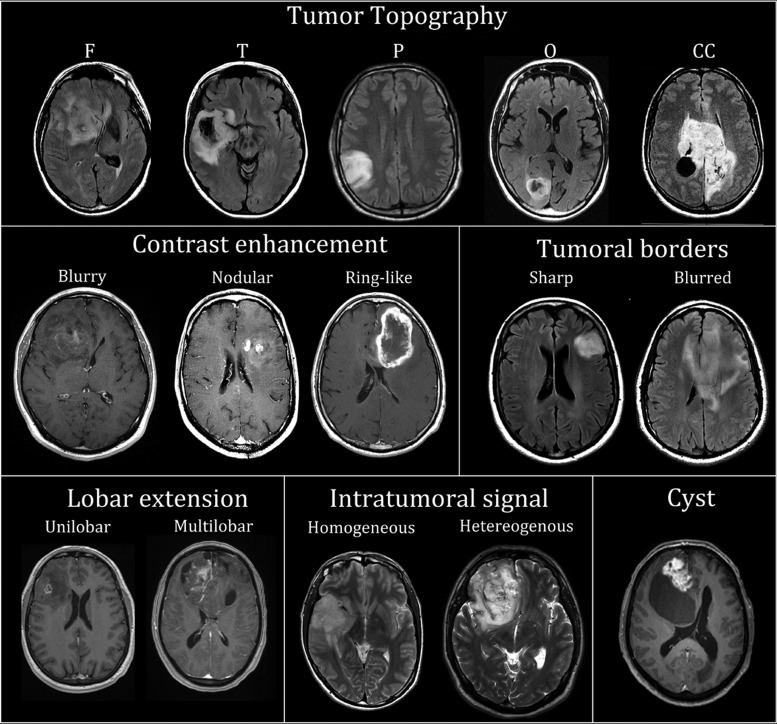
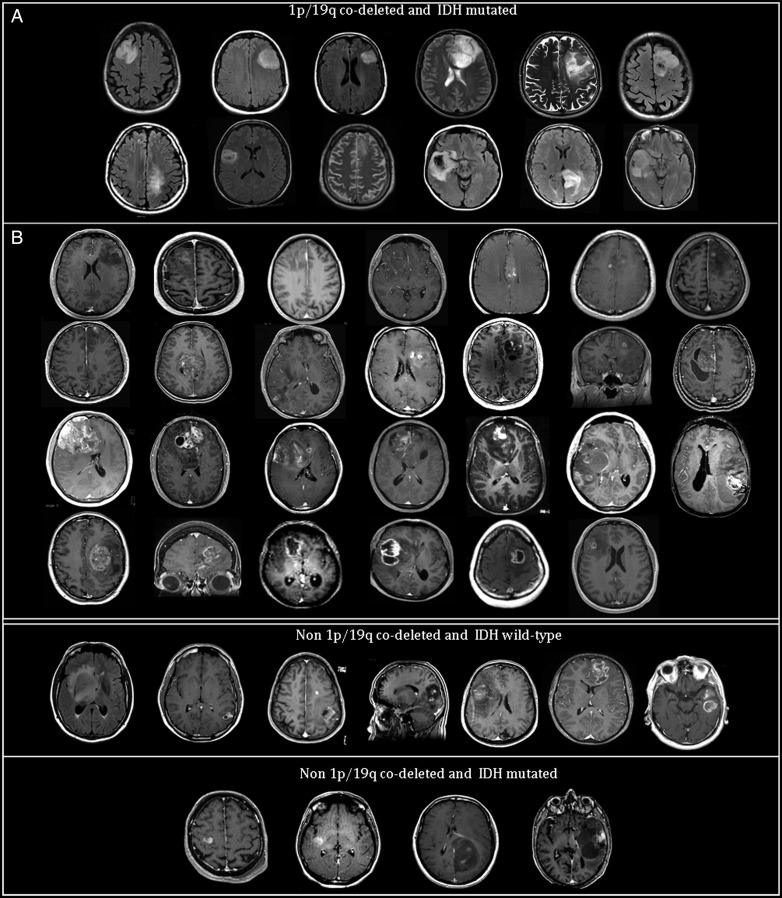
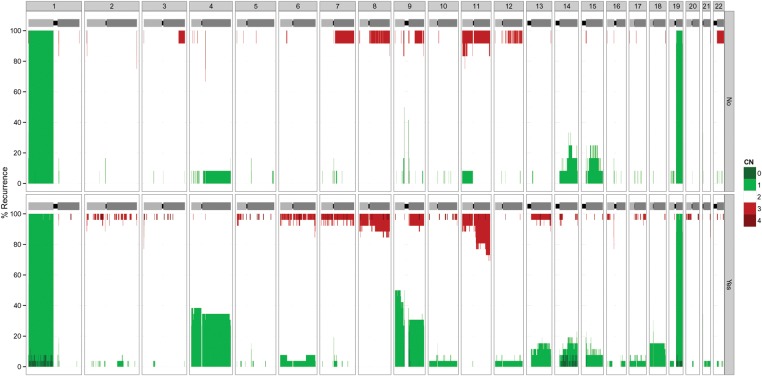
German Reyes-Botero
German Reyes-Botero
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Caroline Dehais
Caroline Dehais
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,✉,
Ahmed Idbaih
Ahmed Idbaih
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Nadine Martin-Duverneuil
Nadine Martin-Duverneuil
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Marion Lahutte
Marion Lahutte
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Catherine Carpentier
Catherine Carpentier
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Eric Letouzé
Eric Letouzé
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Olivier Chinot
Olivier Chinot
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Hugues Loiseau
Hugues Loiseau
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Jerome Honnorat
Jerome Honnorat
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Carole Ramirez
Carole Ramirez
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Elisabeth Moyal
Elisabeth Moyal
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Dominique Figarella-Branger
Dominique Figarella-Branger
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
François Ducray
François Ducray
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1;
POLA Network1,†,
Christine Desenclos
Christine Desenclos
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Henri Sevestre
Henri Sevestre
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Philippe Menei
Philippe Menei
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Sophie Michalak
Sophie Michalak
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Edmond Al Nader
Edmond Al Nader
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Joel Godard
Joel Godard
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Gabriel Viennet
Gabriel Viennet
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Antoine Carpentier
Antoine Carpentier
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Sandrine Eimer
Sandrine Eimer
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Phong Dam-Hieu
Phong Dam-Hieu
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Isabelle Quintin-Roué
Isabelle Quintin-Roué
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Jean-Sebastien Guillamo
Jean-Sebastien Guillamo
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Emmanuelle Lechapt-Zalcman
Emmanuelle Lechapt-Zalcman
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Jean-Louis Kemeny
Jean-Louis Kemeny
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Pierre Verrelle
Pierre Verrelle
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Thierry Faillot
Thierry Faillot
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Claude Gaultier
Claude Gaultier
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Marie Christine Tortel
Marie Christine Tortel
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Christo Christov
Christo Christov
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Caroline Le Guerinel
Caroline Le Guerinel
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Marie-Hélène Aubriot-Lorton
Marie-Hélène Aubriot-Lorton
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Francois Ghiringhelli
Francois Ghiringhelli
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
François Berger
François Berger
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Catherine Lacroix
Catherine Lacroix
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Fabrice Parker
Fabrice Parker
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
François Dubois
François Dubois
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Claude-Alain Maurage
Claude-Alain Maurage
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Edouard-Marcel Gueye
Edouard-Marcel Gueye
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Francois Labrousse
Francois Labrousse
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Anne Jouvet
Anne Jouvet
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Luc Bauchet
Luc Bauchet
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Valérie Rigau
Valérie Rigau
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Patrick Beauchesne
Patrick Beauchesne
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Jean-Michel Vignaud
Jean-Michel Vignaud
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Mario Campone
Mario Campone
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Delphine Loussouarn
Delphine Loussouarn
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Denys Fontaine
Denys Fontaine
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Fanny Vandenbos
Fanny Vandenbos
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Chantal Campello
Chantal Campello
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Pascal Roger
Pascal Roger
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Melanie Fesneau
Melanie Fesneau
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Anne Heitzmann
Anne Heitzmann
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Jean-Yves Delattre
Jean-Yves Delattre
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Selma Elouadhani
Selma Elouadhani
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Karima Mokhtari
Karima Mokhtari
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Marc Polivka
Marc Polivka
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Damien Ricard
Damien Ricard
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Pierre-Marie Levillain
Pierre-Marie Levillain
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Michel Wager
Michel Wager
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Philippe Colin
Philippe Colin
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Marie-Danièle Diebold
Marie-Danièle Diebold
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Dan Chiforeanu
Dan Chiforeanu
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Elodie Vauleon
Elodie Vauleon
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Olivier Langlois
Olivier Langlois
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Annie Laquerriere
Annie Laquerriere
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Marie Janette Motsuo Fotso
Marie Janette Motsuo Fotso
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Michel Peoc'h
Michel Peoc'h
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Marie Andraud
Marie Andraud
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Servane Mouton
Servane Mouton
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Marie-Pierre Chenard
Marie-Pierre Chenard
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Georges Noel
Georges Noel
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Nicolas Desse
Nicolas Desse
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Raoulin Soulard
Raoulin Soulard
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Alexandra Amiel-Benouaich
Alexandra Amiel-Benouaich
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Emmanuelle Uro-Coste
Emmanuelle Uro-Coste
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1,
Frederic Dhermain
Frederic Dhermain
1AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Paris, France (G.R.B., C.D., A.I.); Université Pierre et Marie Curie–Paris 6, Centre de Recherche de l'Institut du Cerveau et de la Moelle Epinière, Paris, France (A.I., C.C.); AP-HP, Groupe Hospitalier Pitié-Salpêtrière, Service de Neuro-radiologie, Paris, France (N.M.-D.); Service de Santé des Armées, Hôpital d'Instruction des Armées, Paris, France (M.L.); Programme Carte d'Identité des Tumeurs, Ligue Nationale Contre le Cancer, Paris, France (E.L.); AP-HM, Hôpital de la Timone, Service de Neuro-Oncologie, Marseille , France (O.C.); CHU Bordeaux, Hôpital Pellegrin, Service de Neurochirurgie, Bordeaux, France (H.L.); Hospices Civils de Lyon, Hôpital Pierre Wertheimer, Service de Neuro-Oncologie, Bron, France (J.H., F.D.); INSERM U1028, CNRS UMR5292, Bron, France (J.H., F.D.); CHU Lille, Hôpital Roger Salengro, Clinique de Neurochirurgie, Lille, France (C.R.); Institut Claudius Regaud, Département de Radiothérapie, Toulouse, France (E.M.); AP-HM, Hôpital de la Timone, Service d'Anatomie Pathologique et de Neuropathologie, Marseille, France (D.F.-B.); Université de la Méditerranée, Aix-Marseille, Faculté de Médecine La Timone, Marseille, France (D.F.B.)
1