Abstract
In this paper, we report the development of rod shaped semiconductor nanocrystals (quantum rods) as fluorescent biological labels. Water soluble biocompatible quantum rods have been prepared by surface silanization and applied for non-specific cell tracking as well as specific cellular targeting. Quantum rods are brighter single molecule probes as compared to quantum dots. They have many potential applications as biological labels in situations where their properties offer advantages over quantum dots.
The challenges of biological imaging demand further development of new molecular probes and contrast agents that have better sensitivity, longer stability, good biocompatibility and minimum invasiveness. The convergence of nanotechnology and biotechnology has created many innovations to meet this challenge. A variety of different approaches in making new nanoprobes have been developed in recent years. For example, nanoparticle-based bio-bar codes were reported for ultrasensitive detection of proteins1; noble metal nanoparticles have been reported as molecular rulers based on plasmon coupling2; and magnetic nanocrystals have been shown as effective contrast agents for magnetic resonance imaging3, 4. Among various nanomaterials developed, semiconductor nanocrystals, also known as quantum dots (QDs), represent one of the most successful new biological probes. Compared to conventional organic fluorophores, QDs have advantageous properties, including tunable emission, exceptional photostability, high multiplexing capability and high brightness 5–8. QDs are now commercially available and used in an ever-widening array of biological applications.
The ability to manipulate the shape of semiconductor nanocrystals has led to rod shaped semiconductor nanocrystals, hereafter referred to as “quantum rods” (QRs) 9–11. QRs are semiconductor nanocrystals with diameters ranging from 2 to 10nm and with lengths ranging from 5 to 100nm. In addition to the properties inherited from QDs, such as size-tunable broad absorption, narrow symmetric emission, and extreme resistance to photobleaching, QRs have many unique properties that make them potentially better probes for some biomedical applications than QDs. For example, QRs have larger absorption cross section 12, faster radiative decay rate 13, bigger Stokes shift 9, and can be functionalized with multiple binding moieties. Furthermore, a single quantum rod exhibits linearly polarized emission unlike the plane-polarized light from a single quantum dot 9. The emission of single QRs can be reversibly switched on-off by external electric fields 14. These unique properties make QRs highly desirable for certain biological applications and bring new possibilities for biological imaging.
In this paper, we report the use of surface modified CdSe/CdS/ZnS core/shell QRs as a biological label, and demonstrate that QRs can be used in a variety of bio-imaging applications. Further, for single molecule fluorescence imaging, they are much brighter than QDs.
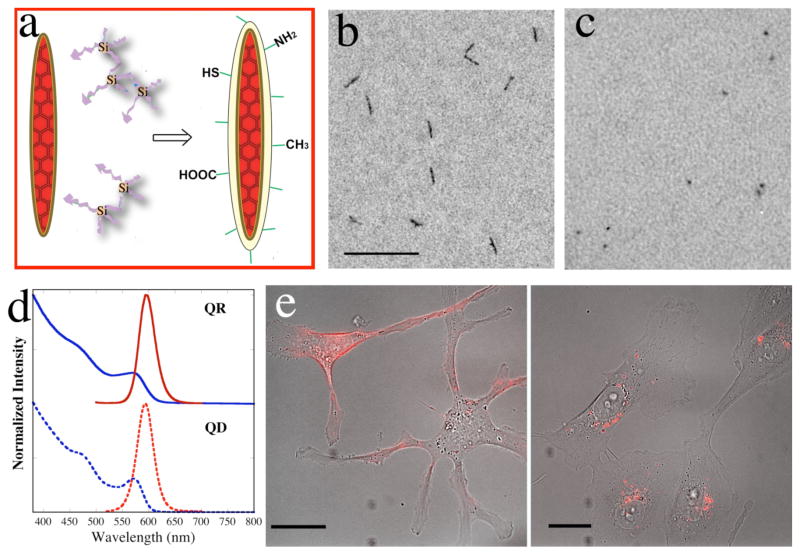
Similar to QDs, high quality QRs as synthesized 10 are only soluble in organic solvents. Hence it is necessary to design a surface coating method to transfer QRs into aqueous solution and render them biofunctional. We developed a coating method for surface silanization of QRs. To overcome the enhanced surface strain from a rod, silane molecules were added in the priming step under a condition that favored condensation (Figure 1a), enabling a well-coated rod surface as compared to single-silane-molecule-priming reported for the silanization of spherical QDs 15. Also, most silanization steps were performed inside a sonicator with temperature control, promoting uniform coating and a highly reproducible process. The silanization procedure thus developed for QRs could be readily applied for making water-soluble QDs and other types of nanoparticles, representing a general method to modify surfaces of nanoparticles. QRs after silanization were stable in aqueous buffer for over 2 years. All experiments in this paper were performed on QRs and QDs going through the same silanization procedure. For comparison purpose, we selected silanized QRs and QDs with the same fluorescence wavelength (emission maxima at 593nm) and the same quantum yield (9%). Transmission electron microscopy (TEM) images of the silanized QRs and QDs are shown in Figure 1b and 1c. Their absorption and emission spectra are shown in Figure 1d.
Figure 1.
(a) A cartoon illustrating silanization of quantum rods. Crosslinked silanes are priming molecules for the surface coating. (b) TEM image of silanized rods in neutral phosphate buffer. Scale bar = 100 nm. (c) TEM image of silanized dots in neutral phosphate buffer. Scale bar = 100 nm. (d) The UV-Vis absorption and emission spectra of silanized QR and QD. The blue curves are the absorption spectra; the red curves are the emission spectra. (e) Silanized QRs are biocompatible and non-toxic to living cells. The red fluorescence in the images is from QRs in human breasts cancer cells MDA-MB-231 after 1h (left) and 24h (right) transfected with Chariot™. These are merged images of transmission and fluorescent micrograms. Scale bar is 20 μm.
QRs after silanization have been proved to be biocompatible. Previously our group demonstrated phagokinetic tracking with QDs 16, 17. When live cells were cultured on a layer of silanized quantum dots, the cells ingested all the dots they passed over, leaving behind a particle free trail which correlates with the metastatic potential of different cell lines 17. Similarly, various live cells could also incorporate silanized QRs as they migrate on a layer of the nanocrystals, without influence on cell division and migration (see Figure S1 of the supporting material). The good biocompatibility of QRs was also evidenced by direct delivery with Chariot™ 18, a peptide non-covalently interacting with QRs and transferring the cargo through the cell membrane (Figure 1e). QRs are accumulated inside intracellular vesicles close to the nucleus over time, with no dramatic difference from the intracellular distribution of QDs 18.
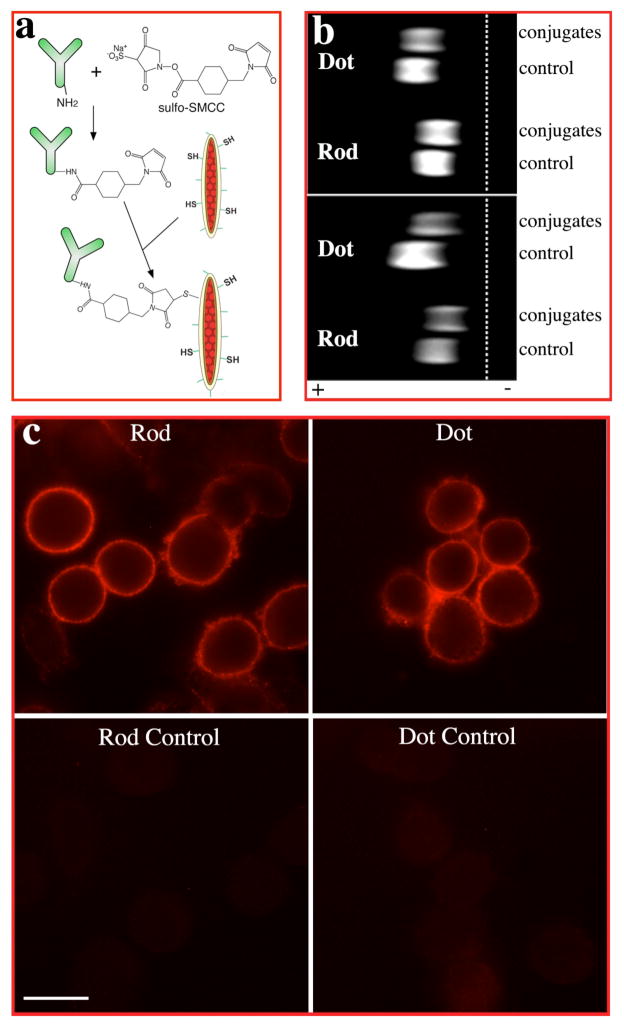
Silanized QRs can be conjugated with various biomolecules through surface amino, mercapto or carboxyl functional groups. Since antibody-antigen affinity is one of the most specific biological interactions and widely used for fluorescence imaging. We tested the conjugation of silanized particles with mercapto surface groups to amino bearing antibodies through a cross linker sulfo-SMCC as schematized in Figure 2a. Conjugation with either whole antibody IgG or antibody fragments were achieved and evidenced by delay of the mobility of conjugates in gel-electrophoresis. To compare the specific cellular labeling efficiency of QRs with QDs, we picked a well-demonstrated system, that is, cancer cell marker Her2 on the surface of human breast cancer cell line SK-BR-319, for specific labeling tests. After incubating the cells with mouse anti-Her2 antibody that binds to the external domain of Her2, we added quantum rod-goat anti-mouse F(ab′)2 and quantum dot-goat anti-mouse F(ab′)2 conjugates with the same OD at 488nm. Specific targeting of the conjugates to cancer marker Her2 was clearly observed in both cases. In ensemble staining experiments, the images obtained with QDs and QRs appear similar, although the same signal intensity can be achieved with a smaller number of QRs.
Figure 2.
(a) Scheme for antibody bioconjugation of quantum rods. (b) Electrophoresis analyses of quantum rods/dots bioconjugation. Top, quantum rods/dots conjugated with F(ab′)2 fragment of goat anti-mouse IgG antibody. Bottom, quantum rods/dots conjugated with whole goat anti-mouse IgG antibody. The conjugates moved slower than the free nanocrystals (control) due to the linkage with antibodies. (c) Immunofluorescence labeling of breast cancer cell marker Her2 on breast cancer cells SK-BR-3. The Her2 marker was labeled with mouse anti-Her2 antibody and goat anti-mouse IgG F(ab′)2 conjugated quantum rods/dots. The bottom images show that there is minimum binding of free nanocrystals to the anti-Her2 antibody treated cells. Scale bar is 20 μm.
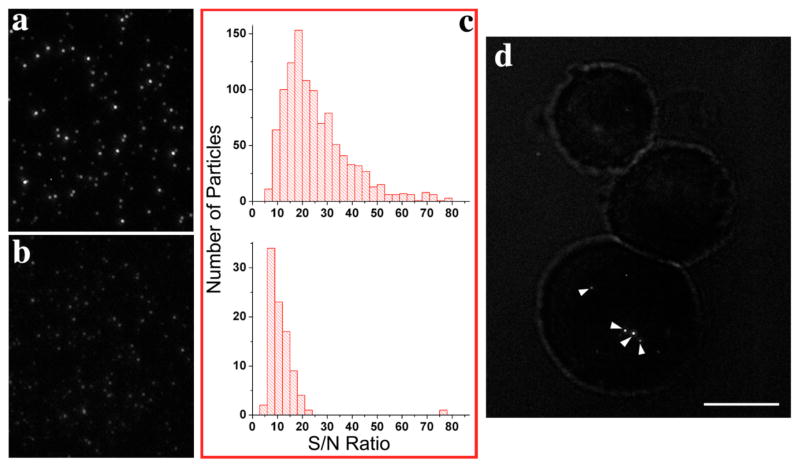
The advantage derived from the enhanced sensitivity of QRs is apparent in single molecule fluorescence imaging, as the signal intensity in ensemble measurement can be improved by increasing the number of labeling particles. The ability to track single molecules is a powerful method to study the dynamic and kinetic behavior of biomolecules inside living cells. Although QDs were shown to be able to image single molecules in living cells 20, 21, the enhanced fluorescence signal from single QR makes them a better probe for single molecule tracking. We compared the fluorescence signals of QRs and QDs at the single molecule level as evidenced by blinking. Under the same excitation and detection conditions, the fluorescence signal of QRs was greatly improved compared to that of QDs (Figure 3a and 3b, also see Movie 1a and 1b of supporting material). To quantitatively compare the fluorescence signals, both rod and dot images were analyzed by automatically collecting fluorescence intensities from a 15-frame image sequence using a self-written Matlab program. Figure 3c shows the histograms of signal to noise ratio (S/N) distribution of both QRs and QDs. The average S/N is 26 for the QRs and 11 for the QDs.
Figure 3.
Fluorescence microscope images show that at the single molecule level, QRs (a) are much brighter than QDs (b). (c) Statistical results of S/N distribution of QRs (top) and QDs (bottom) from 15 image sequences. The mean S/N for single rods is 26, while it is 11 for single dots. (d) Single QRs (indicated by arrows) are still very bright inside live MDA-MB-231 human breast cancer cells. Scale bar is 10 μm.
To demonstrate the ability to detect and track single QRs within living cells, we introduced a small amount of silanized QRs to human breast cancer cell line MDA-MB-231 by the use of streptolysin-O (SLO), a bacterial protein that binds to cholesterol and forms holes in the plasma membrane of animal cells 22. QRs retained their brightness inside living cells (Figure 3d). The tracking at single molecule level was proved by particle blinking (see Movie 2 of the supporting material). Under the same experimental condition, no single QD can be observed inside MDA-MB-231 breast cancer cells.
Although silanization only adds 2 or 3nm of coating thickness to nanocrystals (see Figure. S2 of the supporting material), as rod sizes get bigger, they may interfere with the molecular events that they characterize, hence a balance has to be found between the enhanced brightness of QRs and the disadvantages in terms of their larger size for successful biological applications. However, this should not become an intrinsic limitation, as much bigger particles have been successfully applied in single molecule investigations 23.
The introduction of biocompatible semiconductor QDs in 1998 24, 25 has led to tremendous advances in biotechnologically important applications, including multiplexed in vivo imaging26, 27, long term single molecule tracking20, deep tissue imaging and imaging guided surgery28, as well as hybrid inorganic-bioreceptor based optical sensing 29. In this paper, we have described the development of rod shaped semiconductor nanocrystals for biological imaging. We have overcome the difficulty of rod surface modification and successfully transferred the nanocrystals from organic solvent to biological aqueous solutions by a silanization process. Silanized QRs have good biocompatibility. After further biofunctionization, QRs can be used as immunofluorescent probes. Compared to QDs, QRs are brighter probes, which is demonstrated clearly in single molecule imaging. Other distinct properties of QRs as compared to QDs require further study. These include the possibility of electric field induced switching of the fluorescence and the use of linearly polarized emission to observe orientation. We anticipate biocompatible QRs with properties superior to organic fluorophores and spherical QDs will have a very beneficial impact in many aspects of biomedical imaging and detection schemes.
Supplementary Material
Acknowledgments
This work is supported in part by the DARPA/AFOSR DURINT Program Grant No. F49620-01-1-0474 under subcontract 066995 between the university of Southern California and the University of California Berkeley, and by the NIH National Center for Research Resources through the University of California subaward agreement 0980GFD623 through the U.S. Dept. of Energy under Contract No. DE-AC02-05CH11231, and by the Director, office of Energy Research, Office of Science, Division of Materials Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This work is also funded by the US Department of Energy, Office of Biological and Environmental Research (DE-AC02-05CH11231), the National Center for Research Resources of the National Institutes of Health (P41 RR019664) and the National Institutes of General Medicine, NIH (GM070445).
Footnotes
Supporting Information Available: A detailed information about experimental methods and additional figures and movies. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nam J, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 2.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nature Biotechnology. 2005;23(6):741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 3.Turner JL, Pan D, Plummer R, Chen Z, Whittaker AK, Wooley KL. Advanced Functional Materials. 2005;15(8):1248–1254. [Google Scholar]
- 4.Jun YW, HYM, Choi JS, Lee JH, Song HT, Kim S, Yoon S, Kim KS, Shin JS, Suh JS, Cheon J. J Am Chem Soc. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 5.Alivisatos AP, Gu W, Larabell C. Ann Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 6.Fu A, Gu W, Larabell C, Alivisatos AP. Current Opinion in Neurobiology. 2005;15:568–575. doi: 10.1016/j.conb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nature Materials. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 8.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu JT, Li LS, Yang WD, Manna L, Wang LW, Alivisatos AP. Science. 2001;292(5524):2060–2063. doi: 10.1126/science.1060810. [DOI] [PubMed] [Google Scholar]
- 10.Manna L, Scher EC, Li LS, Alivisatos AP. Journal Of The American Chemical Society. 2002;124(24):7136–7145. doi: 10.1021/ja025946i. [DOI] [PubMed] [Google Scholar]
- 11.Peng X, Manna L, Yang W, Wickham J, Scher E, Kadavanich A, Alivisatos AP. Nature. 2000;404:59–61. doi: 10.1038/35003535. [DOI] [PubMed] [Google Scholar]
- 12.Htoon H, Hollingworth JA, Malko AV, Dickerson R, Klimov VI. Applied Physics Letters. 2003;82(26):4776–4778. [Google Scholar]
- 13.Shabaev A, Efros L. Nano Lett. 2004;4(10):1821–1825. [Google Scholar]
- 14.Rothenberg E, Kazes M, Shaviv E, Banin U. Nano Lett. 2005;5(8):1581–1586. doi: 10.1021/nl051007n. [DOI] [PubMed] [Google Scholar]
- 15.Gerion D, Pinaud F, Williams SC, Parak WJ, Zanchet D, Weiss S, Alivisatos AP. J Phys Chem B. 2001;105:8861–8871. [Google Scholar]
- 16.Parak WJ, Boudreau R, Le Gros M, Gerion D, Zanchet D, Micheel CM, Williams SC, Alivisatos AP, Larabell C. Advanced Materials. 2002;14(12):882–885. [Google Scholar]
- 17.Pellegrino T, Parak WJ, Boudreau R, Le Gros MA, Gerion D, Alivisatos AP, Larabell CA. Differentiation. 2003;71(9–10):542–548. doi: 10.1111/j.1432-0436.2003.07109006.x. [DOI] [PubMed] [Google Scholar]
- 18.Morris MC, Depollier J, Mery J, Heitz F, Divita G. Nature Biotechnology. 2001;19:1173–1176. doi: 10.1038/nbt1201-1173. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Liu H, Liu J, Haley KN, Joseph AT, Larson JP, Ge N, Peale F, Bruchez MP. Natuer Biotechnology. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 20.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Science. 2003;302(5644):442–5. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 21.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Nature Biotechnology. 2004;22(2):198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 22.Giles RV, Grzybowski J, Spiller DG, Tidd DM. Nucleosides & Nucleotides. 1997;16:1155–1163. doi: 10.1080/07328319908044855. [DOI] [PubMed] [Google Scholar]
- 23.Itoh H, Takahashi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 24.Bruchez MJ, Maronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 25.Chan WCW, Nie S. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 26.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. Nature Biotechnology. 2004;22(8):969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 27.Stroh M, Zimmer JP, Duda DG, Levchenko TS, Cohen KS, Brown EB, Scadden DT, Torchilin VP, Bawendi MG, Fukumura D, Jain RK. Nature Medicine. 2005;11(6):678–682. doi: 10.1038/nm1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, et al. Nature Biotechnology. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nature Materials. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.