Abstract
Slow motions of proteins modulate electron-transfer rates during the early stages of photosynthesis.
Effective photosynthesis requires the efficient transfer of electrons across biomembranes (1). Yet, many aspects of how protein structure and dynamics control electron transfer—especially in the early stages of photosynthesis—remain only partly understood. On page 747 of this issue, Wang et al. (2) investigate the rate of the initial photosynthetic electron-transfer reaction in wild-type and mutant photosynthetic reaction centers of Rhodobacter sphaeroides. By combining their data with modeling of protein conformational changes in the photosynthetic reaction center, the authors show that protein motions modulate the electron-transfer rate.
The electron donor in the initial electron-transfer reaction of bacterial photosynthesis is a special pair of chlorophylls. When the latter are photoexcited, an electron transfers through a bridging chlorophyll to a pheophytin acceptor within 2 ps (1 ps = 10–12 s) (see the figure). Experimental studies of this process have focused on understanding the speed and high efficiency of the reaction (an electron is transferred for each photon absorbed) (1). Conventional electron-transfer theory does not fully describe this kind of electron-transfer reaction, because the theory assumes that electron transfer is slow compared to the relaxation of the medium (protein and chromophore), and the medium is therefore assumed to be equilibrated before the electron-transfer step. The protein motion in the initial photosynthetic reaction, however, cannot keep up with the pace of charge separation.
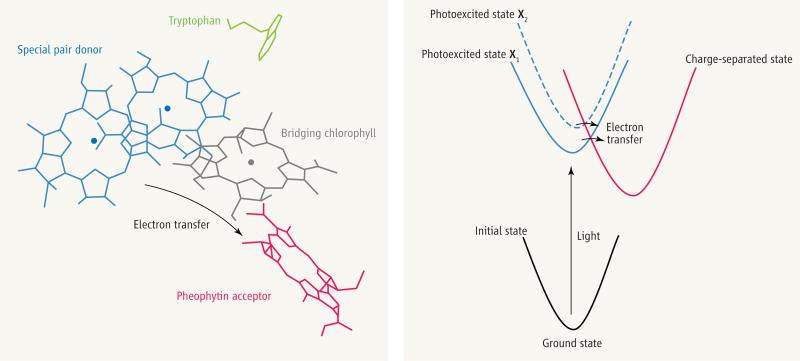
The initial photosynthetic electron-transfer reaction.
(Left) In the photosynthetic reaction center, an electron is transferred from a special pair of chlorophylls via a bridging chlorophyll to the pheophytin acceptor. A nearby tryptophan—1 of 39 in the protein—is also shown. Wang et al. use the time evolution of the protein's tryptophan absorbance to track the protein's response (in the Sumi-Marcus model, motion along the X coordinate) following photoexcitation of the special pair. (Right) Application of the Sumi-Marcus model by Wang et al. (2) to the electron-transfer step shown in the left panel. The parabolas describe the energy of the electron-transfer system as a function of the motions q for the initial state of the system before photoexcitation (black), the photoexcited state before electron transfer (blue), and the final charge-separated state (red).The slow motion X shifts the photoexcited energy surface, thus modulating the speed of the electron-transfer step (horizontal arrows). The colors for the photoexcited and charge-separated energy surfaces correspond to the colors for the electron donor and acceptor moieties in the left panel.
The study of Wang et al. indicates that a model for electron transfer controlled by slow atomic motion, the Sumi-Marcus model (3), produces a satisfactory description of the initial photosynthetic electron-transfer kinetics (2). The Sumi-Marcus model assumes that electron transfer is affected both by fast atomic motions q of the medium that are equilibrated before the electron-transfer step, and by motions X that are slow on the electron-transfer time scale. X modulate the speed of electron transfer. This model is similar in spirit to descriptions of ligand binding to heme proteins, which are also understood in the context of the slow interchange among protein substates (4).
Wang et al. use the absorption spectra of the protein's 39 tryptophan residues to track the medium's response to photoexcitation and initial photosynthetic electron transfer. The spectra are essentially identical in the wild type and in the 14 mutants, with electron-transfer times varying from 2 to tens of ps. Therefore, the protein motion that is tracked by the spectra is not affected by the rate of electron flow. Further, the time evolution of the absorption spectra is multiexponential, with time constants (3, 10, and 190 ps) that are similar to or longer than the time scales of electron transfer.
The authors find that the Sumi-Marcus model describes the observed electron-transfer kinetics if the diffusion constant for the slow coordinate X in the model is derived from the time evolution of the tryptophan absorption. The fit predicts changes in reaction free energy (ΔG) values that are consistent with independent thermo-dynamic data for the mutants, a surprising result given the wide range of electron-transfer rates observed.
It is remarkable that changes in the tryptophan absorption spectra can track reorganizations in the electron-transfer medium that modulate the electron-transfer rate. These reorganizations are not the response of the protein to the arrival of the electron at the pheophytin. Rather, they are probably the protein's response to electron density changes in the donor special pair and the bridging chlorophyll upon photoexcitation. Indeed, the slowness of the protein dynamics on the electron-transfer time scale helps to explain the multiexponential nature of the initial photosynthetic electron-transfer kinetics.
Is the nature of the protein relaxation important for photosynthetic reaction center function? Wang et al. suggest that the slow protein dynamics discussed above may help to overcome reaction barriers produced by membrane potentials or by environmental factors that perturb the photosynthetic reaction center and potentially slow down the electron-transfer rate. Thus, protein motion could overcome reaction barriers produced by cellular factors that might otherwise perturb the electron-transfer kinetics.
The prediction by Wang et al. that slow protein motion modulates electron transfer and generates multiexponential kinetics in the photosynthetic reaction centers should encourage further single-protein studies of biological electron transfer similar to those reported in (5). The emerging physical picture for the photosynthetic reaction center suggests that the temperature dependence for electron transfer should be complex, because temperature variation will change the diffusion among protein substates as well as the access to the activated complex for any given substate. The relation between tryptophan absorption changes and protein reorganizations that affect electron transfer suggests the use of tryptophan transient absorption spectra as a further probe of protein reorganizations linked to protein function.
The experimental data reported by Wang et al. also encourage renewed theoretical attention to the early events in photosynthesis. Models that include quantized nuclear dynamics seem particularly important, because high-frequency quantum modes influence fast electron transfer, producing nonexponential kinetics and unusual temperature dependence (6).
The influence of biomolecular dynamics on electron-transfer rates is not limited to the regime of ultrafast electron-transfer reactions. Recent studies of nonadiabatic electron transfer over long distances show that protein motion also modulates the donor-acceptor electronic coupling interactions (7–9). Furthermore, the internal motions of electron-transfer macromolecules may also change the reaction mechanism from single-step electron tunneling to multistep hopping, a transition that is of great interest and importance—and of some mystery—in the fields of bioenergetics and of DNA damage and repair (10).
Contributor Information
Spiros S. Skourtis, Department of Physics, University of Cyprus, Nicosia 1678, Cyprus. skourtis@ucy.ac.cy
David N. Beratan, French Family Science Center, Departments of Chemistry and Biochemistry, Duke University, Durham, NC 27708, USA. david.beratan@duke.edu
References
- 1.Wakeham MC, Jones MR. Biochem. Soc. Trans. 2005;133:851. doi: 10.1042/BST0330851. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, et al. Science. 2006;316:747. [Google Scholar]
- 3.Sumi H, Marcus RA. J. Chem. Phys. 1986;84:4894. [Google Scholar]
- 4.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 5.Yang H, et al. Science. 2003;302:262. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 6.Jortner J, Bixon M, editors. Electron Transfer—From Isolated Molecules to Biomolecules. 106–107. Wiley; New York: 1999. Advances in Chemical Physics. [Google Scholar]
- 7.Prytkova TR, Kurnikov IV, Beratan DN. Science. 2007;315:622. doi: 10.1126/science.1134862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skourtis SS, Balabin IA, Beratan DN. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3552. doi: 10.1073/pnas.0409047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler KE, et al. J. Am. Chem. Soc. 2007;129:3906. doi: 10.1021/ja067598g. [DOI] [PubMed] [Google Scholar]
- 10.Schuster GB, editor. Topics in Current Chemistry. 236–237. Springer; New York: 2004. Long- Range Charge Transfer in DNA I. [Google Scholar]