Abstract
Cells maintain precise gene expression by balancing transcriptional activation and repression. While much work has focused on elucidating transcriptional activation in the central nervous system (CNS), little is known about transcriptional repression. One means to repress gene expression is to initiate binding of transcription factors to DNA, which then recruit co-repressors as well as other accessory proteins, forming a multi-protein repressor complex. These multi-protein repressor complexes include histone modifying enzymes that trigger processes such as histone acetylation, methylation, and ubiquitylation, altering chromatin structures to impact gene expression. Within these complexes transcriptional repressor proteins per se do not exhibit enzymatic reactions to remodel chromatin structure, whereas histone modifying enzymes lack intrinsic DNA binding activity but have an ability to process post-translational modifications on histones. Thus, the mutual association between transcriptional repressors and histone modifying enzymes is essential to sculpt chromatin to favor transcriptional repression and down regulate gene expression. Additionally, co-repressors are integral components in the context of gene repression as they bridge the association of transcriptional repressors and histone modifying enzymes. In this review, we will discuss the roles of some of the major components of these repressor complex in the CNS as well as their cellular functions that may underlie fundamental behavior in animals.
1. Introduction
Over the past several years studies have started to elucidate the role of transcriptional activators in the central nervous system with many shown to be involved in various aspects of brain function during embryonic and postnatal development, as well as throughout adulthood (Hoch et al., 2009; West and Greenberg, 2011; Wonders and Anderson, 2006). By contrast, the role of transcriptional repressors in the adult nervous system is far less characterized; however, accumulating data indicates they play a critical role in brain function. Mechanisms of transcriptional repression can be divided into three categories; inhibition of the basal transcriptional machinery, ablation of transcriptional activator function, and remodeling of chromatin (Gaston and Jayaraman, 2003). The first category represents the repressors that interfere with the binding of RNA polymerase, TATA box-binding protein, and/or general transcription factors to transcription start sites; thus, preventing transcription initiation. The second mechanism occurs by repressors targeting transcriptional activators and co-activators, resulting in their degradation, nuclear export, or inhibiting their ability to bind DNA. The third mechanism involves recruitment of chromatin remodeling factors, which cause epigenetic alterations on the genome. In addition to the above categories of gene repression, recent studies further establish an emerging role of non-coding RNAs in transcriptional repression by interacting with chromatin modifying factors (Wang et al., 2010).
There has been much interest in investigating the role of individual transcriptional repressors as well as proteins that form part of transcriptional repressor complexes in vivo. In initial studies, constitutive knockout mice were generated to many individual transcriptional repressors as well as proteins that are part of the transcriptional repressor complex, however the loss of these factors often resulted in perinatal, embryonic, or early postnatal lethality precluding in depth characterization of the mice (Perissi et al., 2010). To circumvent this problem, researchers have turned to site-specific recombination technology that allows for targeted gene deletion in a spatially and temporally controlled manner. Conditional knockout mice have started to demonstrate distinct roles of individual transcriptional repressors, as well as for proteins that assemble into repressor complexes, in the adult brain. In this review, we will summarize the current understanding of two of the most extensively studied transcriptional repressors in the brain, namely MeCP2 and REST, as well as individual components of repressor complexes in which they associate, and their impact on primarily adult brain function.
2. Transcriptional repressors
MeCP2
Methyl-CpG-binding protein 2 (MeCP2) is a ubiquitous protein present in both neuronal and non-neuronal tissues and was originally purified from the brain as a heterochromatin protein that binds to DNA containing a single methyl-CpG dinuceotide (Lewis et al., 1992). Subsequently in vitro experiments demonstrated inhibition of transcription from DNA templates in a methylation-dependent manner that occurs through recruitment of a corepressor complex containing Sin3A, histone deacetylase (HDAC)1, and HDAC2 at the target gene promoter; thus, providing the first indication that MeCP2 links two epigenetic mechanisms involved in transcriptional repression, DNA methylation, and histone deacetylation (Jones et al., 1998; Meehan et al., 1992; Nan et al., 1998). Research into the role of MeCP2 in the brain has been further advanced by the identification of mutations in the MECP2 gene in >95% of patients with the neurodevelopmental disorder, Rett syndrome (RTT) (Amir et al., 1999). The disease causing mutations within the MECP2 gene are believed to result in loss of MeCP2 function. Many of the mutations in the MECP2 gene identified in RTT patients are localized within either the DNA binding or transcriptional repression domains, suggesting that loss of MeCP2 activity could alter chromatin architecture by either interfering with the binding to DNA or to the formation of the co-repressor complex, leading to abnormal brain functions and behavioral phenotypes associated with RTT. However, rather intriguingly there are people with MECP2 mutations that do not display the characteristic features of RTT, highlighting the importance of clinical diagnosis for this disorder (Suter et al., 2013).
Rather unexpectedly, data from array comparative genomic hybridization has shown that increased MECP2 copy number can lead to MECP2 duplication syndrome, a neurodevelopmental disorder with some overlapping features of RTT (Van Esch et al., 2005). In order to study the impact of MeCP2 overexpression in the CNS, we recently characterized a mouse line in which MeCP2 overexpression was restricted to neurons and found that these mice recapitulate key behavioral phenotypes of the disorder (Na et al., 2012). We also found that neurons overexpressing MeCP2 have significant reductions in specific aspects of neurotransmission that appear to be due to MeCP2 as a transcriptional repressor. As there have been only a few studies investigating MeCP2 overexpression in animal models (Collins et al., 2004; Na et al., 2012), we will restrict our discussion to MeCP2 loss of function.
To understand the role of MeCP2 loss of function in the brain, several laboratories have generated lines of Mecp2 mutant mice. Constitutive deletion of the Mecp2 gene in mice resulted in a variety of aberrant phenotypes reminiscent of RTT patients, including ataxic gait, hindlimb clasping, and irregular breathing; however, the mice also had a shortened lifespan that prevented a more thorough characterization of them as adults (Chen et al., 2001; Guy et al., 2001; Pelka et al., 2006). Conditional Mecp2 knockout mice, in which the deletion was targeted to specific subsets of neurons, has started to dissect the role of specific neuronal populations in mediating behavioral phenotypes related to RTT, and in some cases has revealed unexpected phenotypes that are presented in atypical RTT patients. For example, our laboratory demonstrated that postnatal deletion of Mecp2 in broad forebrain regions was sufficient to recapitulate some of the core clinical features of RTT, such as impaired motor coordination, heightened anxiety, and deficits in social interaction (Gemelli et al., 2006). In separate work, mice lacking Mecp2 specifically in GABAergic neurons displayed several RTT features including impaired motor coordination, altered social behavior, and deficits in hippocampal-dependent learning and memory (Chao et al., 2010). Additionally, these mice developed stereotypies, self-injury, and compulsive behavior, which are more reminiscent of autistic features but overlap with RTT symptoms. The characterization of additional conditional Mecp2 mouse lines has revealed that some of these lines also recapitulate atypical clinical features, including aggression and obesity, not seen in classical RTT patients (Couvert et al., 2001; Kleefstra et al., 2002). For example, the restricted deletion of Mecp2 in the hypothalamus resulted in increased aggression, hyperphagia, and obesity and was accompanied by an increased physiological response to stress as shown by elevated corticosterone levels in serum (Fyffe et al., 2008). The reader is referred to a recent review by Li and Pozzo-Miller discussing the characterization and phenotypes of Mecp2 mutant mice (Li and Pozzo-Miller, 2012).
While recent work has demonstrated that MeCP2 plays important roles in mediating complex behavior and synaptic function, the molecular mechanisms behind how this seemingly straightforward-acting transcription factor leads to such a wide array of clinical features presented in RTT is still elusive (Chao et al., 2010; Dani et al., 2005; Moretti et al., 2006; Nelson et al., 2006). Given the fact that MeCP2 represses gene transcription in a DNA methylation dependent manner, earlier studies investigated whether the loss of MeCP2 causes genome-wide misregulation of gene expression. Initial attempts to identify MeCP2 target genes from whole-brain gene expression profiling analysis resulted in rather surprisingly, only subtle changes in gene expression and gained little information on the putative genes relevant to RTT (Tudor et al., 2002). Subsequent candidate gene approaches using tissues from specific brain regions have identified promising putative genes regulated by MeCP2, including serum glucocorticoid-inducible kinase, FK506-binding protein 5, corticotropin-releasing hormone, Fxyd1 encoding Na+/K+ ATPase, and protocadherin beta 1, in which MeCP2 binds to methylated promoters and down-regulates their expression (Deng et al., 2007; Miyake et al., 2011; Nuber et al., 2005). Additionally, microarray analysis from hypothalamic RNA revealed down-regulation of a majority of genes in Mecp2 deficient tissue, suggesting that MeCP2 may also function as an activator of gene transcription (Chahrour et al., 2008). Identification of target genes supports the premise that MeCP2 acts at specific gene promoters whose expression changes contribute to overt neurological symptoms seen in RTT patients. Notably, this was contradicted by a recent study demonstrating that MeCP2 binding occurs globally, rather than at specific gene loci, across the genome in the adult brain and that MeCP2 functions to track the density of methylated CpG sites; i.e., MeCP2 binds wherever DNA is methylated (Skene et al., 2010). It is plausible that widespread binding of MeCP2 across the genome could be a critical factor to reduce unnecessary transcriptional noise as well as prevent aberrant transcription from areas such as intergenic regions. Global distribution of MeCP2 in the genome could be a prerequisite condition for activity-dependent neuronal gene transcription. For example, expression of brain-derived-neurotrophic factor (BDNF) is induced upon depolarization of neurons that triggers phosphorylation of MeCP2 at amino acid serine 421 leading to dissociation of MeCP2 from the Bdnf gene promoter (Chen et al., 2003; Martinowich et al., 2003). Thus, neuronal activity may alter MeCP2 occupancy on the chromatin leading to the induction of gene transcription, leading one to postulate that alterations in activity dependent processes may impact MeCP2 dependent gene transcription and underlie aspects of RTT.
A recent study by Li et al indicates a more complex role for MeCP2 in maintaining transcriptional programs, particularly in neurons (Li et al., 2013). Using neurons derived from human embryonic stem cells of RTT patients, unbiased gene expression analysis revealed a drastic decrease in total RNA on a per-cell level, as well as reduction of active transcription in mutant MECP2 neurons. Importantly, this phenomena was coupled to reduced protein translation. This finding suggests that MeCP2 rather facilitates gene transcription, at least in neurons, and supports the previously proposed function of MeCP2 as a transcriptional activator in hypothalamus (Chahrour et al., 2008).
Another important question regarding MeCP2 is whether its loss of function alters epigenetic signatures at target gene promoters or globally in the genome. A study of Mecp2 mutant mice with a stop codon at amino acid 308, Mecp2308, recapitulate many RTT symptoms and demonstrate global hyperacetylation of histones H3 and H4 in the brain (Shahbazian et al., 2002). More recently, Skene et al. confirmed elevated acetylation of histone H3 in neurons but not glia. We also observed increased acetylation of histone H3 following the deletion of Mecp2 specifically in the basolateral amygdala of mice, as well as heightened anxiety and deficits in fear-associated learning and memory (Adachi et al., 2009). Importantly, we were able to demonstrate that chronic infusion of an HDAC inhibitor into the basolateral amygdala produced behavioral deficits similar to those observed in mice with specific knockdown of Mecp2 in this brain region. These studies suggest that MeCP2 potentially shapes epigenetic landscapes through the regulation of HDACs, and that dysfunction of MeCP2 disrupts these processes, which may contribute to the pathophysiology of RTT. The rapid advancements in next generation sequencing technologies such as RNA-Seq and ChIP-Seq will be important to construct epigenetic patterns under normal as well as MeCP2-defective conditions to further investigate these possibilities.
REST
Repressor element-1 silencing transcription factor (REST), also known as neuron-restrictive silencer factor (NRSF), was initially identified as a transcription factor that represses genes containing a repressor element-1/neuron restrictive silencer element (RE1/NRSE) cis-regulatory DNA sequence in their promoters. Earlier investigations revealed predominant expression of REST in non-neuronal tissues and the presence of RE1 in many neuronal genes, such as Nav1.2 sodium channel, SCG10, and synapsin 1, establishing the classical view that REST suppresses neuronal gene in non-neuronal tissues to establish and maintain phenotypic identity of non-neuronal cells (Chong et al., 1995; Schoenherr and Anderson, 1995; Schoenherr et al., 1996). The binding of REST to the RE1 promoter permits the assembly of a corepressor complex containing CoREST, which recruits HDAC1 and HDAC2 (Andres et al., 1999; Ballas et al., 2005). This assembly alters chromatin status by decreasing histone acetylation; thus, favoring a silent transcription state.
In pluripotent stem cells and neural progenitors, REST has been reported to orchestrate a network of genes related to neural development during embryonic and adult neurogenesis (Aoki et al., 2012; Ballas et al., 2005; Covey et al., 2012; Yamada et al., 2010). In undifferentiated neurons, REST expression declines as these cells mature during brain development. Therefore, it was surprising that REST expression is present in the adult brain, including neurons in the hippocampus, pons/medulla, and midbrain (Palm et al., 1998). A couple of recent studies have begun to examine the role of REST in the adult brain. During postnatal development in rodents, one hallmark of synaptogenesis is the developmental switch of NMDA receptor subunit expression from GluN2B to GluN2A, conferring NMDA receptors with different channel properties and kinetics. It was found that at a specific developmental time window, a transient increase in REST expression occurs in the hippocampus leading to increased occupancy of REST at the GluN2B promoter; thus, repressing its gene expression (Rodenas- Ruano et al., 2012). This event correlates with increased epigenetic markers for gene repression at the GluN2B promoter, indicating REST’s ability to repress the target gene via epigenetic remodeling.
Another emerging role of REST in the adult brain is its involvement in ischemia-induced cell death in hippocampal neurons. Ischemic strokes induce not only REST expression in the CA1 subregion of hippocampus, but also enrichment of REST at a discrete subset of genes promoters and leads to subsequent transcriptional repression of genes including those encoding ionotropic glutamate receptors (Noh et al., 2012). Following ischemia, it has been shown the REST-CoREST repressor complex, which also contains MeCP2 and Sin3A, assembles at the Gria2 (AMPA receptor subunit, GluA2) gene promoter. The recruitment of this repressor complex is also accompanied by alterations in epigenetic marks of the GluA2 promoters that represent silencing of gene transcription. It was demonstrated that RNAi-mediated knockdown of Rest in the hippocampus normalized GluA2 expression suppressed by the ischemic strokes and prevented neuronal death. Thus, REST has a variety of functions in the CNS ranging from the regulation of postnatal development to ischemia-associated neurodegeneration, via the remodeling of epigenetic signatures.
In addition to HDAC1 and 2, REST has been co-purified with other chromatin modifiers, such as jumonji AT-rich interactive domain 1C protein (JARID1C)/SMCX histone H3K4 demethylase, suggesting a link between REST-mediated gene repression and tri-methylation of histone H3 (Tahiliani et al., 2007). Co-occupancy of REST and JARID1C/SMCX histone demethylase within the promoters of several REST target genes further supports this notion. Importantly, knockdown of Jarid1C/Smcx leads to decreased tri-methylation of histone H3 at the REST target gene promoters, accompanied by de-repression of their gene expression. In relation to CNS function of REST-JARID1C/SMCX histone demethylase interaction, mutations in the JARID1C/SMCX gene have been found in some patients with X-linked mental retardation. These mutations result in reduced activities of histone demethylase, suggesting that impairment in REST-JARID1C/SMCX mediated chromatin modification and gene repression may underlie the pathophysiology of some mental disorders (Iwase et al., 2007).
3. HDACs and Other Corepressor Proteins
Gene transcription can be modulated by post-translational modifications of amino acids in histone tails. Histone acetylation is a key modulator of chromatin structure and gene transcription and is determined by the balance between histone acetyltransferases (HATs) and histone deacetylases (HDACs) activities. The acetylation of histones occurs at lysine residues of histone tails that relaxes chromatin and allows recruitment of transcription factors; thereby, favoring active gene transcription. In contrast, histone deacetylation is the removal of the acetyl group that results in chromatin compaction and favors transcriptional repression. However, the role of HATs and HDACs is complex in that recent genome wide analyses has mapped the presence of both HATs and HDACs at transcriptionally active loci which are bound by RNA polymerase, indicating tight equilibrium of histone acetylation levels in maintaining gene transcription in non-neuronal cells (Wang et al., 2009). Consistent with this observation, in rodent hippocampus an HDAC inhibitor preferentially, but not globally, targets its action on the transcriptionally active loci (Lopez-Atalaya et al., 2013). In addition to histone modifications some of the HDACs, in particular those that belong to the Class II HDAC family, have been shown to act on many other non-histone substrates in the nucleus, suggesting they have complex roles in impacting gene transcription that is not yet fully understood. While there is little known about the role of individual HATs in the adult CNS, recent data has started to elucidate the role of individual HDACs in the brain.
Inhibition of HDACs
Recent studies have suggested that HDACs may play an important role in learning and memory. A role for HDACs in learning and memory was initially based on the finding that increased histone H3 acetylation in the CA1 subregion of the hippocampus occurs during consolidation of memory formation in the fear conditioning paradigm (Levenson et al., 2004). This study also showed that systemic injection of sodium butyrate, a broad histone deacetylase inhibitor, prior to the fear conditioning enhanced long-term memory formation and long-term potentiation (LTP) at Schaffer-collateral synapses in the CA1 region. In subsequent studies, intrahippocampal infusion of additional HDAC inhibitors, such as Trichostatin A, valproic acid, and suberoylanilide hydroxamic acid (SAHA), were shown to enhance learning and memory in a variety of behavioral tests (Bredy and Barad, 2008; Guan et al., 2009; Haettig et al., 2011; Hawk et al., 2011; Vecsey et al., 2007). Furthermore, increased histone acetylation in response to memory formation has been linked to specific gene promoters, including, Bdnf, Creb, Zif268, and Nr4a orphan nuclear receptor genes (Guan et al., 2009; Hawk et al., 2012; McNulty et al., 2012; Vecsey et al., 2007). These data suggest that inhibition of HDAC activities facilitates learning and memory via remodeling of chromatin landscapes within a limited number of genes. In the following sections, we will discuss the contribution of individual HDACs in the brain with an emphasis on their role in learning and memory.
The HDAC gene family
The mammalian genome encodes eleven individual HDAC genes, which are categorized into three subclasses I, II, and IV, based on amino acid identity and subcellular localization (Haberland et al., 2009; Yang and Seto, 2008). The class I HDAC family consists of HDAC1, 2, 3, and 8 and are expressed predominantly in the nuclei of many cell types. The class II HDAC family consists of HDAC4, 5, 6, 7, 9, and 10. The class IV HDAC family currently consists of only HDAC11, and little is known about its function despite its enrichment in a variety of tissues including the brain (Liu et al 2008). Recent work has only begun to delineate the role of individual HDACs in the adult brain, and these studies have largely concentrated on the class I HDACs in the context of learning and memory (for review see, Morris and Monteggia, 2013), which will be briefly discussed below.
Class I HDACs
The members of the class I HDAC family share high homology in the deacetylase domain; yet, each HDAC member forms a variety of corepressor complexes depending on cellular context. For example, HDAC1 and HDAC2 are often found to interact with one other and associate with Sin3A, CoREST, and the nucleosome remodeling and histone deacetylation (NURD). In contrast, HDAC3 is found in a complex containing nuclear receptor corepressor (NCoR) and silencing mediator of retinoic and thyroid hormone receptor (SMRT). To date, the identity of corepressor complex for HDAC8 is not fully understood.
HDAC1 and HDAC2
HDAC1 and HDAC2 have a high degree of sequence homology (86% identity) and differ only within short stretches of their N- and C-termini, suggesting functional redundancy. However, constitutive deletion of either Hdac1 or Hdac2 in mice resulted in lethality. Constitutive Hdac1 knockout mice displayed severe proliferation defects and general growth retardation, leading to embryonic death (Lagger et al., 2002; Montgomery et al., 2007). Null Hdac2 mice were unable to survive beyond 24 hrs after birth due to severe cardiac malformations (Montgomery et al., 2007; Trivedi et al., 2007). These observations indicate that HDAC1 and HDAC2 possess distinct roles and do not necessarily share full functional redundancy during development despite their high amino acid sequence identity.
To delineate the cellular functions of HDAC1 and HDAC2 in the CNS, we investigated their impact on synaptic transmission using primary hippocampal cultures and found distinct contributions to excitatory neurotransmission that were dependent on the developmental stage of the neurons (Akhtar et al., 2009). In immature hippocampal neurons, loss of both Hdac1 and Hdac2 together, but not individually, resulted in an increase in spontaneous excitatory neurotransmission that was accompanied by increased synapse numbers indicating facilitated maturation of excitatory synapses. In contrast, Hdac2 deletion in mature hippocampal neurons, but not Hdac1, results in a decrease in basal excitatory neurotransmission without alterations in synapse numbers or cell viability. These in vitro data indicate a developmental switch in the functions of HDAC1 and HDAC2 in that HDAC1 and HDAC2 in immature neurons are negative drivers for the maturation of excitatory synaptic networks, whereas HDAC2, but not HDAC1, in mature neurons is necessary to maintain basal neurotransmission.
As discussed earlier, while pharmacological approaches in vivo suggested that HDACs were negative regulators of long term memory formation, these studies do not provide a clear view as to whether individual HDACs contribute different or redundant roles in hippocampal-dependent learning and memory. In a recent work, Guan et al. demonstrated that mice overexpressing a 2–3 fold increase in HDAC2 displayed impaired memory formation in the fear conditioning paradigm and the Morris water maze, while conversely conditional Hdac2 knockout mice had facilitated memory formation in these behavioral tests. The conditional Hdac2 mice were also shown to have increased dendritic spine density and enhanced LTP, suggesting possible correlates to the behavioral enhancement of learning and memory. In contrast, overexpression of HDAC1 did not alter learning and memory or the cellular correlates, suggesting a specificity of HDAC2 to the learning and memory phenotypes. We recently generated conditional Hdac2, as well as Hdac1, single gene knockout mice lacking the respective gene in broad forebrain regions during postnatal development. We tested these mice in an array of behavioral tests to assess whether all types of learning and memory were enhanced in the Hdac2 knockout mice or whether there was specificity to the behavioral effect, as well as further characterizing the conditional Hdac1 knockout mice. We found that conditional Hdac2 knockout mice, but not conditional Hdac1, had enhancements in fear associated learning and memory, as well as in extinction learning and conditioned taste aversion learning (Morris et al., 2013). However, the conditional Hdac2 knockout mice had normal episodic memory in spatial and novel object recognition tasks, suggesting that inhibition of HDAC2 is sufficient to facilitate performances in specific forms of learning and memory. While these data demonstrate that HDAC2 is involved in hippocampal dependent learning and memory, the mechanisms underlying this cognitive enhancement are unknown. The earlier observation that elevated histone H3 acetylation occurred transiently during consolidation of memory implicates a relaxed chromatin structure and subsequent activation of genes (Levenson et al., 2004). It is reasonable to speculate that HDAC2 inhibition, may alter chromatin structure in a similar manner and activate gene expression. Supporting this notion, hyper-acetylation of histones at the Egr1 and Creb promoter regions were documented concomitant with increased protein expression of the respective genes in the hippocampus of Hdac2 knockout mice (Guan et al., 2009). Intriguingly, a recent study demonstrated elevated levels of HDAC2 expression in the CA1 subregion of hippocampus in a mouse model of neurodegeneration, which coincided with increased occupancy of HDAC2 and decreased histone acetylation at promoters of synaptic plasticity related genes, as well as decreases in their mRNA expression (Graff et al., 2012). Importantly, a manipulation to reduce elevated HDAC2 levels normalized the molecular alterations observed in the mouse model of neurodegeneration and alleviated memory deficits. Thus, selective inhibition of HDAC2 may hold promise as a therapeutic target to counteract cognitive impairments presented in neurodegenerative disorders.
HDAC3
The HDAC3 gene is also a member of the class I family although its role in the brain has only recently started to be examined. HDAC3 has been shown to form repressor complexes with two closely related corepressors, nuclear receptor corepressor (NCoR) and the silencing mediator of retinoic acid and thyroid hormone receptors (SMRT) (Yang and Seto, 2008). This contrasts to HDAC1 and HDAC2, which are generally found in Sin3A, CoREST, NuRD-mediated complexes. Thus, it is plausible that differential corepressor association drives specificity of individual HDACs in the class I family. In recent studies, McQuown et al. reported that mice with localized deletion of Hdac3 in the CA1 subregion of the hippocampus displayed enhanced long-term memory in spatial object recognition paradigm, indicating HDAC3 as well may also act as a negative regulator of learning and memory (McQuown et al., 2011). In addition, the localized deletion of Hdac3 resulted in increased acetylation of histone H4 at the lysine 8 residue as well as increased expression of the Nr4a2 gene in the hippocampus. The authors went on to show that intrahippocampal delivery of siRNA targeting the Nr4a2 gene in mice lacking Hdac3 in the CA1 subregion attenuated the long-term memory enhancement. These data suggest that HDAC3 may regulate long-term memory formation by altering chromatin environments and fine-tuning expression of Nr4a2 gene.
In addition to the role of HDAC3 in hippocampal learning and memory, HDAC3 has been implicated as a negative regulator for the formation of cocaine associated memory formation (Rogge et al., 2013). Regional deletion of Hdac3 in the nucleus accumbens facilitated acquisition of cocaine-induced conditioned place preference, and increased Nr4a2 gene expression, which likely resulted from a decrease in Nr4a2 gene promoter occupancy by HDAC3 as well as an increase in histone H4 lysine 8 acetylation. Taken together, these recent reports suggest that HDAC3 is involved in hippocampal long-term as well as psychomotor stimulants induced memory formation by altering chromatin status at the specific gene promoter, Nr4a2.
HDAC8
HDAC8 is also a member of the class I HDAC family, however the components of the repressor complex containing HDAC8 are not as well delineated and little is known about the role of this HDAC in CNS function. HDAC8 is widely expressed in the brain, and differs in sequence identity much more than the rest of members in the class I family, suggesting it may have a distinct role in the brain compared to HDAC1, 2 and 3. Future work is necessary to elucidate the role of this HDAC in the CNS.
Class II HDAC
A few studies have started to examine the role of class II HDACs in the CNS; thus, we will briefly summarize their functions in this review. Recently generated conditional knockout mice with a brain-specific deletion of Hdac4, which shares strong amino acid sequence identity with Hdac5, were characterized and found to have impairments in hippocampal-dependent memory formation as well as deficits in LTP without impacting basal synaptic transmission (Kim et al., 2012). In the same study, constitutive Hdac5 knockouts were shown to have normal locomotor activity, motor coordination, anxiety, and learning and memory as well as intact basal synaptic transmission, suggesting distinct roles for these HDACs in learning and memory. In separate work, HDAC5 has been linked to the action of the psychomotor stimulant, cocaine. Chronic cocaine has been shown to induce phosphorylation of HDAC5 and trigger its nuclear export (Renthal et al., 2007). Transient overexpression of HDAC5 in the nucleus accumbens attenuates the locomotor response to chronic cocaine treatments, whereas loss of HDAC5 results in hypersensitivity to chronic cocaine. Collectively, this data suggests that HDAC5 does not play a significant role in learning and memory as well as several other behavioral measures under normal conditions but may be an important factor in certain disease states such as addiction.
Corepressors
Five corepressor proteins, Sin3a/3b, NCoR, SMRT, CoREST, and NURD have been reported to interact with the class I HDAC family, with their biochemical associations well characterized (Perissi et al., 2010; Yang and Seto, 2008). These corepressors do not possess DNA binding or chromatin remodeling activities, rather they serve as scaffolding proteins to help stabilize assembly of each repressor complex. Constitutive deletion of Sin3a/3b, NCoR, or SMRT in mice results in embryonic lethality and defective cardiovascular and erythrocyte development during embryogenesis, indicating the distinct and necessary roles of these nuclear scaffolding proteins during development (Dannenberg et al., 2005; David et al., 2008; Jepsen et al., 2008; Jepsen et al., 2000). More recently, NCoR and SMRT were reported to participate in cell fate determination of neural stem cells (Hermanson et al., 2002). In the current issue, Schoch and Able discuss the roles of these corepressors in the adult brain and whether chromatin-based alterations are underlying mechanisms for adult brain function in the context of activity-dependent transcription and cognition.
4. Conclusions
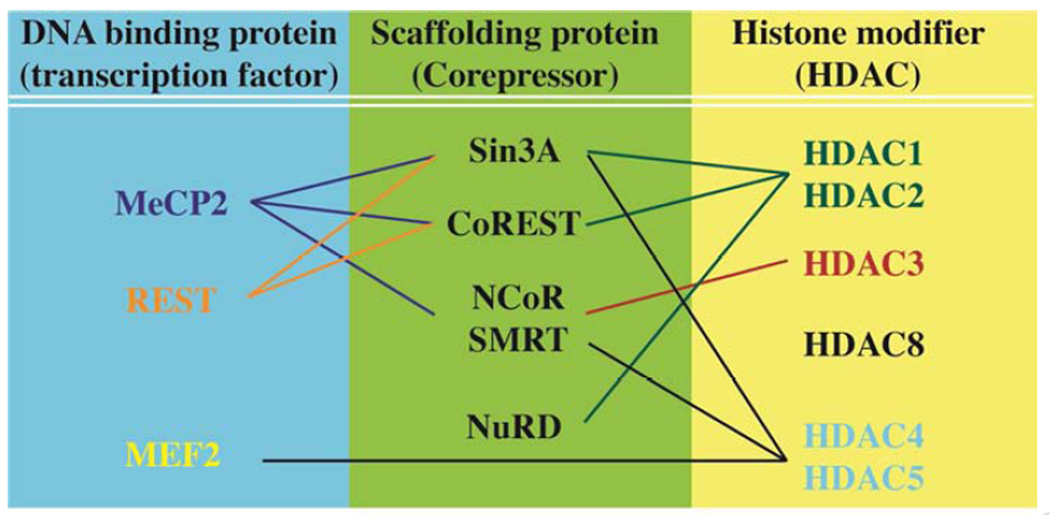
Accumulating evidence has uncovered essential roles for transcription repressors and the components of their repressor complexes in many aspects of brain function. Transcriptional repression takes place only when DNA-binding proteins, chromatin-remodeling enzymes, and corepressors are assembled together at target gene promoters. However, when the gene encoding each component in a repressor complex has been individually deleted, the resultant phenotypes were not necessarily similar and in some cases have been quite distinct. One such example involves HDAC2 and MeCP2, which are the components of a Sin3a-containing repressor complex. Mice lacking Hdac2 in the broad-forebrain display enhanced performance in many forms of learning and memory without affecting other behavioral phenotypes, whereas Mecp2-deficient mice in the same brain region manifest a variety of behavioral deficits (Gemelli et al., 2006; Morris et al., 2013). In particular, the Mecp2-deficient mice performed worse in cue-dependent fear conditioning (amygdala-dependent learning) with a trend in impaired context-dependent fear conditioning (hippocampal-dependent learning). One possible reason for this disparity could be the corepressor-dependent action of each factor. Each behavioral phenotype might be attributed to a particular set of changes in gene expression defined by a distinct corepressor. HDAC2 is found not only with Sin3a, but also in NURD and CoREST-mediated repressor complexes (Figure 1) (Ballas et al., 2001; Tong et al., 1998; You et al., 2001; Zhang et al., 1998). Similarly, some studies report the presence of MeCP2 with CoREST in addition to Sin3A (Ballas et al., 2005; Lunyak et al., 2002). Moreover, recent studies demonstrated association of MeCP2 with NCoR and SMRT complexes (Ebert et al., 2013; Lyst et al., 2013). This latter association is functionally significant as mice expressing a mutant MeCP2 that either lacks the ability to bind NCoR or results in constitutive binding of NCoR, reproduced some RTT-like phenotypes. Therefore, careful consideration of transcriptional repressors and their associated chromatin modifiers as unique corepressor units may unveil clearer roles of these complexes in the adult CNS.
Figure 1.
Composition of repressor complexes in the adult brain
Alternatively, we cannot exclude the possibility that the ability of HDACs to target non-histone substrates may attribute to phenotypes of Hdac2 knockout, which are distinct from MeCP2-deficient animals. Acetylation of non-histone proteins, such as transcription factors, NF-kB, GATA, and p53, is a functionally significant post-translation modification that regulates their transcriptional activity. Loss of HDAC1 and HDAC2 have been shown to alter levels of acetylation of the non-histone proteins, leading to abnormal phenotypes in a wide array of tissues, including cardiomyocyte proliferation, epidermal differentiation, and Schwann cell differentiation (Kelly and Cowley, 2013). Thus, examination of non-histone proteins in the context of adult brain lacking HDAC2 and MeCP2 may tease out their specific roles.
Recent mouse studies strongly support HDAC2 playing a predominate role in the regulation of learning and memory among HDACs in the class I family. Despite the fact that HDAC1 and HDAC2 share high sequence identity, it is somewhat surprising that HDAC1 does not possess functional redundancy to HDAC2, at least in the realm of learning and memory. Notably, HDAC1 is expressed in both neurons and glia and HDAC2 predominantly in neurons in the adult rodent brain (MacDonald and Roskams, 2008). In separate studies, western blot analysis revealed similar levels of expression of HDAC1 and HDAC2 in specific brain regions of adult mice (Guan et al., 2009; Morris et al., 2013). Considering the high sequence similarity and the presence of both HDAC1 and HDAC2 in the brain, the preferential role of HDAC2 in the learning and memory may depend on the association of distinct corepressors such as Sin3A, NCoR, CoREST, NURD, with the individual HDACs (Figure 1). An HDAC2 repressor complex composed of a specific combination of factors may give rise to specific gene regulation essential for enhanced learning and memory. Given that all HDACs are widely expressed at varying levels in the adult brain, it will be important to investigate the role for each individual HDAC in behavioral and cognitive functions.
In light of the therapeutic potential for HDAC inhibitors in enhancing learning and memory, and as putative therapeutic targets for neurodegenerative diseases, it is important to assess if chronic treatment with HDAC inhibitors produces aversive unwanted side effects (Chuang et al., 2009). Many inhibitors target the catalytic domain of HDACs and are likely to block the activity of multiple HDACs, rather than a specific HDAC subtype. In mature hippocampal neurons, chronic inhibition of HDAC activity results in impaired excitatory neurotransmission (Akhtar et al., 2009). In separate work, chronic infusion of SAHA, a class I HDAC inhibitor, to the amygdala led to behavioral abnormalities including deficits in learning and memory as well as heightened anxiety (Adachi et al., 2009). Additionally, specificity of the HDAC inhibitors is a key issue for therapeutic application. While studies from transgenic mice indicate the beneficial role of HDAC2 and more recently HDAC3 in memory formation, Hdac4 knockout mice revealed surprisingly deficits in learning and memory. Taken together, these data suggest that the therapeutic application of HDAC inhibitors, in particular those requiring chronic treatments, should be carefully examined as they may potentially produce undesirable psychiatric side effects.
As further research elucidates the role of individual components of transcriptional repressor complexes in the CNS, this information will hopefully start to provide information relevant to drug discovery to specific disorders. The identification of these transcriptional processes in the brain as well as in the disease state may provide a more complete approach towards processes that go awry during certain disorders as well as allow one to hone in on individual factors that may prove to be therapeutic targets. The work outlined in this review represents an exciting approach to understanding transcriptional repression in the brain and sets the groundwork for delineating these processes further.
Highlights.
-
-
Transcriptional repressors serve as a foundation on DNA for forming repressor complexes.
-
-
Repressor complexes carry enzymatic activity to alter chromatin environment.
-
-
Components of repressor complexes displays diverse functions in the CNS.
-
-
Disruption of transcriptional repression mechanism may contribute to disease.
-
-
Histone deacetylases play a critical role in cognition but not all are beneficial.
Acknowledgements
The authors would like to thank members of the Monteggia laboratory for helpful discussions and comments on the manuscript. This work was supported MH081060 (LMM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi M, Autry AE, Covington HE, 3rd, Monteggia LM. MeCP2-mediated transcription repression in the basolateral amygdala may underlie heightened anxiety in a mouse model of Rett syndrome. J Neurosci. 2009;29:4218–4227. doi: 10.1523/JNEUROSCI.4225-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar MW, Raingo J, Nelson ED, Montgomery RL, Olson EN, Kavalali ET, Monteggia LM. Histone deacetylases 1 and 2 form a evelopmental switch that controls excitatory synapse maturation and function. J Neurosci. 2009;29:8288–8297. doi: 10.1523/JNEUROSCI.0097-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H, Hara A, Era T, Kunisada T, Yamada Y. Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development. 2012;139:667–677. doi: 10.1242/dev.072272. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15:39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David Sweatt J, Zoghbi HY. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Couvert P, Bienvenu T, Aquaviva C, Poirier K, Moraine C, Gendrot C, Verloes A, Andres C, Le Fevre AC, Souville I, Steffann J, des Portes V, Ropers HH, Yntema HG, Fryns JP, Briault S, Chelly J, Cherif B. MECP2 is highly mutated in X-linked mental retardation. Hum Mol Genet. 2001;10:941–946. doi: 10.1093/hmg/10.9.941. [DOI] [PubMed] [Google Scholar]
- Covey MV, Streb JW, Spektor R, Ballas N. REST regulates the pool size of the different neural lineages by restricting the generation of neurons and oligodendrocytes from neural stem/progenitor cells. Development. 2012;139:2878–2890. doi: 10.1242/dev.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH, Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Grandinetti KB, Finnerty PM, Simpson N, Chu GC, Depinho RA. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci U S A. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng V, Matagne V, Banine F, Frerking M, Ohliger P, Budden S, Pevsner J, Dissen GA, Sherman LS, Ojeda SR. FXYD1 is an MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum Mol Genet. 2007;16:640–650. doi: 10.1093/hmg/ddm007. [DOI] [PubMed] [Google Scholar]
- Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, Greenberg ME. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013;499:341–345. doi: 10.1038/nature12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, McGill BE, Goulding EH, Sullivan E, Tecott LH, Zoghbi HY. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008;59:947–958. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston K, Jayaraman PS. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell Mol Life Sci. 2003;60:721–741. doi: 10.1007/s00018-003-2260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemelli T, Berton O, Nelson ED, Perrotti LI, Jaenisch R, Monteggia LM. Postnatal loss of methyl-CpG binding protein 2 in the forebrain is sufficient to mediate behavioral aspects of Rett syndrome in mice. Biol Psychiatry. 2006;59:468–476. doi: 10.1016/j.biopsych.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, Su SC, Samiei A, Joseph N, Haggarty SJ, Delalle I, Tsai LH. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haettig J, Stefanko DP, Multani ML, Figueroa DX, McQuown SC, Wood MA. HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learn Mem. 2011;18:71–79. doi: 10.1101/lm.1986911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME, Kroener BT, Manglesdorf DJ, Abel T. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J Clin Invest. 2012;122:3593–3602. doi: 10.1172/JCI64145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Florian C, Abel T. Post-training intrahippocampal inhibition of class I histone deacetylases enhances long-term object-location memory. Learn Mem. 2011;18:367–370. doi: 10.1101/lm.2097411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Hoch RV, Rubenstein JL, Pleasure S. Genes and signaling events that establish regional patterning of the mammalian forebrain. Semin Cell Dev Biol. 2009;20:378–386. doi: 10.1016/j.semcdb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Gleiberman AS, Shi C, Simon DI, Rosenfeld MG. Cooperative regulation in development by SMRT and FOXP1. Genes Dev. 2008;22:740–745. doi: 10.1101/gad.1637108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick SM, Mandel G, Glass CK, Rose DW, Rosenfeld MG. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans. 2013;41:741–749. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- Kim MS, Akhtar MW, Adachi M, Mahgoub M, Bassel-Duby R, Kavalali ET, Olson EN, Monteggia LM. An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J Neurosci. 2012;32:10879–10886. doi: 10.1523/JNEUROSCI.2089-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Yntema HG, Oudakker AR, Romein T, Sistermans E, Nillessen W, van Bokhoven H, de Vries BB, Hamel BC. De novo MECP2 frameshift mutation in a boy with moderate mental retardation obesity and gynaecomastia. Clin Genet. 2002;61:359–362. doi: 10.1034/j.1399-0004.2002.610507.x. [DOI] [PubMed] [Google Scholar]
- Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification sequence and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li W, Pozzo-Miller L. Beyond Widespread Deletions to Model Rett Syndrome: Conditional Spatio-Temporal Knockout Single-Point Mutations and Transgenic Rescue Mice. Autism Open Access. 2012;2012:5. doi: 10.4172/2165-7890.S1-005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Loven J, Kwok SM, Feldman DA, Bateup HS, Gao Q, Hockemeyer D, Mitalipova M, Lewis CA, Vander Heiden MG, Sur M, Young RA, Jaenisch R. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Genomic targets and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res. 2013;41:8072–8084. doi: 10.1093/nar/gkt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, Guy J, Kastan NR, Robinson ND, de Lima Alves F, Rappsilber J, Greenberg ME, Bird A. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev Dyn. 2008;237:2256–2267. doi: 10.1002/dvdy.21626. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- McNulty SE, Barrett RM, Vogel-Ciernia A, Malvaez M, Hernandez N, Davatolhagh MF, Matheos DP, Schiffman A, Wood MA. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learn Mem. 2012;19:588–592. doi: 10.1101/lm.026385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2 a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K, Hirasawa T, Soutome M, Itoh M, Goto Y, Endoh K, Takahashi K, Kudo S, Nakagawa T, Yokoi S, Taira T, Inazawa J, Kubota T. The protocadherins PCDHB1 and PCDH7 are regulated by MeCP2 in neuronal cells and brain tissues: implication for pathogenesis of Rett syndrome. BMC Neurosci. 2011;12:81. doi: 10.1186/1471-2202-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis growth and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Mahgoub M, Na ES, Pranav H, Monteggia LM. Loss of histone deacetylase 2 improves working memory and accelerates extinction learning. J Neurosci. 2013;33:6401–6411. doi: 10.1523/JNEUROSCI.1001-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Monteggia LM. Unique functional roles for class I and class II histone deacetylases in central nervous system development and function. Int J Dev Neurosci. 2013;31:370–381. doi: 10.1016/j.ijdevneu.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na ES, Nelson ED, Kavalali ET, Monteggia LM. The impact of MeCP2 loss- or gain-of-function on synaptic plasticity. Neuropsychopharmacology. 2012;38:212–219. doi: 10.1038/npp.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nelson ED, Kavalali ET, Monteggia LM. MeCP2-dependent transcriptional repression regulates excitatory neurotransmission. Curr Biol. 2006;16:710–716. doi: 10.1016/j.cub.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Noh KM, Hwang JY, Follenzi A, Athanasiadou R, Miyawaki T, Greally JM, Bennett MV, Zukin RS. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci U S A. 2012;109:E962–E971. doi: 10.1073/pnas.1121568109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, Bird A. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:2247–2256. doi: 10.1093/hmg/ddi229. [DOI] [PubMed] [Google Scholar]
- Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka GJ, Watson CM, Radziewic T, Hayward M, Lahooti H, Christodoulou J, Tam PP. Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain. 2006;129:887–898. doi: 10.1093/brain/awl022. [DOI] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Rodenas-Ruano A, Chavez AE, Cossio MJ, Castillo PE, Zukin RS. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci. 2012;15:1382–1390. doi: 10.1038/nn.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge GA, Singh H, Dang R, Wood MA. HDAC3 is a negative regulator of cocaine-context-associated memory formation. J Neurosci. 2013;33:6623–6632. doi: 10.1523/JNEUROSCI.4472-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci U S A. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Treadwell-Deering D, Zoghbi HY, Glaze DG, Neul JL. Brief Report: MECP2 Mutations in People Without Rett Syndrome. J Autism Dev Disord. 2013 doi: 10.1007/s10803-013-1902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Esch H, Bauters M, Ignatius J, Jansen M, Raynaud M, Hollanders K, Lugtenberg D, Bienvenu T, Jensen LR, Gecz J, Moraine C, Marynen P, Fryns JP, Froyen G. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77:442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol. 2010;3:a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Aoki H, Kunisada T, Hara A. Rest promotes the early differentiation of mouse ESCs but is not required for their maintenance. Cell Stem Cell. 2010;6:10–15. doi: 10.1016/j.stem.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]