure optimization can be fast enough to generate such structures on a large scale. Here, we summarize the efforts made on the large-scale applications of the algorithms that are presented in the preceding sections.
As one of the earliest examples, the MULTIPROSPECTOR dimeric threading method was applied to the yeast proteome, yielding 7,321 predicted interactions, comparing favorably to other sequence-based computational interaction prediction methods [69]. Later, Aloy et al. constructed the structures for 42 out of 102 known yeast protein-protein complexes using a homology-based search [30]; the authors recently developed Interactome3D which collects structural information for 12,000 protein-protein interactions in 8 model organisms, associated with the pathway databases [65•]. The PRISM suite, which starts with monomerto- complex structure comparisons, has been applied to structures in the PDB and generated >60,000 putative interactions among 6,170 target proteins [46]. The algorithm has recently been used to assign structural information to interactions on a key cancer and inflammation pathway [70]. Similarly, both HOMBACOP [25] and an earlier method [71] were used to generate homology models that were integrated into the GWIDD database, a complex structure resource containing both experimental and predicted structures for ~25,000 interactions within 771 proteomes [72•]. Notably, Zhang et al recently developed PrePPI which was used to predict 30,000 binary interactions within the yeast, and 300,000 interactions within the human proteome, with the accuracy impressively comparable to highthroughput experimental methods [20••]. Other notable efforts include Coev2Net, whose primary purpose is to assign confidence levels to experimentally found interactions, which has been applied to the human MAPK interactome [47•]; the same group also extended their algorithms (Struct2Net and iWrap) to the interaction predictions within the human, fly, and yeast proteomes [26][27]; using HOMCOS, Fukuhara et al predicted the structures of all yeast heterodimers [49]; and Tyagi et al. [38] recently generated 13,217 interaction predictions between 3,614 human proteins. Most recently, the SPRING method was extended to the interactome of E coli. By integrating the high-throughput experimental data, SPRING generated structural models for 46,033 interactions between 4,280 target proteins; for interaction prediction, this method has a Matthews correlation coefficient higher than either high-throughput experimental methods or pure computational prediction according to tests performed on a benchmark set (Guerler, Warner, Zhang, submitted). Overall, these remarkable efforts seem to converge in approaching the desirable goal of creating a detailed atlas of protein-protein interactions [3].
Most template-based complex modeling approaches focus on predicting dimer structures, judging that extending the technique to higher oligomers is straightforward. Many of the key molecular machines in the cell are, however, multimolecular complexes, and assembling models for them is essential for their functional annotation. In one of the few attempts, Aloy et al. [30] fully assembled 42 yeast protein complexes that were identified by tandem affinity purification. Multiprotein complexes were computationally assembled from pair-wise complexes by superposition, using electron microscopy images when available to aid in the reconstruction. In addition, models were constructed by this method for many transient complexes that are created by transitory interactions between complexes, which are likely candidates for interactions between biological functional pathways (cross-talk). Sali and coworkers have made significant efforts to construct models for several large macromolecular complexes by integrating comparative modeling with experimental data from cryoEM, X-ray crystallography, chemical cross-linking, and proteomics techniques; but the focus of this modeling is on the low-resolution molecular architecture rather than on the structure of the complexes at atomic resolution [59–62].
Structure-based prediction of whether two proteins interact
Numerous computational methods have been developed to predict whether two proteins interact (reviewed in [3,73–74]). Most of these methods are sequence-based which use various information sources such as orthology, gene co-expression, co-localization, etc. to predict interacting partners. There are also structure-based approaches which deduce the protein interactions using 3D structural templates or structural features. Here, we use the term “structure-based” for a wide-range of approaches utilizing various structure information of target proteins, compared to the term “template-based” which refers specifically to the methods that deduce predictions from a template library.
Several methods infer the existence of an interaction between two proteins simply from the existence of a known complex structure whose chains are homologous to the query proteins [38,71,75]. However, to improve the specificity of the prediction, most of the structure-based approaches also evaluate the interface in a putative complex model, e.g. by using an interfacial potential [21••,24,26–28,44], or scoring the interface by various features (see subsection “Interface evaluation”) [20••,23,25,46–47•]. A recent method, iLoop, predicts interactions based on the presence of structural features such as certain types of loops in the query proteins [76]. Often, these structure-based methods to predict interactions are not used by themselves but are integrated with information from experiments or other types of computational prediction to increase the confidence of the predictions [20••,47•]. It is important to note that the error rate of some experimental protein-protein interaction detections is very high. For example, it was estimated that the yeast two hybrid system, a common method of detecting protein-protein interactions, has a 70% false positive rate, and only 50% of the interactions in the DIP-YEAST database are reliable (see for example, [77]). Training and testing these methods requires gold standard data sets. The construction of a high-confidence negative data set, i.e. with protein pairs that are known not to interact, is of critical importance [78–80].
Concluding remarks
Significant progress has been achieved in the structural modeling of protein-protein interactions, largely due to the rapid blooming of the concept of template-based modeling (TBM) in the past few years. Extending the methods of protein tertiary structure prediction, template-based modeling of complex structures has primarily focused on the detection of homologous templates [24,30]. Structure-based alignment and superposition of monomer and complex structures have proven useful for increasing alignment coverage of homology-based template construction [18•,21••] and for assisting interaction framework detection in templatebased docking [19•–20••,22•–23]. In particular, the structural alignment of the interface regions has been shown to significantly enhance the accuracy of the resulting complex models [21••,29]. Due to the high speed of template identification and the fact that models can be constructed from sequences alone (in contrast to conventional rigid-body docking which starts from unbound monomer structures), TBM methods have achieved impressive success in genome-wide applications for constructing complex models for the interactomes of various organisms [20••,26,47•,65•,69]. When used to predict interactions, some TBM methods perform with accuracy comparable to that of high-throughput experiments [20••] (Guerler, Warner, Zhang, submitted).
Despite the encouraging progress, serious bottlenecks exist in both TBM method development and the high-resolution genome-wide applications. First, the current complex structure library is far from complete in covering the quaternary structure space of nature [51–52], which essentially limits the range of proteins that can be modeled by TBM approaches. Although studies have shown that the interface structure space is close to complete [53••], how to exploit interface similarity to model global quaternary structures remains a largely unsolved issue. Recent data have shown that structural analogs can be found among the solved complex structures in the PDB for all monomer structures, a finding analogous to an earlier claim stating that the PDB library is nearly complete in the tertiary structure space [81–82]; this seems to suggest that the current PDB can provide templates for docking all interactions with known component structures [22•]. However, the accuracy of template-based docking is low (~23% when at least one of the chains has no homologous templates with a sequence identity >40% to the target [22•]), which is probably still due to the low coverage of the quaternary structure space by the template library, i.e. there is no analogous interaction template in the PDB to guide the template-based docking procedure in those failed cases.
Another bottleneck comes from the limited ability of the current TBM methods to detect distant homologous templates. For threading-based methods [18•,21••,24], the query-template alignment accuracy sharply decreases in the twilight-zone region (e.g. a sequence identity <25%), since even alignment methods using advanced profiles or hidden Markov models are still essentially built on a presumed evolutionary relationship between the target and template proteins. At this point, the template-based docking method seems a promising approach to detect non-homologous templates by structural alignment. However, the data resulting from such approaches also demonstrated a somewhat unexpected dependence on homologous templates, i.e. the majority of the successful docking models are for the targets with templates with a sequence identity >40% to the targets [22•,83], which partly reflects the inherent correlation between the evolutionary relationship and the structural similarity between different protein complexes.
Third, we still lack efficient full-length complex structure refinement methods. Currently, the quality of the initial templates essentially dictates the correctness of the final structural models, although local structural improvements have been reported [28]. Combining multiple template alignments with advanced ab initio binding site predictions within extensive fragment reassembly simulations might be a promising avenue for larger scale model refinement.
Overall, while template-based protein complex structure prediction is still in wait for a more complete structure set of protein-protein interactions, the protein interaction oriented structural genomics projects should play an increasing role in enlarging the coverage of quaternary structure space [4]. Forthcoming efforts of computational TBM approaches should focus on increasing the sensitivity of detecting distant homologous and non-homologous templates to maximize the usefulness of the currently available PDB database, while a combination of multiple-chain threading and template-based docking with an emphasis on interface similarity might be a promising direction to go. Meanwhile, efficient methods for full-length complex structure construction and model refinement will be in high demand with the progress of template recognition approaches. Finally, the integration of current modeling approaches with low-resolution structure and proteomics data, together with appropriate validation from high-resolution experimental data, will be essential to increase the usefulness of genome-wide complex structure modeling efforts, especially for systems biology and the functional annotation of protein interactomes.
Highlights.
The most accurate complex models are built by copying close homologous templates
Non-homologous templates can be detected by interface structure comparisons
Complex template libraries are extended by recombining monomeric structures
Template-based modeling is complementary to rigid-body docking approach
Genome-wide complex structure prediction is generated by template-based modeling
Acknowledgments
We are grateful to Jeffrey Brender for critical reading of the manuscript and Srayanta Mukherjee in generating data for Figure 3. This work was supported in part by the National Institute of General Medical Sciences (GM083107, GM084222) and the Hungarian Scientific Research Fund (OTKA K105415).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 3.Mosca R, Pons T, Ceol A, Valencia A, Aloy P. Towards a detailed atlas of protein-protein interactions. Curr Opin Struct Biol. 2013 doi: 10.1016/j.sbi.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Montelione GT. The Protein Structure Initiative: achievements and visions for the future. F1000 Biol Rep. 2012;4:7. doi: 10.3410/B4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein A, Mosca R, Aloy P. Three-dimensional modeling of protein interactions and complexes is going 'omics. Curr Opin Struct Biol. 2011;21:200–208. doi: 10.1016/j.sbi.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J, Murali T, Finley RL., Jr Assigning confidence scores to protein-protein interactions. Methods Mol Biol. 2012;812:161–174. doi: 10.1007/978-1-61779-455-1_9. [DOI] [PubMed] [Google Scholar]
- 9.Vajda S, Camacho CJ. Protein-protein docking: is the glass half-full or half-empty? Trends Biotechnol. 2004;22:110–116. doi: 10.1016/j.tibtech.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Moreira IS, Fernandes PA, Ramos MJ. Protein-protein docking dealing with the unknown. J Comput Chem. 2010;31:317–342. doi: 10.1002/jcc.21276. [DOI] [PubMed] [Google Scholar]
- 11.Janin J. Protein-protein docking tested in blind predictions: the CAPRI experiment. Mol Biosyst. 2010;6:2351–2362. doi: 10.1039/c005060c. [DOI] [PubMed] [Google Scholar]
- 12.Lensink M, Wodak S. Docking and scoring protein interactions: CAPRI 2009. Proteins. 2010 doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 13.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 15.Ginalski K. Comparative modeling for protein structure prediction. Curr Opin Struct Biol. 2006;16:172–177. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y. Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79(Suppl 10):37–58. doi: 10.1002/prot.23177. [DOI] [PubMed] [Google Scholar]
- 18. Mukherjee S, Zhang Y. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;19:955–966. doi: 10.1016/j.str.2011.04.006. This work develops the COTH method for binding-site guided multiple-chain threading. It shows that a combination of tertiary and quaternary template libraries can increase the alignment coverage and interface contact accuracy of complex structure modeling.
- 19. Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein-protein interfaces. Proteins. 2010;78:3235–3241. doi: 10.1002/prot.22812. This paper introduces a template-based docking approach to protein complex structure modeling, where complex structure models are generated by partial, local superposition of the monomer models to interface templates.
- 20. Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. The work introduces a structure-based, protein interaction prediction pipeline (PrePPI) and predicts 30,000 binary interactions in yeast and 300,000 in human, with structure models provided for each interaction.
- 21. Guerler A, Govindarajoo B, Zhang Y. Mapping Monomeric Threading to Protein- Protein Structure Prediction. J Chem Inf Model. 2013;53:717–725. doi: 10.1021/ci300579r. The work introduces a novel template-based complex structure prediction algorithm which uses tertiary threading template alignments to retrieve complex structural framework from associate oligomer proteins. The algorithm allows detection of multiple binding modes from homologous complexes.
- 22. Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proc Natl Acad Sci U S A. 2012;109:9438–9441. doi: 10.1073/pnas.1200678109. By structurally aligning monomer structures with protein complexes, this paper examines the problem of whether structural templates are available in the PDB to model all complexes for which the component structures are known
- 23.Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: an algorithm for the prediction of protein-protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 25.Kundrotas PJ, Lensink MF, Alexov E. Homology-based modeling of 3D structures of protein-protein complexes using alignments of modified sequence profiles. Int J Biol Macromol. 2008;43:198–208. doi: 10.1016/j.ijbiomac.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Singh R, Park D, Xu J, Hosur R, Berger B. Struct2Net: a web service to predict protein-protein interactions using a structure-based approach. Nucleic Acids Res. 2010;38:W508–W515. doi: 10.1093/nar/gkq481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosur R, Xu J, Bienkowska J, Berger B. iWRAP: An interface threading approach with application to prediction of cancer-related protein-protein interactions. J Mol Biol. 2011;405:1295–1310. doi: 10.1016/j.jmb.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Skolnick J. M-TASSER: an algorithm for protein quaternary structure prediction. Biophys J. 2008;94:918–928. doi: 10.1529/biophysj.107.114280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kundrotas PJ, Vakser IA. Global and local structural similarity in protein-protein complexes: Implications for template-based docking. Proteins. 2013 doi: 10.1002/prot.24392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 31.Wu S, Zhang Y. MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72:547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skolnick J, Kihara D, Zhang Y. Development and large scale benchmark testing of the PROSPECTOR 3.0 threading algorithm. Protein. 2004;56:502–518. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- 33.Lu H, Lu L, Skolnick J. Development of unified statistical potentials describing protein-protein interactions. Biophys J. 2003;84:1895–1901. doi: 10.1016/S0006-3495(03)74997-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic. Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrey D, Honig B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 36.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 37.Vakser IA. Low-resolution structural modeling of protein interactome. Curr Opin Struct Biol. 2013 doi: 10.1016/j.sbi.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyagi M, Hashimoto K, Shoemaker BA, Wuchty S, Panchenko AR. Large-scale mapping of human protein interactome using structural complexes. EMBO Rep. 2012;13:266–271. doi: 10.1038/embor.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vreven T, Hwang H, Pierce B, Weng Z. Evaluating template-based and template-free protein–protein complex structure prediction. Briefings in Bioinformatics. 2013 doi: 10.1093/bib/bbt047. in press. This work made a systematic examination of the strengths and weaknesses of the template-free docking method versus two template-based (threading and structure alignment) methods in modeling protein complex structures.
- 40.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Rekha N, Machado SM, Narayanan C, Krupa A, Srinivasan N. Interaction interfaces of protein domains are not topologically equivalent across families within superfamilies: Implications for metabolic and signaling pathways. Proteins. 2005;58:339–353. doi: 10.1002/prot.20319. [DOI] [PubMed] [Google Scholar]
- 42.Aloy P, Pichaud M, Russell RB. Protein complexes: structure prediction challenges for the 21st century. Curr Opin Struct Biol. 2005;15:15–22. doi: 10.1016/j.sbi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 43.Liang SD, Zhang C, Liu S, Zhou YQ. Protein binding site prediction using an empirical scoring function. Nucleic Acids Research. 2006;34:3698–3707. doi: 10.1093/nar/gkl454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aloy P, Russell RB. InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics. 2003;19:161–162. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 45.Fukuhara N, Kawabata T. HOMCOS: a server to predict interacting protein pairs and interacting sites by homology modeling of complex structures. Nucleic Acids Res. 2008;36:W185–W189. doi: 10.1093/nar/gkn218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keskin O, Nussinov R, Gursoy A. PRISM: protein-protein interaction prediction by structural matching. Methods Mol Biol. 2008;484:505–521. doi: 10.1007/978-1-59745-398-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hosur R, Peng J, Vinayagam A, Stelzl U, Xu J, Perrimon N, Bienkowska J, Berger B. A computational framework for boosting confidence in high-throughput protein-protein interaction datasets. Genome Biol. 2012;13:R76. doi: 10.1186/gb-2012-13-8-r76. It proposes a method to assign confidence levels to experimentally determined interactions based on the coevolutionary relationships at protein interfaces
- 48.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci U S A. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukuhara N, Go N, Kawabata T. Prediction of interacting proteins from homology-modeled complex structures using sequence and structure scores. BIOPHYSICS. 2007;3:13–26. doi: 10.2142/biophysics.3.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh R, Xu J, Berger B. Struct2net: integrating structure into protein-protein interaction prediction. Pac Symp Biocomput. 2006:403–414. [PubMed] [Google Scholar]
- 51.Aloy P, Russell RB. Ten thousand interactions for the molecular biologist. Nat Biotechnol. 2004;22:1317–1321. doi: 10.1038/nbt1018. [DOI] [PubMed] [Google Scholar]
- 52.Garma L, Mukherjee S, Mitra P, Zhang Y. How Many Protein-Protein Interactions Types Exist in Nature? PLoS One. 2012;7:e38913. doi: 10.1371/journal.pone.0038913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gao M, Skolnick J. Structural space of protein-protein interfaces is degenerate, close to complete, and highly connected. Proc Natl Acad Sci U S A. 2010;107:22517–22522. doi: 10.1073/pnas.1012820107. Through a sequence-order independent comparison of protein interface structures, this work argues that the library of protein interfaces is close to complete and comprised of roughly 1,000 distinct interface types. Thus, one could in principle exploit the completeness of protein interfaces to predict most dimeric quaternary structures.
- 54. Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc Natl Acad Sci U S A. 2010;107:10896–10901. doi: 10.1073/pnas.1005894107. This work shows that protein-protein interfaces are often significantly conserved across remote structural neighbors.
- 55.Gunther S, May P, Hoppe A, Frommel C, Preissner R. Docking without docking: ISEARCH--prediction of interactions using known interfaces. Proteins. 2007;69:839–844. doi: 10.1002/prot.21746. [DOI] [PubMed] [Google Scholar]
- 56.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: a structural classification of protein-protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pulim V, Berger B, Bienkowska J. Optimal contact map alignment of protein-protein interfaces. Bioinformatics. 2008;24:2324–2328. doi: 10.1093/bioinformatics/btn432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sinha R, Kundrotas PJ, Vakser IA. Protein docking by the interface structure similarity: how much structure is needed? PLoS One. 2012;7:e31349. doi: 10.1371/journal.pone.0031349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lasker K, Velazquez-Muriel JA, Webb BM, Yang Z, Ferrin TE, Sali A. Macromolecular assembly structures by comparative modeling and electron microscopy. Methods Mol Biol. 2012;857:331–350. doi: 10.1007/978-1-61779-588-6_15. [DOI] [PubMed] [Google Scholar]
- 60.Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lasker K, Phillips JL, Russel D, Velazquez-Muriel J, Schneidman-Duhovny D, Tjioe E, Webb B, Schlessinger A, Sali A. Integrative structure modeling of macromolecular assemblies from proteomics data. Mol Cell Proteomics. 2010;9:1689–1702. doi: 10.1074/mcp.R110.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3.0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mosca R, Ceol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Methods. 2013;10:47–53. doi: 10.1038/nmeth.2289. The work introduces a structural resource containing structural information (both experimental and from predictions) for a large number of interactions from a range of interactomes.
- 66.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrey D, Xiang Z, Tang CL, Xie L, Gimpelev M, Mitros T, Soto CS, Goldsmith-Fischman S, Kernytsky A, Schlessinger A, et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53(Suppl 6):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 68.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu L, Arakaki AK, Lu H, Skolnick J. Multimeric threading-based prediction of protein-protein interactions on a genomic scale: application to the Saccharomyces cerevisiae proteome. Genome Res. 2003;13:1146–1154. doi: 10.1101/gr.1145203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guven Maiorov E, Keskin O, Gursoy A, Nussinov R. The structural network of inflammation and cancer: Merits and challenges. Semin Cancer Biol. 2013;23:243–251. doi: 10.1016/j.semcancer.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 71.Kundrotas PJ, Alexov E. Predicting 3D structures of transient protein-protein complexes by homology. Biochim Biophys Acta. 2006;1764:1498–1511. doi: 10.1016/j.bbapap.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 72. Kundrotas PJ, Zhu Z, Vakser IA. GWIDD: a comprehensive resource for genomewide structural modeling of protein-protein interactions. Hum Genomics. 2012;6:7. doi: 10.1186/1479-7364-6-7. GWIDD (Genome Wide Docking Database) combines available experimental data with models built by docking techniques
- 73.Skrabanek L, Saini HK, Bader GD, Enright AJ. Computational prediction of protein-protein interactions. Mol Biotechnol. 2008;38:1–17. doi: 10.1007/s12033-007-0069-2. [DOI] [PubMed] [Google Scholar]
- 74.Lees JG, Heriche JK, Morilla I, Ranea JA, Orengo CA. Systematic computational prediction of protein interaction networks. Phys Biol. 2011;8 doi: 10.1088/1478-3975/8/3/035008. 035008. [DOI] [PubMed] [Google Scholar]
- 75.Hue M, Riffle M, Vert JP, Noble WS. Large-scale prediction of protein-protein interactions from structures. BMC Bioinformatics. 2010;11:144. doi: 10.1186/1471-2105-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Planas-Iglesias J, Bonet J, Garcia-Garcia J, Marin-Lopez MA, Feliu E, Oliva B. Understanding protein-protein interactions using local structural features. J Mol Biol. 2013;425:1210–1224. doi: 10.1016/j.jmb.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 77.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 78.Smialowski P, Pagel P, Wong P, Brauner B, Dunger I, Fobo G, Frishman G, Montrone C, Rattei T, Frishman D, et al. The Negatome database: a reference set of non-interacting protein pairs. Nucleic Acids Res. 2010;38:D540–D544. doi: 10.1093/nar/gkp1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trabuco LG, Betts MJ, Russell RB. Negative protein-protein interaction datasets derived from large-scale two-hybrid experiments. Methods. 2012;58:343–348. doi: 10.1016/j.ymeth.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 80.Chen XW, Jeong JC, Dermyer P. KUPS: constructing datasets of interacting and non-interacting protein pairs with associated attributions. Nucleic Acids Res. 2011;39:D750–D754. doi: 10.1093/nar/gkq943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc. Natl. Acad. Sci. USA. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Skolnick J, Zhou HY, Brylinski M. Further Evidence for the Likely Completeness of the Library of Solved Single Domain Protein Structures. Journal of Physical Chemistry B. 2012;116:6654–6664. doi: 10.1021/jp211052j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kundrotas PJ, Vakser IA, Janin J. Structural templates for modeling homodimers. Protein Sci. 2013 doi: 10.1002/pro.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]