SUMMARY
Telomeres are protected from nonhomologous end-joining (NHEJ) to avoid deleterious chromosome fusions, yet they associate with the Ku heterodimer that is principal in the classical NHEJ (c-NHEJ) pathway. T-loops have been proposed to inhibit Ku’s association with telomeric ends, thus inhibiting c-NHEJ; however, deficiencies in the t-loop model suggest additional mechanisms are in effect. We demonstrate that TRF2 interacts with Ku at telomeres and via residues in Ku70 helix 5 (α5), which are vital for NHEJ. We show that Ku’s interaction with a TRF2 mutant that induces telomeric fusions is significantly impaired. Additionally, we demonstrate that Ku70 α5 is required for Ku self-association in live cells, which can bridge DNA ends. Together, these findings lead us to propose a model in which telomeres are directly protected from c-NHEJ via TRF2 impeding Ku’s ability to synapse telomere ends.
INTRODUCTION
Cells continuously suffer DNA double-strand breaks (DSBs) that, if left unrepaired, threaten genomic stability. Nonhomologous end-joining (NHEJ) is the major pathway devoted to the repair of such breaks (Lieber, 2010), operating efficiently throughout the cell cycle, including G1, when homologous recombination, the other major pathway of DSB repair, is restricted (Rothkamm et al., 2003). Simultaneously, the natural ends of linear chromosomes pose potential and ever-present substrates for NHEJ. These ends, however, are protected from engagement by the telomeric nucleoprotein complex. When such protection fails, NHEJ-dependent chromosome end-to-end fusions occur, leading to cessation of cell growth, presumably due to the inability to segregate the resultant multicentric chromosomes at mitosis (Celli and de Lange, 2005).
Two NHEJ pathways have been described, referred to as the classical (or canonical) (c-NHEJ) and alternative (alt-NHEJ) pathways (Mladenov and Iliakis, 2011). One of the factors that distinguishes these pathways is Ku, a heterodimeric complex, which initiates and is required for c-NHEJ and suppresses alt-NHEJ both at DSBs and telomeres (Bombarde et al., 2010; Fattah et al., 2010; Sfeir and de Lange, 2012; Wang et al., 2006). Ku is comprised of the Ku70 and Ku80 subunits, which upon heterodimerization form a high-affinity DNA binding ring that allows Ku to thread onto DNA ends independent of sequence (Walker et al., 2001). Interestingly, Ku is associated with telomeric chromatin across species and has integral roles in telomere structure and function (Fisher and Zakian, 2005). As a result of studies in Saccharomyces cerevisiae, the “two-face” model of Ku was proposed to explain Ku’s paradoxical roles in both NHEJ and at telomeres, where the outward face, which predominantly includes Ku70 residues, is essential for DNA repair and the inward face, which predominantly includes Ku80 subunits, is responsible for telomeric functions (Ribes-Zamora et al., 2007). Ku is essential in human cells, not for its role in NHEJ but rather for its role in inhibiting catastrophic telomere loss, presumably through homology- directed repair (HDR) (Wang et al., 2009). Thus, as functional telomeres are precluded from c-NHEJ, mechanisms must exist that inhibit Ku’s c-NHEJ functions while leaving its protective properties intact.
Mammalian telomeric DNA is comprised of tandem repeats of G-rich sequence and terminates in a 3′ overhang. Telomeric ends are protected by shelterin (Palm and de Lange, 2008), a telomere-specific complex comprised of TRF1, TRF2, Rap1, TIN2, TPP1, and POT1 (Palm and de Lange, 2008). At the forefront is TRF2, which both inhibits the DNA damage response (DDR) at telomeres and plays the dominant role in preventing c-NHEJ, as evidenced by the massive Ku-dependent fusions observed upon loss of TRF2 (Celli et al., 2006; Okamoto et al., 2013). Although inhibition of the DDR is crucial to TRF2-mediated inhibition of telomeric fusions, such fusions are not completely abrogated by the absence of ATM or 53BP1 in TRF2 knockout cells (Denchi and de Lange, 2007; Sfeir and de Lange, 2012). Conversely, telomeres retain some protection from fusions in the presence of TRF2 mutants that have lost the ability to inhibit the DDR (Okamoto et al., 2013). These results indicate that there is a level of inhibition of telomeric NHEJ that is independent of the DDR.
The promotion of t-loop formation by TRF2 is also thought to contribute to the inhibition of c-NHEJ (Griffith et al., 1999). This telomeric architecture, in which the telomere end loops back and invades the upstream duplex telomeric sequence, is proposed to prevent the loading of Ku onto the telomeric end where it must reside to mediate c-NHEJ (de Lange, 2010). However, experimental evidence indicating that t-loops provide a protective function in cells has yet to be demonstrated, and it is unclear whether all telomeres adopt the t-loop structure (Griffith et al., 1999). In addition, t-loops must be resolved to allow for proper telomere replication (Vannier et al., 2012). Ku might be prevented from loading onto the telomere end at that time by the coating of the single-stranded overhang by POT1. However, POT1 has been shown to come off telomeres in G2 at a time when the ends become accessible to modifying enzymes and are transiently recognized as DSBs (Verdun et al., 2005). Given Ku’s association with functional telomeres (Hsu et al., 1999), its high affinity for DNA ends, and its abundance, it seems unlikely to never have access to telomere ends, especially at a point in the cell cycle such as this. Indeed, recent data in S. cerevisiae indicate that Ku must load onto the telomeric end to perform functions required for normal telomere structure and function (Lopez et al., 2011). Therefore, it is likely that shelterin provides an additional continuous mechanism for blocking Ku at functional telomeres.
c-NHEJ is achieved through a series of steps (Lieber, 2010), any of which could be targeted to inhibit the ultimate ligation of telomeric ends. Ku is the first responder in the c-NHEJ pathway (Mari et al., 2006) and, following DNA end-binding, recruits DNA-PKcs to the DSB to form the major kinase regulator of c-NHEJ, the DNA-PK holoenzyme (Gottlieb and Jackson, 1993). DNA-PKcs-binding results in the displacement of Ku inward, along more internal tracks of DNA (Yoo and Dynan, 1999); DNA-PKcs molecules at each end of the break then dimerize to form a synaptic bridge across the DSB that holds the two ends together (DeFazio et al., 2002; Spagnolo et al., 2006). In addition to DNAPKcs and the associated nuclease Artemis, Ku bound to DNA leads to the recruitment of a number of factors utilized in NHEJ including the ligation complex formed by XLF, XRCC4, and DNA ligase IV (Lieber, 2010).
Although current models of c-NHEJ place DNA-PKcs as the major bridging factor between the two ends of DNA (Dobbs et al., 2010; Llorca, 2007), there are some data to indicate a role for Ku as well. Early studies with recombinant Ku indicated that it was able to self-associate in vitro. Ku-Ku interactions were first proposed following atomic force and electron microscopy experiments that demonstrated Ku-mediated DNA looping (Cary et al., 1997) and later supported by coprecipitation of radiolabeled DNA with biotinylated DNA in the presence of recombinant Ku, indicating Ku-Ku interactions could bridge DNA ends (Ramsden and Gellert, 1998). Ku-dependent linking of DNA molecules has also been shown to be promoted in vitro by DNA ligase IV/XRCC4, which is known to stabilize Ku’s association with DNA ends (Zhang et al., 2007). Nonetheless, Ku heterotetramers have never been shown in vivo and how Ku-Ku association would occur or whether this association is essential for NHEJ has not been demonstrated. Thus, the putative role of Ku heterotetramerization in bridging DNA ends for NHEJ remains to be elucidated.
Ku has been shown to interact individually with three of the shelterin members, TRF1, TRF2, and Rap1 (Hsu et al., 2000; O’Connor et al., 2004; Song et al., 2000), all of which have been directly implicated in inhibiting telomeric c-NHEJ (Bae and Baumann, 2007; Celli and de Lange, 2005; Martínez et al., 2009; Sarthy et al., 2009). TRF1 and TRF2 anchor the shelterin complex to telomeres via their high affinity for the telomeric sequence (Sfeir and de Lange, 2012); Rap1 is specifically anchored by TRF2 (Li et al., 2000). Although the interaction of Ku with these three shelterin components has been established, little else has been determined about the localization or functional significance of these interactions. As Ku has many binding partners that contribute to its multitude of functions, it is highly plausible that its telomeric protein interactions negatively regulate Ku to maintain genomic stability.
In this study, we sought to determine the role of Ku heterotetramerization in NHEJ and shelterin’s involvement in inhibiting Ku’s NHEJ function at human telomeres. We present data to support a mechanism by which TRF2 directly protects telomere ends from engaging in c-NHEJ. We localize Ku-TRF2 and TRF1 interactions to telomeres. TRF2, but not TRF1 or Rap1, were found to interact with Ku via an essential NHEJ region, Ku70 α5, directly implicating TRF2 in regulating Ku’s ability to initiate c-NHEJ at telomeres. Additionally, Ku70 had a decreased association with the TRF2ΔBΔM mutant, which is known to induce NHEJ-dependent telomeric fusions. Furthermore, we demonstrate that Ku70 α5 mediates Ku heterotetramerization in vivo. Thus, these findings suggest a model of telomere end protection whereby Ku’s ability to initiate NHEJ via synapsing DNA ends is blocked at telomeres by its interaction with TRF2.
RESULTS
Ku70 Localizes to Telomeres when Interacting with TRF1 and TRF2
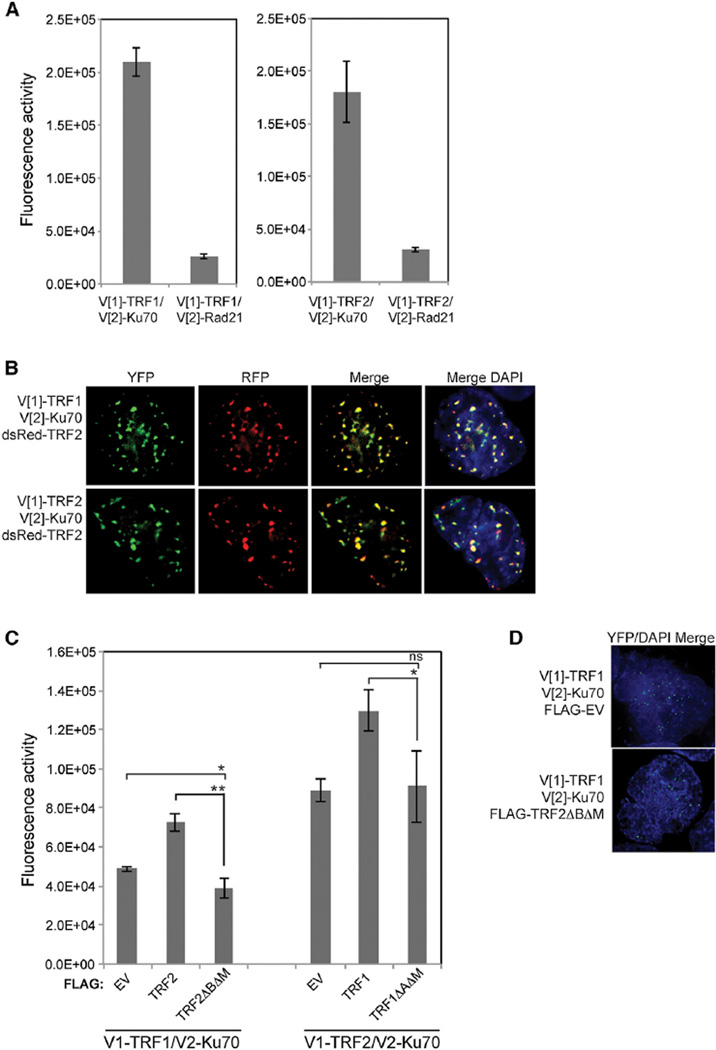
Whereas the interaction of Ku with TRF1 and TRF2 is well-established, the localization of these interactions had not been determined and warranted further assessment as Ku can be found throughout the cell (Muller et al., 2005). Additionally, TRF1 and TRF2 are found to some extent at interstitial telomeric sequences (Martinez et al., 2010; Yang et al., 2011). To analyze the interaction of Ku70 with TRF1 and TRF2 in living cells, we employed fluorescent protein-fragment complementation assays (PCA) using N- (Venus [1] [V[1]]) and C- (V[2]) terminal fragments of the yellow fluorescent protein (YFP) variant Venus (Remy et al., 2004). Whereas the V[1] and V[2] fragments do not come together when expressed alone, when fused to interacting proteins, there is the potential, depending on their proximity and spatial orientation, for them to properly fold together and reform the fluorophore (Remy et al., 2004). Ku70, TRF1, and TRF2 were, therefore, N-terminally tagged with the V[1] or V[2] fragments. As anticipated by the prior interaction studies (Hsu et al., 2000; Song et al., 2000), we found that coexpression of V[2]-Ku70 with either V[1]-TRF1 or V[1]-TRF2 resulted in robust fluorescence (Figure 1A), signifying an interaction. Both proteins were found to produce a weak fluorescence with Rad21, a nonspecific, nuclear protein control (Figure 1A), indicating the specificity of Ku70’s interactions with these shelterin components. Importantly, the YFP fluorescence in the PCAs was not diffuse, as might be expected if the interaction between Ku70 and the TRFs were nonspecific and simply driven by expression levels; instead, the YFP fluorescence manifested as a punctate nuclear signal, suggesting the interaction was occurring at telomeres (Figure 1B). To confirm this, we determined whether the foci colocalized with DsRed-TRF2. Following triple transfection of V[1]-TRF1 or V[1]-TRF2, V[2]-Ku70, and DsRed-TRF2 (Barrientos et al., 2008), we observed co-localization of the V[1]-TRF1/V[2]-Ku70 and the V[1]-TRF2/V[2]-Ku70 signals with the similarly punctate DsRed-TRF2 signals. TRF2-Rap1 interaction was used as a positive control, showing telomeric localization for these two known binding partners in a PCA as expected (Figure S1A).
Figure 1. Ku70 Interacts with TRF1 and TRF2 at Telomeres.
(A) Fluorescence quantification of PCA for Ku70 or Rad21 with either TRF1 or TRF2. Shown are the averages for three independent transfections. Error bars represent SD of the mean.
(B) Colocalization of PCA for Ku70 with TRF1 or TRF2, YFP channel, with the DsRed-TRF2 telomeric marker, RFP channel, and (merge) in HEK293T cells.
(C) Fluorescence quantification of PCA for Ku70 and TRF1 or Ku70 and TRF2 with coexpression of either FLAG-tagged empty vector (EV), TRF1 or TRF2, and TRF1ΔAΔM or TRF2ΔBΔM. p values were determined by Student’s two-tailed unpaired t test with *, p < 0.05; **, p < 0.01; and ns, not significant. For V[1]-TRF1/V[2]-Ku70: p = 0.0345 for EV versus TRF2ΔBΔ Mand p = 0.0011 for TRF2 versus TRF2ΔBΔM. For V[1]-TRF2/V[2]-Ku70: p = 0.8566 for EV versus TRF1ΔAΔM and p = 0.0343 for TRF1 versus TRF1ΔAΔM. The error bars represent SD of the mean.
(D) Colocalization of PCA for Ku70 with TRF1 with coexpression of either FLAG-EV or FLAG-TRF2ΔBΔM (merge) in HEK293T cells.
See also Figure S1.
Although TRF1 and TRF2 do not directly interact, they coexist within the shelterin complex. Therefore, it was possible that the interaction of endogenous TRF2 and the Ku70 PCA construct within shelterin may have positioned the Ku70 protein to allow for fluorescence reconstitution with the TRF1 PCA construct and vice versa with endogenous TRF1 and the Ku70 and TRF2 PCA constructs. To test these possibilities, we examined the Ku-TRF interactions in the presence of dominant negative TRF mutant proteins, TRF2ΔBΔM (van Steensel et al., 1998) and TRF1ΔAΔM (van Steensel and de Lange, 1997), which titrate the respective TRF protein off the telomere. Relative to the coexpression of the vector control, the Ku70-TRF1 fluorescence was 20% lower when TRF2ΔBΔM was coexpressed, whereas the Ku70-TRF2 fluorescence was not affected by coexpression of TRF1ΔAΔM, despite equivalent protein expression levels (Figures 1C and S1B). This could reflect the Ku70-TRF1 PCA fluorescence being partly due to the TRF2 interaction with Ku70. Alternatively, it could be due to a release of Ku70 from TRF1 secondary to the widespread telomere deprotection induced by TRF2ΔBΔM. Although, the Ku70-TRF1 fluorescence was decreased in the presence of the TRF2ΔBΔM mutant, the signal remained punctate (Figure 1D). Expression of the opposing full-length TRF resulted in an increase in Ku70 fluorescence with either TRF1 or TRF2, indicating a stabilizing effect when the opposing TRF is bound to the telomere. Collectively, these results are consistent with Ku70 interacting with both TRF1 and TRF2 at telomeres in vivo.
TRF2, but Not Rap1 or TRF1, Interacts with a Region of Ku70 Required for NHEJ
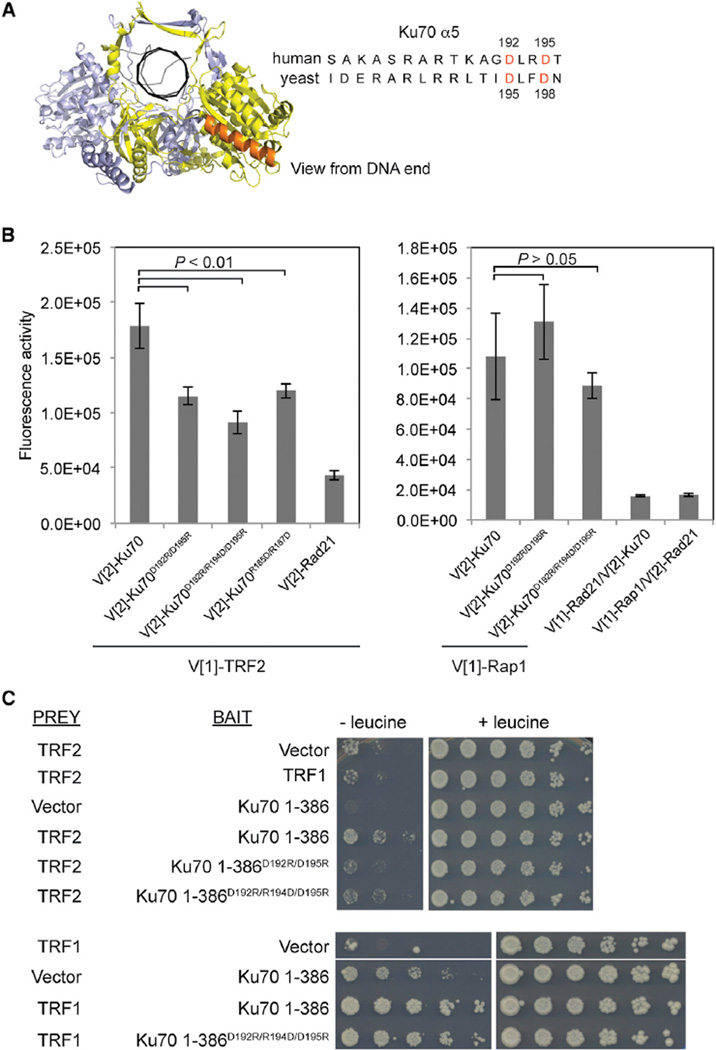
As TRF2, TRF1, and Rap1 have been implicated in the inhibition of NHEJ at telomeres, we considered the possibility that their interactions with Ku70 served as a mechanism to inhibit Ku’s NHEJ function. We therefore turned our attention to the conserved helix 5 in the Ku70 N terminus (Ku70 α5) (Figure 2A), which has been shown to be essential for NHEJ in budding yeast and human cells (Fell and Schild-Poulter, 2012; Ribes-Zamora et al., 2007). Consistent with previous reports, we found that, despite similar expression levels, transient transfection of Ku70−/− mouse embryonic fibroblast (MEF) cells with a mutant of human Ku70 α5, Ku70D192R/D195R, did not restore colony survival to wild-type levels after gamma irradiation (Figure S2A).
Figure 2. TRF2, but Not Rap1 or TRF1, Interacts with the Ku70 NHEJ Domain.
(A) Left, frontal view of Ku’s outward face. Ku70 and Ku80 are depicted in yellow and blue, respectively; Ku70 α5 in orange; and DNA in black. Right, alignment of human and budding yeast Ku70 residues located in α5. Residues mutagenized in Ku70D192R/D195R are depicted in red.
(B) Fluorescence quantification of PCA for the designated Ku70 mutants or Rad21 with either TRF2 or Rap1. Shown are the averages for three independent transfections. Error bars represent SD of the mean. p values were obtained using the Student’s two-tailed unpaired t test. p values for pairs including TRF2 are as follows: p = 0.0072 for Ku70 versus Ku70D192R/D195R, p = 0.0026 for Ku70 versus Ku70D192R/R194D/D195R, and p = 0.0092 for Ku70 versus Ku70R185D/R187D. p values for pairs including Rap1 are as follows: p = 0.3540 for Ku70 versus Ku70D192R/D195R and p = 0.3162 for Ku70 versus Ku70D192R/R194D/D195R.
(C) Yeast two-hybrid analysis for ADH1pLexA-DBD-TRF1 or the designated ADH1pLexA-DBD-Ku70 1–386 alleles (bait) with either GAL1pB42-AD-TRF2 or GAL1pB42-AD -TRF1 (prey). Serial dilutions were grown on Gal-His-Trp-Ura media without (−) or with (+) of leucine. Positive interaction is determined by growth in the absence of leucine.
See also Figure S2.
We therefore asked whether TRF1, TRF2, and/or Rap1 interacted with Ku70 via helix 5. To do this, we used three Ku70 α5 mutants: Ku70D192R/D195R, Ku70D192R/R194D/D195R, and Ku70R185D/R187D. Overall, these mutations impaired the association of Ku70 with TRF2 up to 50% (Figure 2B). As the proteins in this assay were expressed at levels at least comparable to wild-type, the decreased fluorescence could not be attributed to decreased protein expression (Figure S2B). The impact of the Ku70 α5 mutations on Ku70-TRF2 interaction was corroborated by yeast two-hybrid analysis using a Ku70 C-terminal truncation, Ku70 1–386, encompassing the domain that includes helix 5. In this yeast two-hybrid analysis, an interaction between the TRF2 prey and Ku70 1–386 bait was demonstrated by their expression resulting in enhanced growth in the absence of leucine compared to expression of the TRF2 prey alone or the TRF2 prey in the presence of a noninteracting bait (TRF1; Figure 2C, top panel). Both Ku70 mutants exhibited a 5- to 10-fold decreased interaction with TRF2, as evidenced by a reduction in growth in the absence of leucine compared to wild-type Ku70 1–386 with TRF2 (Figures 2C, top panel, and S2C). An enhanced effect of the Ku70 mutants seen in the two-hybrid compared to the PCA are explained by the inherent irreversibility of the YFP fragments folding in the PCA, which we reason lessens the degree of disruption of the Ku70-TRF2 interaction caused by the mutations.
Although Ku70 showed a robust interaction with Rap1, in contrast to the effect on TRF2 interaction, mutations in Ku70 α5 did not impact the Ku70-Rap1 PCA, further arguing for the specificity of the influence of Ku70 α5 on the Ku70-TRF2 interaction (Figures 2B and S2D). Because the results in Figure 1C show that TRF2 may influence the Ku70-TRF1 PCA interaction, we used yeast two-hybrid to analyze the effect of the Ku70 mutants on the TRF1 interaction. The Ku70D192R/R194D/D195R mutant did not show a decrease in interaction with TRF1, despite a robust interaction and comparable protein expression levels (Figures 2C, bottom panel, and S2E). Taken together, these results demonstrate that TRF2, but not Rap1 or TRF1, interacts with a region of Ku70 required for NHEJ.
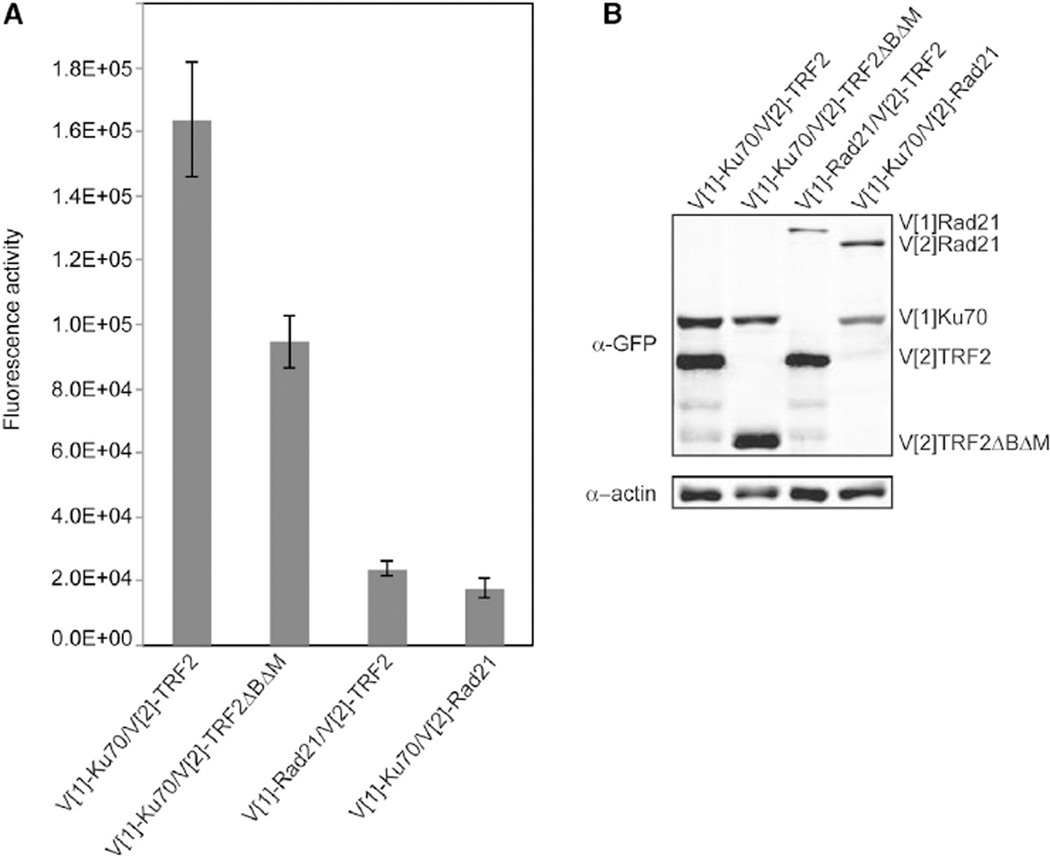
As the binding of TRF2 to Ku70 involved α5, it could provide a mechanism by which c-NHEJ is directly inhibited at telomeres. We next determined whether TRF2ΔBΔM mutant, which leads to c-NHEJ-dependent telomeric fusions (Smogorzewska et al., 2002; van Steensel et al., 1998), is impaired for the interaction. Consistent with this, we found that TRF2ΔBΔM had a 45% lower fluorescence in the PCA with Ku70 as compared to wild-type TRF2 (Figure 3). This opens the possibility that loss of Ku-TRF2 interaction may contribute to the telomere fusion phenotype observed in cells expressing TRF2ΔBΔM.
Figure 3. Ku70 Has Decreased Association with TRF2ΔBΔM.
(A) Fluorescence quantification of PCA for Ku70 and either TRF2 or TRF2ΔBΔM. Rad21 was used as a nonspecific control for interaction. The error bars represent SD of the mean.
(B) Protein levels resulting from transfections in (A) detected by immunoblot analysis of WCE using a GFP antibody. β-actin was used as a loading control.
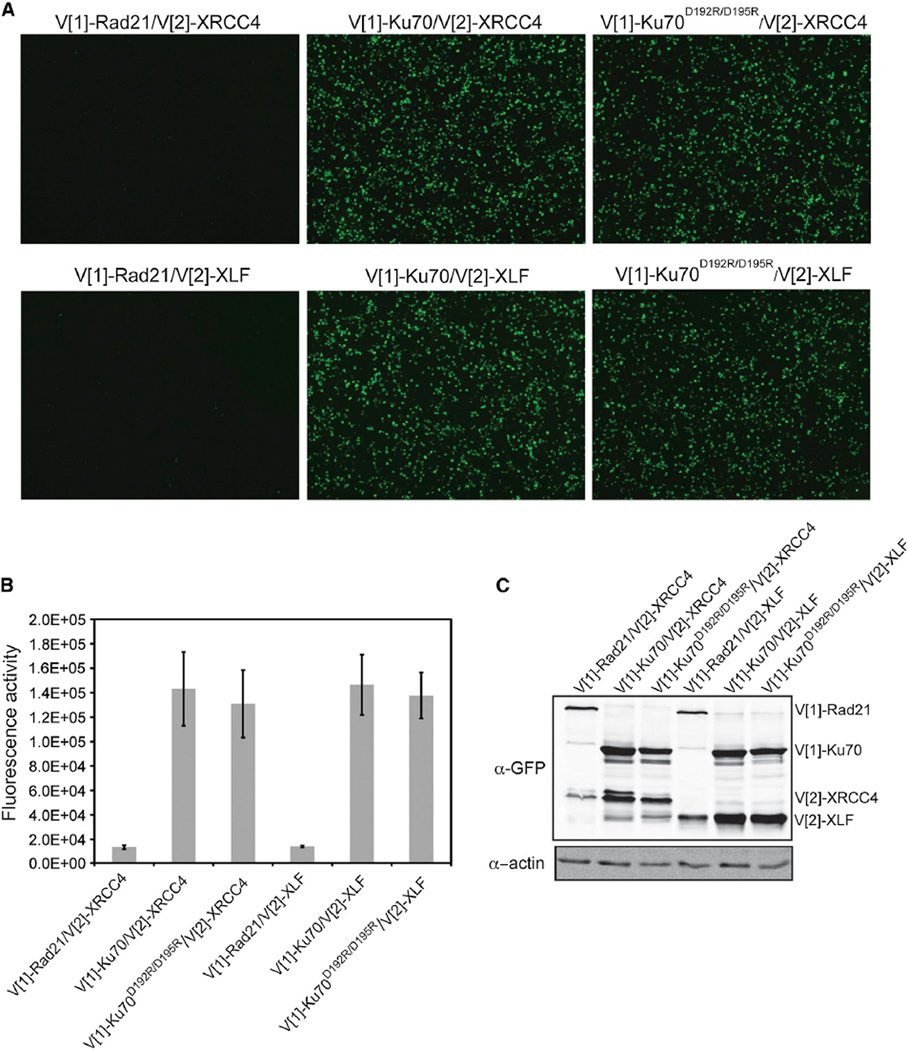
Ku70 α5 Is Not Required for Ku70’s Association with XRCC4 or XLF
Having established that TRF2 interacted with Ku via a region required for NHEJ, we next sought to determine the role of Ku70 α5 in NHEJ. One possibility was that Ku70 α5 is required for Ku’s ability to interact with one of its NHEJ binding partners. Whereas Ku’s interaction with DNA-PKcs has been localized to the Ku80 C-terminal domain (Singleton et al., 1999), the region(s) required for its interactions with key NHEJ factors XRCC4 (Mari et al., 2006) and XLF (Yano et al., 2008) have not been defined. Whereas PCA demonstrated an interaction between Ku70 and XRCC4 and XLF, we found that V[1]-Ku70D192R/D195R was able to restore fluorescence with V[2]-XRCC4 and V[2]-XLF to the same extent as wild-type V[1]-Ku70 (Figure 4); therefore, the radiation sensitivity of Ku70D192R/D195R was not due to the loss of its ability to interact with XRCC4 or XLF. Additionally, the lack of impact of V[1]-Ku70D192R/D195R on fluorescence with V[2]-XRCC4 and V[2]-XLF as compared to wild-type further supports the specificity of the reduction in fluorescence observed when the mutant was expressed with V[2]-TRF2 (Figure 2B).
Figure 4. Ku70 α5 Is Not Required for Ku’s Association with XRCC4 or XLF.
(A) Visualization of PCA using Ku70 or Ku70D192R/D195R with XLF or XRCC4. Images are at 4× magnification.
(B) Fluorescence quantification of PCA shown in (A). The error bars represent SD of the mean.
(C) Protein levels resulting from transfections in (A) detected by immunoblot analysis of WCE using a GFP antibody. β-actin was used as a loading control.
Ku Self-Associates Independently of DNA-PKcs
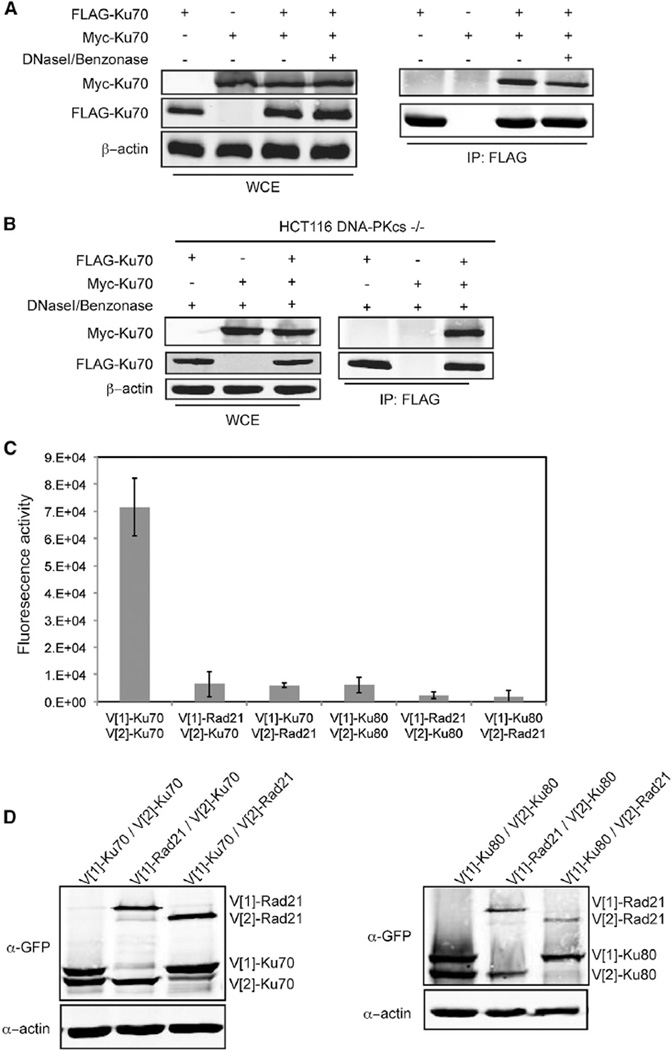
Given the localization of Ku70 α5 to the outward face of Ku as opposed to the face oriented inward along the DNA continuum (Figure 2A), we next considered the possibility that this region contributed to the Ku-Ku interactions that had been previously observed in vitro with recombinant proteins (Cary et al., 1997; Ramsden and Gellert, 1998). First, we wanted to determine whether Ku-Ku interactions could be detected in vivo; therefore, we performed coimmunoprecipitation (coIP) assays using whole-cell extracts (WCEs) prepared from cells cotransfected with differentially tagged versions of Ku70. Consistent with prior studies indicating Ku self-association, we detected Myc-Ku70 in FLAG-Ku70 immunoprecipitates (Figure 5A) and, conversely, FLAG-Ku70 in Myc-Ku70 immunoprecipitates (Figure 6A, middle panel). Similar results were obtained when using differentially tagged versions of Ku80 (Figure S3A). Because Ku bound to DNA forms a complex with DNA-PKcs, which has been implicated in bridging DNA ends (DeFazio et al., 2002), we tested the possibility that the coIP of differentially tagged Ku subunits was the result of DNA-PKcs interactions and not direct Ku interactions. To do this, we repeated the coIP assays in HCT116 DNA-PKcs−/− cells (Ruis et al., 2008). We found that differentially tagged Ku70 subunits associated in the absence of DNA-PKcs (Figure 5B). We also found that the coIP of the differentially tagged Ku70 or Ku80 subunits was not the result of different heterodimers being bound to opposing ends of the same DNA fragment because the interaction was not eliminated by treatment of the WCEs with the combination of DNaseI and Benzonase nuclease prior to immunoprecipitation (Figures 5A, 5B, and S3A). As neither Ku70 nor Ku80 homodimerize, and both are unstable when not heterodimerized (Errami et al., 1996; Gu et al., 1997), these results are consistent with the differentially tagged Ku heterodimers forming multimers.
Figure 5. Ku Heterodimers Self-Associate via the Ku70 Subunit.
(A) Coimmunoprecipitation of Myc-Ku70 with FLAG-Ku70. Immunoprecipitations with FLAG antibody were performed using WCEs untreated (−) or treated (+) with DNaseI/Benzonase. FLAG and Myc immunoblots were performed on the WCEs (left) and immunoprecipitates (IP) (right, IP:FLAG). β-actin represents a loading control.
(B) Coimmunoprecipitation of Myc-Ku70 with FLAG-Ku70 in DNaseI/Benzonase treated WCEs from HCT116 DNA-PKcs−/− cells.
(C) Fluorescence quantification of PCA using the indicated combinations of Ku70, Ku80, or Rad21. The error bars represent SD of the mean.
(D) Protein levels resulting from the transfections in (C) detected by immunoblot analysis of WCEs using a GFP antibody. β-actin was used as a loading control.
See also Figure S3.
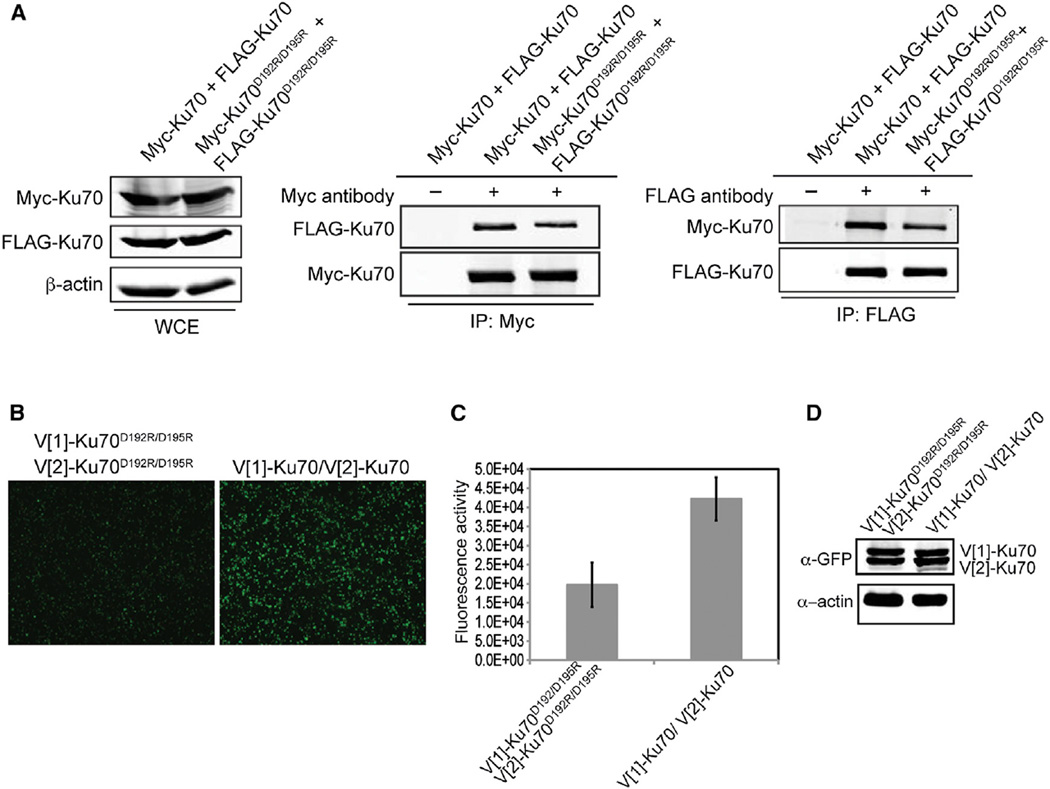
Figure 6. Ku70 α5 Is Required for Heterotetramerization of Ku.
(A) Differentially tagged Ku70 or Ku70D192R/D192 were transiently coexpressed in 293T cells as indicated. Immunoprecipitations with anti-FLAG, anti-Myc, or no antibody (−) were performed. FLAG and Myc immunoblots were performed on the WCEs (left) and the Myc (middle) and FLAG (right) immunoprecipitates (IP). β-actin immunoblot of the WCEs was performed as a loading control.
(B) Visualization of PCA for Ku self-association using either Ku70D192R/D195R or Ku70. Images are at 4× magnification.
(C) Fluorescence quantitation of PCA shown in (B). The error bars represent SD of the mean.
(D) Protein levels resulting from the transient transfections in (A) detected by immunoblot analysis of WCE using a GFP antibody. β-actin was used as a loading control.
See also Figure S4.
To investigate Ku-Ku interactions further in living cells, we performed PCA. We found that transiently transfected V[1]-Ku70 reconstituted fluorescence robustly when coexpressed with either V[2]-Ku80 or V[2]-Ku70 but only weakly with V[2]-Rad21, a nonspecific control (Figures 5C and S3B). In contrast, V[1]-Ku80 did not reconstitute fluorescence with V[2]-Ku80 (Figure 5C). V[1]-Ku80 did, however, strongly reconstitute fluorescence with V [2]-Ku70 (Figure S3B), indicating the functionality of this fusion protein. Reversing V [1] and V[2] fragments in each protein gave comparable results (Figures 5C and S3B). Although there was some variation in steady-state protein levels, each of the proteins was readily detected and the differences in fluorescence intensities could not be attributed to differences in protein levels (Figure 5D). The N termini of Ku70 and Ku80 map to opposite faces of the Ku heterodimer, and Ku binds DNA ends with a polarity that places the Ku70 N terminus facing outward (Walker et al., 2001). Thus, the findings that coexpressed N-terminally tagged Ku70 proteins were able to reconstitute fluorescence in the PCA but coexpressed N-terminally tagged Ku80 could not are consistent with Ku heterodimers interacting with each other in an orientation bringing the outward faces together.
Ku70 α5 Is Required for Heterotetramerization
Because interactions involving the outward faces of two Ku heterodimers might contribute to the synapsis of the two ends of a DSB during NHEJ, we then asked whether Ku70 α5 might contribute to Ku heterotetramerization (i.e., the association of two Ku heterodimers) and thereby facilitate NHEJ. Expression of differentially tagged Ku70D192R/D195R mutants resulted in an approximately 50% reduction in the amount of FLAGKu70D192R/D195R recovered in the Myc-Ku70D192R/D195R immunoprecipitates when compared to wild-type (Figure 6A). Similar reductions were observed in the amount of Myc-Ku70D192R/D195R detected in FLAG-Ku70D192R/D195R immunoprecipitates with respect to wild-type. Importantly, expression of Myc-Ku70D192R/D195R pulled down wild-type levels of FLAG-Ku80, indicating the effect of the mutation was not due to destabilization of the heterodimer (Figure S4A).
Similar to the coIP results, V[1]-Ku70D192R/D195R showed a significant reduction in its ability to reconstitute fluorescence in the PCA with V[2]-Ku70D192R/D195R when compared with wild-type V[1]-Ku70 and V[2]-Ku70 (Figures 6B and 6C) despite comparable steady-state protein levels (Figure 6D). The Ku70D192R/R194D/D195R and Ku70R185D/R187D mutants exhibited a decreased self-interaction in the PCA as well (Figure S4B). Thus, Ku70 α5 is utilized for both heterotetramerization, where the two outward faces in each heterodimer interact with each other, and NHEJ, suggesting NHEJ requires Ku heterotetramerization.
DISCUSSION
A fundamental question regarding telomeres is how they are protected from engagement in c-NHEJ. The t-loop model is simple and elegant; however, it is likely that additional mechanisms exist to confer this protection. Our findings provide evidence to support the hypothesis that TRF2, the primary inhibitor of c-NHEJ at telomeres, directly inhibits c-NHEJ by negatively regulating Ku. Here, we show that TRF2 interacts with Ku via Ku70 α5, a region that maps to Ku’s outward face and is necessary for NHEJ. The PCA method, which requires the split Venus fragments to come into such close proximity to fold together, allowed us to determine that TRF2 and Ku70 interact at telomeres. Furthermore, we show that Ku70 α5 contributes to Ku- Ku association, which has been shown to bridge DNA ends in vitro (Ramsden and Gellert, 1998). These data lead us to propose a model in which TRF2 directly inhibits NHEJ at telomeres by blocking Ku from bridging two telomere ends in preparation for ligation (Figure 7). Whereas Ku may be excluded from the telomeric end via mechanisms promoted by TRF2 (e.g., via t-loop formation), the ability of TRF2 to bind a region of Ku required for Ku-Ku interaction and NHEJ may add an additional layer of protection, particularly should Ku gain access to the end.
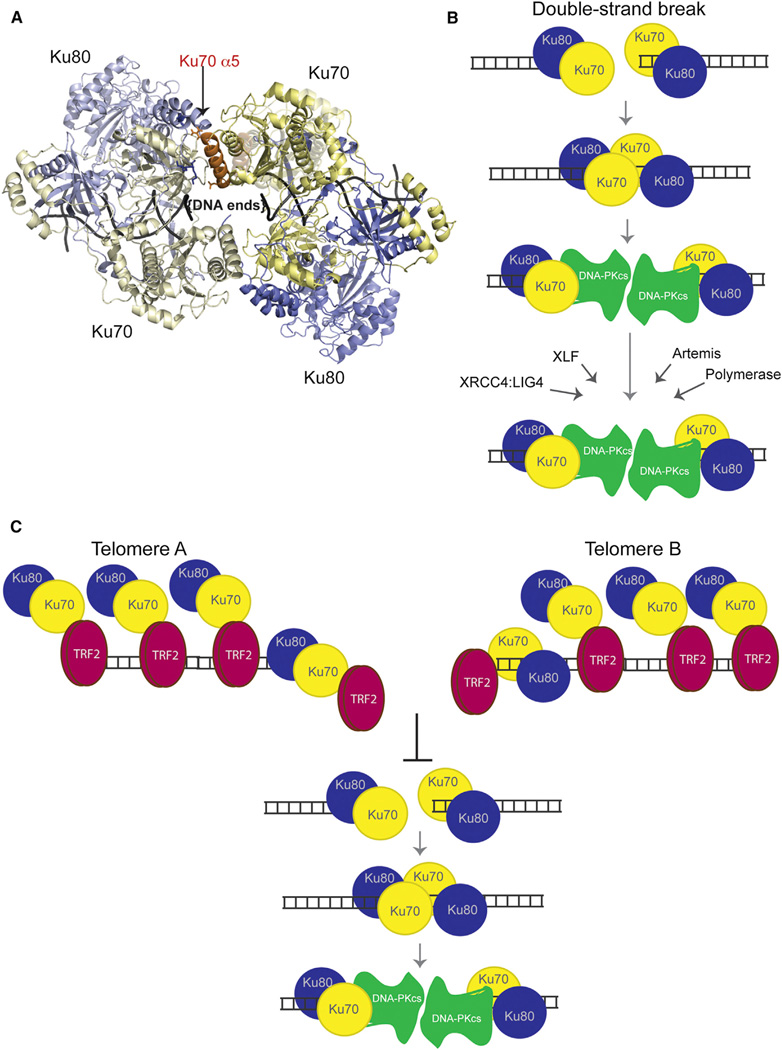
Figure 7. Models for Ku Heterotetramerization and Its Inhibition at Telomeres by TRF2.
(A) Top view of a hypothetical Ku tetramer obtained via protein docking using HADDOCK web server. Yellow: Ku70. Blue: Ku80. Orange: Ku70 α5. Residues R185, D192, and D195 from Ku70 are shown in stick representation. The vWA domain from Ku70 would have to move to accommodate this model of heterotetramerization. Its original position with respect to its native heterodimer is shown in “ghost” (semitransparent) representation.
(B) Mechanistic model for the role of Ku’s heterotetramerization during NHEJ. We propose that Ku heterotetramerization is required in the initial NHEJ steps to synapse the DNA ends of a DSB prior to the DNA-PKcs recruitment. Formation of the DNA-PK complex displaces Ku away from the end, which effectively disassociates the Ku-Ku interaction and allows DNA-PKcs to replace Ku in synapsing the two ends. Later roles of Ku in recruiting NHEJ factors to DSBs would not require heterotetramer formation.
(C) Mechanistic model for the inhibition of Ku’s heterotetramerization by TRF2 at telomeres. We propose that TRF2’s interaction with the Ku70 α5 helix effectively inhibits Ku heterotetramerization at telomeres and the synapsis of telomeric ends, thereby blocking telomeric NHEJ.
We additionally found that the TRF2ΔBΔM dominant-negative mutant has a diminished association with Ku70. As engagement in NHEJ presumably requires single Ku heterodimers bound to each DNA end, then even a moderate release of Ku’s inhibition has the potential to lead to telomere fusions when the ends are accessible. Thus, the catastrophic telomere fusion phenotype seen in this TRF2 mutant may be promoted by the liberation of Ku70’s NHEJ ability. Even though functional telomeres are recognized as DNA damage in G2 (Verdun et al., 2005), TRF2 is still telomeric and presumed capable of inhibiting Ku, which may explain why fusions do not occur at this time. As the Ku70 α5 mutant is defective for NHEJ but presumably proficient for DNA end-binding (Pfingsten et al., 2012), it would be expected to significantly suppress the TRF2ΔBΔM fusion phenotype without allowing alt-NHEJ fusions to occur. The identification of Ku mutants that are impaired for TRF2 binding, but not c-NHEJ, or TRF2 mutants that are still able to localize to telomeres but are defective for Ku70 binding would be valuable for this determination. Such mutants would provide important, direct evidence in support of our proposed model. However, if Ku70 interacts with multiple domains in TRF2, then we anticipate it will be difficult, if not impossible, to identify a separation-offunction mutation that would eliminate Ku70 binding yet preserve TRF2’s interaction with both telomeric DNA and its telomeric binding partners (e.g., Rap1, TIN2, and Apollo). Additionally, our model argues that it might not be possible to identify a separation-of-function allele for Ku where NHEJ is fully functional and TRF2 binding is eliminated if the residues required for TRF2 binding completely overlap with those required for NHEJ.
There are conflicting data as to whether TRF2 itself or its binding partner Rap1 leads to the inhibition of NHEJ (Bae and Baumann, 2007; Martinez et al., 2010; Sarthy et al., 2009; Sfeir et al., 2010). Although the artificial tethering of Rap1 to the telomeric tract in HeLa cells bypassed the requirement of TRF2 for the prevention of telomere fusions (Sarthy et al., 2009), MEFs lacking Rap1 or expressing an allele of TRF2 that cannot bind Rap1 did not result in a DNA damage response or exhibit a telomere fusion phenotype (Martinez et al., 2010; Okamoto et al., 2013; Sfeir et al., 2010). Therefore, Rap1 is dispensable for the protection of telomeres from NHEJ, at the very least, in the mouse model system. Whereas our data suggest a direct role for TRF2 in c-NHEJ inhibition in human cells, we were not able to implicate Rap1, as it did not associate with the NHEJ region of Ku70.
There are also conflicting data for the involvement of TRF1 in the inhibition of NHEJ at telomeres. Whereas a TRF1 knockout mouse model showed notable levels of telomere fusions (Martínez et al., 2009), a conditional TRF1 knockout in MEFs resulted in telomere fusions in only 2% of cells (Sfeir et al., 2009). These two studies also differ in whether the deletion of TRF1 resulted in solely ATR activation or combined ATM/ATR activation, which could further explain the disparities seen in telomere fusions. Our results are consistent with TRF1’s interaction with Ku70 not playing a major role in inhibiting telomeric c-NHEJ in human cells.
In this study, we also address the mechanism of Ku that is specifically blocked at telomeres. We show here that interactions between Ku heterodimers can be detected in living cells. We found that Ku70 α5 mutations disrupted heterotetramer formation and DNA repair, suggesting that Ku heterotetramerization is required for c-NHEJ. Together, our data lead us to propose a model in which Ku heterotetramerization occurs via an interaction of the two outward faces of each Ku heterodimer with Ku70 α5 at the interface (Figure 7A). In the context of a DSB, Ku heterotetramerization would mediate the synapsis of the broken ends (Figures 7A and 7B). Whereas the current mechanistic model for c-NHEJ places DNA-PKcs as the protein responsible for bridging two DNA ends of a DSB (Dobbs et al., 2010; Llorca, 2007), we propose a role for Ku via heterotetramerization, which would occur right after Ku loads onto each end and prior to the recruitment of DNA-PKcs (Figure 7B). Thus, Ku heterotetramerization would play the initial role in restraining the two broken ends. Subsequently, DNA-PKcs recruitment to the Ku-bound DSB would lead to the separation of the heterotetramers and displacement of Ku internally along the DNA. DNA-PKcs would then replace Ku in the task of synapsing the two ends of the DSB. Further c-NHEJ functions of Ku beyond this point, namely recruitment of other c-NHEJ factors, should occur, therefore, in the absence of heterotetramers. This is consistent with our finding that Ku mutations that disrupt heterotetramerization do not disrupt binding with either XRCC4 or XLF.
Although our PCA data do not exclude the possibility that the restored fluorescence for Ku70-Ku70 was caused by one or more proteins bridging the differentially tagged heterodimers, this is unlikely. First, Ku-Ku interactions have been detected in vitro in the absence of other proteins (Cary et al., 1997; Ramsden and Gellert, 1998). Second, our differentially tagged coIP associations were detected in the absence of DNA or DNA-PKcs, two factors known to bridge Ku heterodimers. Third, although the maximal distance between Venus YFP fragments that allows them to come together and fold has not been experimentally determined, the relatively short flexible peptide linkers (ten amino acids [aa] in length) between Ku70 and Venus fragments would indicate that the two Ku70 N termini must allow the Venus fragments to come in very close proximity. For example, the same peptide linker robustly supported dihydrofolate reductase fragments coming together in a PCA if they were separated by 39Å (~10 aa in length) but only weakly if they were separated by 73AÅ (Remy et al., 1999). Therefore, we strongly favor the conclusion that restored fluorescence observed in the Ku70-Ku70 PCA was the result of direct interaction between Ku heterodimers.
If TRF2 interaction with Ku70 can inhibit Ku’s ability to initiate NHEJ, why then would TRF2 inhibit Ku at telomeres and not at DSBs? TRF2 has a high affinity for telomeric repeats and is found to be fully chromatin-bound (Takai et al., 2010), the vast majority being telomeric with additional detection at interstitial sequences (Simonet et al., 2011; Yang et al., 2011). Consistent with a role in telomeric function, we localized Ku70’s interaction with TRF2 to telomeres. Because the TRF2-Ku70 interaction we observed in the PCA was punctate and robust, we speculate that it may be facilitated by TRF2 binding to telomeric DNA. This could also explain why the growth for the yeast two-hybrid TRF2-Ku70 interaction, although present, was not as robust, as it would not recapitulate TRF2 bound to telomeric DNA. In vitro binding experiments with purified proteins in the presence and absence of telomeric repeat-containing DNA are being pursued to address this question.
Importantly, in a prior study, TRF2 was only found to fully block NHEJ in vitro on substrates that contained at least 12 telomeric repeats within 25 nucleotides of the end (Bae and Baumann, 2007). We speculate that this represents a defined threshold of TRF2 molecules needed within the immediate vicinity of a DNA end to block Ku70 α5 from heterotetramerizing. That it is end-associated TRF2’s interaction with Ku70 that is inhibitory reconciles the findings that, although TRF1 also interacts with Ku70, TRF1 itself does not contribute significantly to the protection from c-NHEJ.
Direct inhibition of Ku would provide an additional mechanism by which telomeres are protected from fusions via TRF2. Ku’s interaction with TRF2 may not only inhibit its c-NHEJ function but may additionally promote the execution of its protective telomeric functions, which are to inhibit alt-NHEJ and HDR (Bombarde et al., 2010; Sfeir and de Lange, 2012) and prevent dramatic telomere deletions in human cells, an essential function (Wang et al., 2009). Notably, however, even in the absence of shelterin in mouse cells, Ku protects telomeres from alt-NHEJ indicating that it can mediate this protective capacity independently of shelterin (Sfeir and de Lange, 2012). The future challenges will be to demonstrate that disruption of the direct interaction between Ku70 and TRF2 is sufficient to elicit telomeric NHEJ in vivo, clarify if blocking of Ku’s outward face is only required during telomere replication when telomeres become uncapped, and determine whether Ku’s interactions with TRF2 or other shelterin components promotes its protective functions.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
N-terminal Myc- and/or FLAG-tagged versions of Ku70, Ku80, TRF1, TRF1ΔAΔM, TRF2, and TRF2ΔBΔM were produced by cloning each gene in either CS2-MT or pFLAG-CMV2 vectors. Ku70, Ku80, Rad21, XRCC4, XLF, TRF1, TRF2, and Rap1 were fused at their N terminus with either Venus [1] (V[1]) or Venus [2] (V[2]) fragments by replacing the leucine zipper in either Venus [1]-GCN4-leucine zipper or Venus [2]-GCN4-leucine zipper constructs (provided by S. Michnick). List of plasmids, cloning details, and oligo sequences can be provided upon request. Mutations in Ku70 α5 were generated using oligonucleotide single-stranded mutagenesis. Rap1 was subcloned from pLPC hRap1-FL, a gift from Titia de Lange (Addgene no. 12542). V[2]-TRF2ΔBΔM was generated by PCR of TRF2 and encodes aa 45–455 and contains three additional aa, WRE, at the N terminus.
CoIP Assays
For Myc-Ku70 or Myc-Ku80 immunoprecipitations, 5 × 106 human embryonic kidney 293T cells (HEK293T) or HCT116 DNA-PKcs−/− cells (donated by E. Hendrickson) were cotransfected with plasmids containing FLAG-Ku70 or FLAG-Ku80 using Lipofectamine and Plus Reagents (Invitrogen) according to manufacturer’s protocol. WCEs were prepared with radio immunoprecipitation assay (RIPA) buffer (50 mM Tris HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% Na deoxycholate, 0.1% SDS, and Protease Inhibitor Cocktail III [Calbiochem]) 48 hr after transfection. For each cotransfection, 1.5 mg of WCE was diluted in 500 µl and, where indicated, DNA was removed by adding 10 µl of DNaseI (New England BioLabs), 5 µl of Benzonase nuclease (Novagen), and 2.5 µl of MgCl2 for 1 hr on ice. Extracts were precleared with 150 µl of Protein G Plus-Agarose beads (Calbiochem) for 1 hr at 4°C. After centrifugation and removal of beads, extracts were mixed with 10 µg of α-Myc antibody (Sigma), rotated for 1 hr at 4°C, and then added to 150 µl of Protein G Plus-Agarose beads followed by overnight rotation at 4°C. Beads were pelleted and washed four times with RIPA buffer, and proteins were eluted by heat. The same procedure was followed for FLAG-Ku70 or FLAGKu80 immunoprecipitations but adding 150 µl of FLAG-conjugated agarose beads slurry (Sigma) instead of α-Myc antibody followed by Protein G beads. WCEs and immunoprecipitates were subjected to SDS-PAGE on 10% gels and analyzed by western blotting with either Myc, FLAG, or β-actin (Sigma) primary antibodies and IRDye 800CW-conjugated goat anti-mouse secondary antibody (Li-Cor). Fluorescence was visualized using the Li-Cor Odyssey Infrared Imaging System, and quantitation was performed using ImageQuant software (Molecular Dynamics).
Fluorescent PCA
Plates containing 5 × 105 HEK293T cells were cotransfected with 500 ng of each indicated plasmid, and 48 hr after transfection, cells were washed with PBS and pictures were taken using a magnification of 4× on an Olympus 1×71 fluorescence microscope with 500 ms exposure and ISO200. Deconvolution images were obtained using the same transfection process as above, adding either 200 ng of DsRed-TRF2 (donated by C. Counter) in Figure 1B or 500 ng of FLAG-EV, TRF2, or TRF2ΔBΔM in Figure 1E, with coverslips in the wells. At 48 hr posttransfection, the cells were fixed in 4% paraformaldehyde and mounted in DAPI containing Vectashield mounting medium (Vector Laboratories). All images were taken at 100× magnification unless otherwise specified, using DeltaVision (Deconvolution) Image Restoration Microscope. Each transfection was performed in triplicate, and fluorescence was quantified in 96 well plates containing 1 × 105 cells per well per transfection and using plate reader DTX 800 (Beckman-Coulter) with 485 nm excitation and 535 nm emission filters. Fluorescence was measured for each transfection three times and the fluorescence activity averaged. Remaining cells from each transfection for each V[1]-V[2] combination were pooled and their WCEs subjected to immunoblot analysis using a GFP antibody (Abcam), anti-Ku70 (Lab Vision), anti-TRF2 (Santa Cruz), anti-FLAG (Sigma), or anti-β–actin (Sigma).
Yeast Two-Hybrid Assays
EGY48 was transformed with a prey plasmid containing TRF1 (pAB884) or TRF2 (pAB879) and then a bait plasmid containing either TRF1 (pAB 650), Ku70 1–386 truncation (pAB683), Ku70 1–386D192R/D195R (pAB877), or Ku70 1–386D192R/R194D/D195R (pAB869). Interaction between the bait and prey constructs was determined by growth on Gal-Leu-His-Trp-Ura plates.
Protein Docking
To construct the model shown in Figure 7A, protein docking was performed using HADDOCK web server (Dominguez et al., 2003). All simulations were done in the absence of DNA, which was later modeled into the final complex. The Ku70-Ku80 dimer configuration was taken from the complex deposited in Protein Data Bank as 1JEY (Walker et al., 2001). In the first production run, the von Willebrand antigen (vWA) domain from Ku70 was docked onto the Ku dimer region crystallized with DNA (human Ku70 35–534 and Ku80 6–545 with N-terminal of Ku70 present in the structure) further shortened by six residues. The docking assumed that the flexible tail could move out of the way during interface formation. The region 436–446 in Ku80 was treated as fully flexible in all stages of the docking. The configuration that brought the DNA ends to the smallest distance from each other was then modified by removing vWA Ku70 domain from the dimer, and the remaining construct (dimer without Ku70 vWA + Ku70 vWA from the other dimer docked onto Ku80 residues) was then docked onto itself, using the 5 Å footprint that vWA leaves on the rest of the dimer in the crystallized structure as the docking target, with C2 symmetry imposed on the docked configuration. The rest of the parameters were used at their default values. The resulting heterotetrameric complex with the best overall HADDOCK score is discussed in the text.
See the Extended Experimental Procedures for more information.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Counter (Duke University), E. Hendrickson (University of Minnesota), T. de Lange (Rockefeller University), and S. Michnick (University of Montreal) for sharing reagents; the Dan L. Duncan Cancer Center of Baylor College of Medicine and the Texas Children’s Hospital Cancer and Hematology Centers flow cytometry staff for technical support; and members of the Bertuch lab for useful discussions and comments on the manuscript.
This project was supported by the Integrated Microscopy Core at Baylor College of Medicine with funding from the National Institutes of Health (NIH) grants HD007495, DK56338, and CA125123; the Dan L. Duncan Cancer Center; the John S. Dunn Gulf Coast Consortium for Chemical Genomics; and NIH grants R01GM077509 (to A.A.B.), F31AG034764 (to S.M.I.), and K12GM084897 (to A.R.-Z.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.08.040.
REFERENCES
- Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Barrientos KS, Kendellen MF, Freibaum BD, Armbruster BN, Etheridge KT, Counter CM. Distinct functions of POT1 at telomeres. Mol. Cell. Biol. 2008;28:5251–5264. doi: 10.1128/MCB.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarde O, Boby C, Gomez D, Frit P, Giraud-Panis MJ, Gilson E, Salles B, Calsou P. TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 2010;29:1573–1584. doi: 10.1038/emboj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb. Symp. Quant. Biol. 2010;75:167–177. doi: 10.1101/sqb.2010.75.017. [DOI] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst.) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Errami A, Smider V, Rathmell WK, He DM, Hendrickson EA, Zdzienicka MZ, Chu G. Ku86 defines the genetic defect and restores X-ray resistance and V(D)J recombination to complementation group 5 hamster cell mutants. Mol. Cell. Biol. 1996;16:1519–1526. doi: 10.1128/mcb.16.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell VL, Schild-Poulter C. Ku regulates signaling to DNA damage response pathways through the Ku70 von Willebrand A domain. Mol. Cell. Biol. 2012;32:76–87. doi: 10.1128/MCB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher TS, Zakian VA. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst.) 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Blackburn EH, Chen DJ. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O. Electron microscopy reconstructions of DNA repair complexes. Curr. Opin. Struct. Biol. 2007;17:215–220. doi: 10.1016/j.sbi.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Lopez CR, Ribes-Zamora A, Indiviglio SM, Williams CL, Haricharan S, Bertuch AA. Ku must load directly onto the chromosome end in order to mediate its telomeric functions. PLoS Genet. 2011;7:e1002233. doi: 10.1371/journal.pgen.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, Verkaik NS, Brüggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Carlos AR, Gómez-López G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 2010;12:768–780. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat. Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Muller C, Paupert J, Monferran S, Salles B. The double life of the Ku protein: facing the DNA breaks and the extracellular environment. Cell Cycle. 2005;4:438–441. doi: 10.4161/cc.4.3.1565. [DOI] [PubMed] [Google Scholar]
- O’Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J. Biol. Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pfingsten JS, Goodrich KJ, Taabazuing C, Ouenzar F, Chartrand P, Cech TR. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell. 2012;148:922–932. doi: 10.1016/j.cell.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I, Wilson IA, Michnick SW. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- Remy I, Montmarquette A, Michnick SW. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 2004;6:358–365. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
- Ribes-Zamora A, Mihalek I, Lichtarge O, Bertuch AA. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat. Struct. Mol. Biol. 2007;14:301–307. doi: 10.1038/nsmb1214. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Krüger I, Thompson LH, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruis BL, Fattah KR, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol. Cell. Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet T, Zaragosi LE, Philippe C, Lebrigand K, Schouteden C, Augereau A, Bauwens S, Ye J, Santagostino M, Giulotto E, et al. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res. 2011;21:1028–1038. doi: 10.1038/cr.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton BK, Torres-Arzayus MI, Rottinghaus ST, Taccioli GE, Jeggo PA. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 1999;19:3267–3277. doi: 10.1128/mcb.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- Song K, Jung D, Jung Y, Lee SG, Lee I. Interaction of human Ku70 with TRF2. FEBS Lett. 2000;481:81–85. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J. Biol. Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc. Natl. Acad. Sci. USA. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011;21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.