Abstract
Purpose
To evaluate the role of DiGeorge Critical Region 8 (DGCR8), a key component of miRNA biogenesis pathway in ovarian cancer.
Methods
The expression of DGCR8 in ovarian cancer was detected by immunostaining and DGCR8 knockdown in ovarian cancer cells was achieved using lentiviral shRNA. Differential expression of miRNAs was determined using Nanostring miRNA arrays and validated by real-time RT-PCR.
Results
DGCR8 was highly expressed in ovarian cancer. Knockdown of DGCR8 expression inhibits cell proliferation, migration, and invasion, as well as sensitizes cells to apoptosis induced by the chemotherapeutic drug cisplatin. Cellular survival pathways including ERK1/2 mitogen-activated protein kinase and phosphatidylinositol 3-kinase/AKT were attenuated in DGCR8 knockdown cells. DGCR8 knockdown resulting in dysregulated miRNA gene expression. miR-27b was identified as the most highly down-regulated miRNA in DGCR8 knockdown cells and promoted cell proliferation in ovarian cancer cells.
Conclusions
DGCR8 functions as an oncogene in ovarian cancer, which is in part mediated by miR-27b.
Keywords: DGCR8, miRNA, ovarian cancer
INTRODUCTION
Ovarian cancer is a major cause of cancer-related mortality in women. In 2012 more than 20000 new cases were diagnosed in the US and 15000 women died of this disease. DGCR8 is a double-stranded RNA binding protein and its monoallelic deletion on the human chromosome 22q11.2 region is associated with Digeorge Syndrome (DGS). As a key component of miRNA biogenesis pathway, DGCR8 interacts with the RNase III enzyme Drosha and forms a microcomplex in the nucleus to process primary miRNA (pri-miRNA) into precursor miRNA (pre-miRNA). In the cytoplasm the RNAase III enzyme Dicer processes the pre-miRNAs into mature miRNAs, which subsequently regulate gene expression through the RNA induced silencing complex (RISC) (1-3). miRNAs negatively regulate target gene expression at the posttranscriptional level by binding to the 3 untranslated region of target mRNAs. miRNAs may function as tumor suppressors or oncogenes in a variety of human cancers.
Drosha and Dicer have been extensively investigated in a variety of cancers and their expression is associated with cancer metastasis (4-9). The expression of Dicer and Drosha is highly dependent on the specific cancer type, and therefore they may have distinct functions in different cancers. Dicer and Drosha are downregulated in ovarian cancer and their low expression level is correlated with tumor developmental stage. Missense mutations of Dicer or Drosha genes have also been detected (10,11). Dicer is upregulated in ovarian serous carcinoma and associated with specific clinicopathological features (12). DGCR8, a double-stranded RNA binding protein, has been shown to be upregulated in cervical squamous cell carcinoma and salivary gland pleomorphic adenoma (13-15). However, the role of DGCR8 in ovarian cancer is completely unknown. To determine whether DGCR8 plays a role in ovarian cancer, we determined the expression of DGCR8 in clinical samples of ovarian cancer, and silenced DGCR8 expression in SKOV3 ovarian cancer cells by transduction with lentiviral vectors. Our studies indicatethat silencing DGCR8 reduces cell proliferation, migration, and invasion in vitro, and inhibits tumor growth in vivo. miR-27b at least in part contributes to the oncogenic properties of DGCR8.
MATERICALS AND METHODS
Clinical sections of ovary from healthy persons (n=10) and ovarian cancer patients (n=10) were provided by the Department of Pathology of the Third Affiliated Hospital of Zhengzhou University, P.R.China. This study was approved by Institutional Ethic committee of Zhengzhou University.
Cell culture
The ovarian cancer SKOV3 cell line was purchased from ATCC and cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS (Hyclone; Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). HEK293 FT cells were cultured in DMEM media with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 1% glutamine, 1% nonessential amino acid, and geneticin with a final concentration of 1 μg/ml.
Cell proliferation assay
5×104 SKOV3 cells transduced with scramble(SC) or DGCR8 shRNA were plated in triplicate and counted daily for 4 days.
Cell colony formation assay
400 cells transduced with DGCR8 or SC shRNAs were plated in triplicate wells of 6-well plates and cultured for two weeks, then stained with 0.1% crystal violet and cell colonies were counted.
MTT assay
8000 cells transduced with DGCR8 and SC shRNAs were plated per well in 96-well plates and cultured for 24 hr. Thereafter,10 ul of MTT reagent was added to each well and incubated for ~4 h. The reaction was terminated by the addition of 100ul detergent reagent, the plates were incubated at 22°C in the dark for 2 h, and then the absorbance (OD) at 570nm was measured.
Wound healing assay
5×105 SKOV3 cells transduced with DGCR8 and SC shRNAs were plated into triplicate wells of 6-well plates and grown overnight. The well surface was scratched with a sterile 20 μl-pipette-tip and the wells were washed 3 times with PBS. Fresh growth medium was added for additional 24 h. The migration rate was calculated using the formula (area of the wound area at 0 h- the wound area at 24h) / the wound area at 0 h.
Cell invasion assay
1×105 cells transduced with DGCR8 and SC shRNA in 1% of DMEM media were seeded into Matrigel coated upper chambers of transwell plates (BD Sciences, San Jose, CA). 10% DMEM was added to the lower chamber and the plates incubated for 24 h. The cells in the upper membrane of the transwell that did not invade were removed with a cotton swab, and the invaded cells were stained with propidium iodide, visualized by fluorescent microscopy and counted.
Cell apoptosis assay
8000 SKOV3 cells transduced with DGCR8 and SC shRNA were plated into triplicate wells of 96-well plates and incubated at 37°C for 24h. The cells were washed with PBS, starved for 1 h in serum-free media, and then treated with different doses of cisplatin for an additional 24h. The caspase 3/7 activities were determined using the Caspase-Glo 3/7 assay kit (Promega, Madison, WI ) according to the manufacturer.
Cell cytotoxicity assay
5×104 SKOV3 cells transduced with DGCR8 and scramble shRNA were seeded into triplicate wells of a 96–well plate and incubated for 24 h. Cells were treated with different doses of cisplatin and incubated for additional 24h at 37°C. To determine the cell viability following treatment, cells were incubated with10 ul of WST-1 (Roche Applied Science, Indianapolis, IN) for 3 h, and the absorbance was measured at OD 450 nm.
Soft agar assay
1.2% Noble agar was mixed with 2XDMEM containing 10% FCS and added into 6-well plates. After the agar solidified, 2×106 cells transduced with DGCR8 shRNA and in 0.7% Noble agar were placed on top of the agar underlayer. After the top agar layer solidified, DMEM containing 10% FCS was added to the plates. The cells were refed with fresh growth medium every five days, and after two weeks colonies were counted under a light microscope.
Nanostring nCounter miRNA Array
Total RNA extracted from DGCR8 and scramble shRNA transduced cells was subjected to miRNA expression profile analysis using The Nanostring nCounter system (NanoString Technologies, Seattle, WA). In brief, total RNA was annealled, ligated,purified, and then hybridized with probes tailored to each miRNA at 65°C overnight. The unbound probes were removed and the purified ternary complexes were bound to the image surface, elongated, immobilized and counted using the nCounter Prep Station and Digital Analyzer. NanoString nCounter microRNA raw data were normalized to the average counts for all control spikes in each sample and analyzed using nSolver software.
Lentiviral Vector Production
The lentiviral DGCR8 and scramble shRNA vectors were purchased from Addgene. Lentivirus was produced as described previously (16).
miRNA expression by Real-Time RT-PCR
Total RNA was extracted from SKOV3 cells transduced with DGCR8 or scramble shRNA. PolyA tailing real-time RT-PCR was performed as described previously (17). Forward primers for individual miRNAs are listed in supplemental Table 1 (Table S1). The SYBR Green-based real-time RT-PCR was performed using a LightCycler 4800 real-time PCR instrument (Roche Applied Science; Indianapolis, IN). Melting curve analysis was performed to examine the PCR product specificity. The relative expression was normalized to U6 small nuclear RNA by the ΔΔCt method, and expressed as mean ± SE.
Western Blot
Ovarian cancer SKOV3 cells transduced with DGCR8 or scramble shRNA were collected in RIPA buffer (Thermo Scientific; Rockford, IL) containing 1% Halt Proteinase inhibitor Cocktail (Thermo Scientific; Rockford, IL). An equal amount of protein (40 μg/lane) was loaded on 8% SDS-PAGE gels and transferred to nitrocellulose membranes. The membrane was blocked with 5% non-fat milk for 1 h and incubated with primary antibodies against DGCR8 (Santa Cruz; Santa Cruz, CA); β-actin, GAPDH (Sigma; St. Louis, MO), pERK1/2 and pAKT (Cell Signaling; Danvers, MA).
Immunofluorescence
Deparaffinized sections were rehydrated, and antigen retrieval was performed by incubation of the slides for 30 min at 95-100°C in 10 mM sodium citrate, 0.05% Tween 20 (pH 6.0). The sections were treated with blocking buffer (5% normal goat serum, 3% bovine serum albumin, and 0.1% Triton-X 100 in PBS) for 1 h. To detect protein expression, sections were incubated with DGCR8 and PCNA primary antibody at 4°C overnight. After three rinses for 5 min with 0.05% Tween 20 in PBS (PBS-T), the sections were incubated with Alexa 594 conjugated goat anti-rabbit or mouse secondary antibody (Invitrogen, 1:200 in PBST) for 1 h at room temperature. After three washes, the sections were mounted with Vectashield medium containing DAPI or PI (Vector Laboratories Inc.; Burlingame, CA) and visualized under fluorescent microscope.
Tumor xenografts in mice
Animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of the University of Tennessee Health Science Center. Immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) female mice (Jackson Laboratory) were injected into the flanks with 106 SKOV3 (DGCR8 knockdown and scramble) cells. Tumors were measured weekly with a handheld caliper.
Statistical Analysis
Data shown represent the mean ± standard deviation (SD) from at least three different experiments. The differences were analyzed using Student's t-test. P values < 0.05 were considered significant.
RESULTS
DGCR8 is upregulated in ovarian cancer
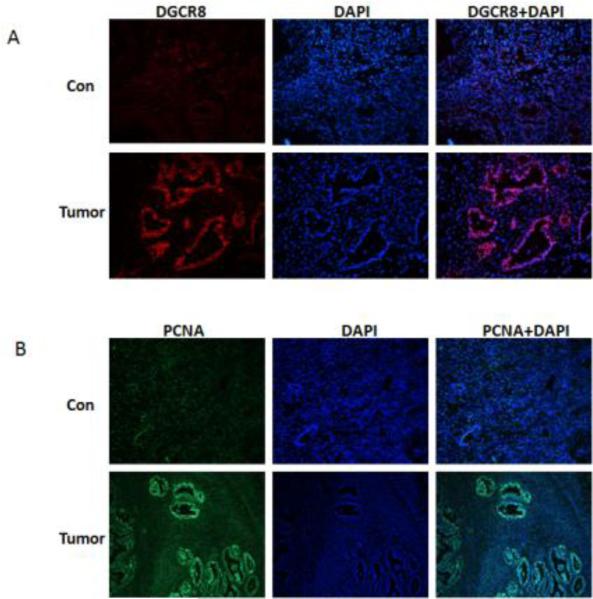
To determine DGCR8 expression in ovarian cancer, we examined sections of 10 ovarian cancer and control tissue by using immunostaining. Ovarian cancer samples were diagnosed by H &E staining as serous adenocarcinoma (Fig. S1). DGCR8 was highly stained in cell nuclei in ovarian cancer when compared to control tissue samples (Fig.1A), indicating that DGCR8 expression is upregulated in ovarian cancer. To examine the proliferative status of samples, sections of ovarian cancer and control tissue were stained using antibody against the cell proliferation marker PCNA. As shown in Figure 1B, PCNA was highly stained in the ovarian tumors when compared to control.
Figure 1. DGCR8 is highly expressed in ovarian cancer.
A. The expression of DGCR8 in immunostained sections of ovarian cancer and normal ovary tissue B. Cell proliferations in sections of ovarian cancer and normal ovary were stained for the proliferation marker PCNA.
To address the role of DGCR8 in ovarian cancer, DGCR8 expression was silenced in the SKOV3 ovarian cancer cell line using lentiviral shRNA (Fig.S2A). SKOV3 ovarian cancer cells were transduced with puromycin-resistant lentiviral vectors that also contained DGCR8 or scramble shRNA, and selected with 3 ug/ml of puromycin. DGCR8 expression was reduced ~75% in DGCR8 knockdown (KD) cells when compared to cells transduced with scramble shRNA, as detected by real-time RT-PCR (Fig. S2B). DGCR8 knockdown was further confirmed by immunofluorescent staining, in which DGCR8 was strongly stained in the nuclei of cells transduced with scramble shRNA, but only weak staining of DGCR8 was observed in the DGCR8 KD cells (Fig.S2B).
Silencing DGCR8 inhibits cell proliferation and colony formation
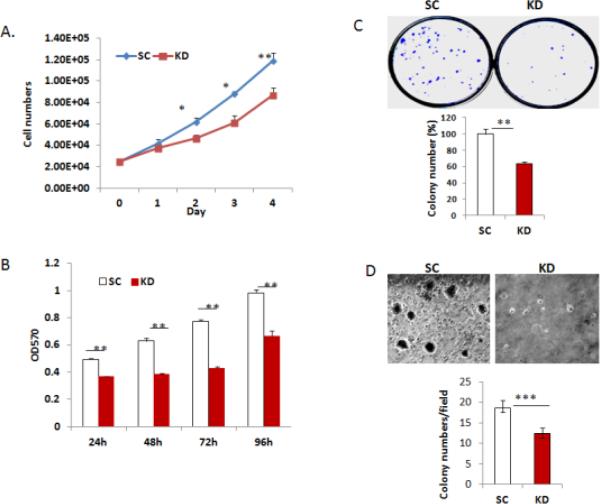
To determine the functional role of DGCR8, we examined the proliferation of SKOV3 cells transduced with DGCR8 or scramble shRNA by cell counting. Silencing DGCR8 significantly inhibits the proliferation of SKOV3 cells within 48 h of plating as compared to cells transduced with scramble shRNA (Fig. 2A). The inhibitory effect of DGCR8 KD on cell growth was also observed in MTT assays (Fig. 2B). We also performed colony formation assays on DGCR8 KD and control cells and found that silencing DGCR8 leads to significantly reduction in colony formation as compared to controls (Fig.2C). In addition, silencing DGCR8 attenuated anchorage-independent growth as determined by soft agar assay (Fig.2D).
Figure 2. Silencing DGCR8 in ovarian cancer cells reduces cell proliferation.
A. Cell growth curves in DGCR8KD and control cells were determined by cell counting (* p<0.05; ** p<0.01). B. Cell proliferation was examined by using MTT assay in DGCR8KD and control cells(**p<0.01). C. Colony formation by DGCR8KD and control cells (**p<0.01). D. Soft agar colony formation assay was performed in a 6-well plate in triplicates. Colonies were photographed and counted after 3 weeks (***p<0.001).
Silencing DGCR8 results in reduced cell migration and invasion
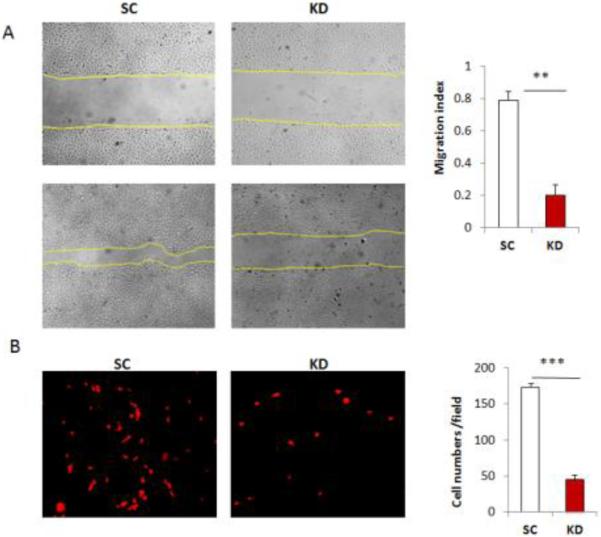
Cancer cell migration and invasion are important events in metastasis. To determine whether silencing DGCR8 affected cell migration, we examined cell migration in wound healing assays. The migration rate is significantly reduced in DGCR8 KD compared with control cells (Fig.3A). We further examined cell invasion using transwell cell invasion assay. Silencing DGCR8 in SKOV3 cells resulted in significant reduction in the number of cells that invade through Matrigel basement membrane (Fig.3B).
Figure 3. Silencing DGCR8 inhibits cell migration and invasion in ovarian cancer cells.
A. Wound healing assays were performed to examine the migration rate of SKOV3 cells transduced with DGCR8 shRNA and scramble control. Photographs were taken at 0 and 24 h following initial scratch. The migration rates were quantified by measuring the injured area from triplicates. Three separate experiments were performed ( **p<0.01). B. Cell invasion assay was performed with Matrigel transwell plates and cells migrating through Matrigel after 24h were stained with PI, photographed after fluorescent microscopy and counted. Data were collected from three separate experiments and analyzed using student T-tests (***p<0.001).
Silencing DGCR8 sensitizes chemotherapeutic drug induced apoptosis and reduces cell viability
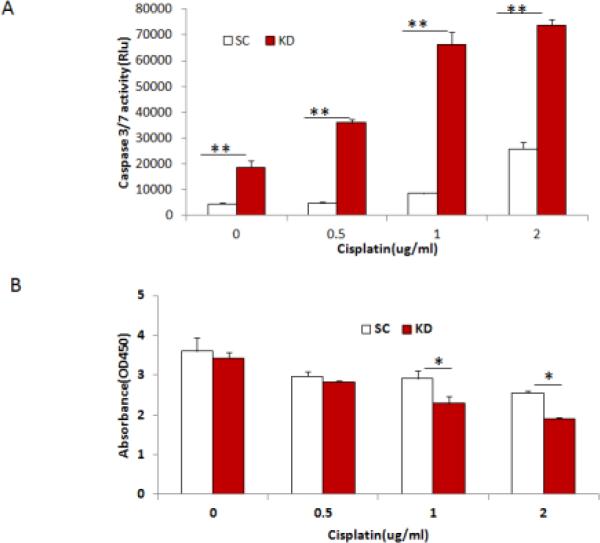
To determine whether DGCR8 regulates cellular sensitivity to apoptotic-inducing agents, we treated SKOV3 cells transduced with DGCR8 or scramble shRNA with different concentrations of the chemotherapeutic drug cisplatin. Apoptosis was determined by measuring caspase-3/7 activity at 24 h following cisplatin exposure. The activities of caspase 3/7 were significantly increased by exposure to increased cisplatin concentrations in both control and DGCR8 KD cells (Fig.4A). Most importantly there was a marked increase in apoptosis at all cisplatin concentrations in DGCR8 KD cells, indicating that knockdown of DGCR8 sensitized cells to apoptosis induced by cisplatin. The cell viability of DGCR8 KD cells was significantly reduced upon treatment with 1 and 2 ug/ml of cisplatin (Fig.4B).
Figure 4. Knockdown of DGCR8 sensitizes cells to cisplatin induced cell apoptosis.
A. DGCR8KD and control SKOV3 cells were treated with different doses of cisplatin and apoptosis was examined at 24h following the treatment by measuring caspase3/7 activity. Data were presented from one representative experiment of three experiments (**p<0.01). B. Cell viabilities were examined in DGCR8 KD and control SKOV3 cells following treatment with various doses of cisplatin . Three separate experiments were repeated (*p<0.05).
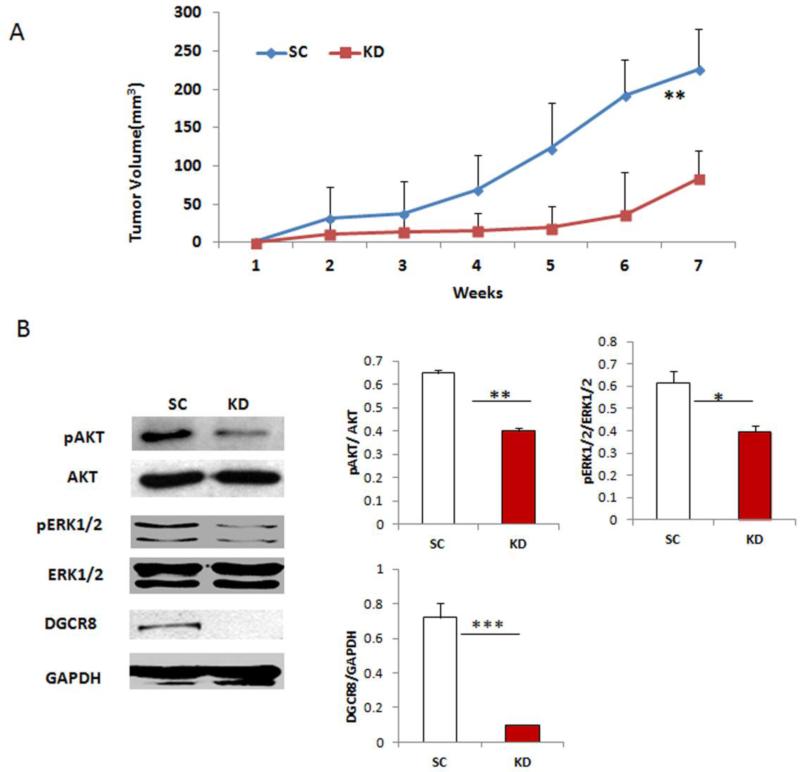
Silencing DGCR8 inhibits tumor growth in vivo
To determine whether DGCR8 affects tumor growth in vivo, NSG mice were injected subcutaneously with 1×106 DGCR8 KD and scramble-transduced SKOV3 cells. Tumor volume was determined weekly by caliper measurement. As shown in Figure 5A, tumors continuously grew in mice injected with control SKOV3 cells during the 7-week experimental period. In marked contrast, tumor growth in mice injected with DGCR8 KD cells was significantly delayed, as early as two weeks after injection of cells. These results suggest that silencing DGCR8 significantly inhibits tumor growth in vivo.
Figure 5. Knockdown of DGCR8 attenuates cellular survival pathways.
A. Tumors were continuously grown in mice injected with control cells whereas tumor significantly were reduced in mice injected with DGCR8KD cells(n=4,**p<0.01). B. DGCR8, phospho and total AKT, phospho and totalERK1/2 and GAPDH expression in DGCR8KD and control SKOV3 cells were detected by Western blot. Protein bands were quantified by densitometry (**p<0.01, ***p<0.001).
Silencing DGCR8 attenuates cellular survival pathway
The miRNA biogenesis pathway contributes to miRNA maturation in cells, and thus regulates cellular homeostasis by controlling the target gene expression. We next examined whether miRNA biogenesis affects two important cellular survival pathways, ERK1/2 mitogen-activated protein kinase and phosphatidylinositol 3-kinase/AKT. Silencing DGCR8 significantly attenuates both ERK1/2 and PI3K/AKT pathways in SKOV3 cells (Fig.5B), indicating that silencing DGCR8 dependent miRNA biogenesis pathway attenuates cellular survival signaling pathways in ovarian cancer cells.
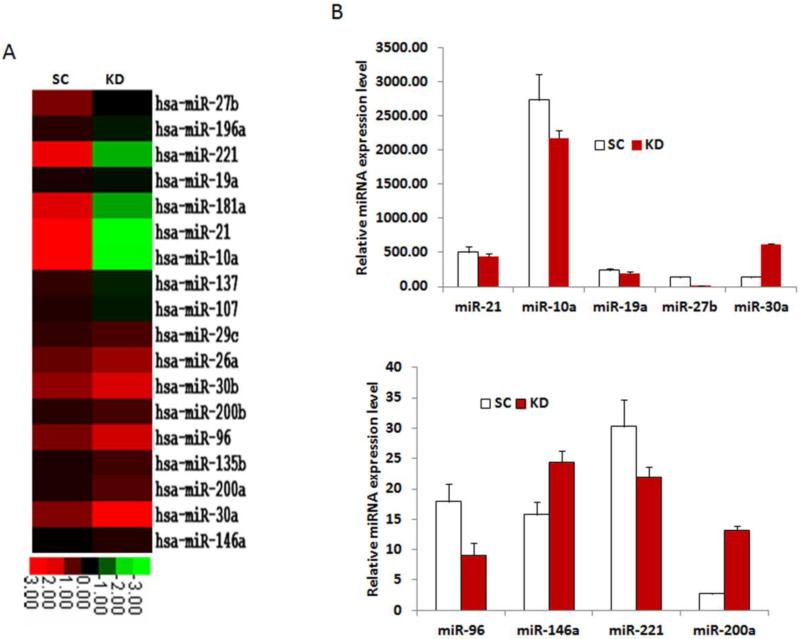
Silencing DGCR8 in ovarian cancer cells selectively affects miRNA gene expression
DGCR8 interacts with Drosha and controls miRNA maturation, and thus silencing DGCR8 disrupts miRNA expression. To determine the role of DGCR8 on the expression of miRNAs in SKOV3 cells, we performed Nanostring miRNA array analysis on DGCR8 KD and control cells. Of 654 human miRNA arrayed, 213 miRNAs were expressed at detectable levels; 18 showed significant downregulation, 85 were not significantly altered, and 355 were upregulated in DGCR8 KD compared with control cells. The array data showed that the loss of DGCR8 had selective effects on miRNA expression as illustrated in heat-maps (Fig.6A). The most downregulated miRNAs in SKOV3 DGCR8 KD cells include miR-27b, 196a, 221, 19a, 181a,10a and miR-21. The upregulated miRNAs are miR-26a, 30b, 200b, 96, 200a, 30a and 146a. We further validated the expression of these differentially regulated miRNAs using polyA tailing real-time RT-PCR and found that miR-27b was approximately 8-fold dowregulated upon DGCR8 KD, while miR-30a and miR-200a were upregulated 4.5 and 4.9-fold upon DGCR8 KD, respectively (Figure 6B,C).
Figure 6. Knockdown of DGCR8 in SKOV3 cells leads to dysregulation of miRNA expression.
A. miRNA expressions inDGCR8KD and SC SKOV3 cells were detected in duplicate using Nano-string miRNA array, and the averaged signaling intensities were displayed in heat-maps for the highest up and down-regulated miRNAs (>2 fold) in DGCR8KD compared to SC control cells. B. Selected miRNA expressions were verified by polyA tailing real-time RT-PCR.
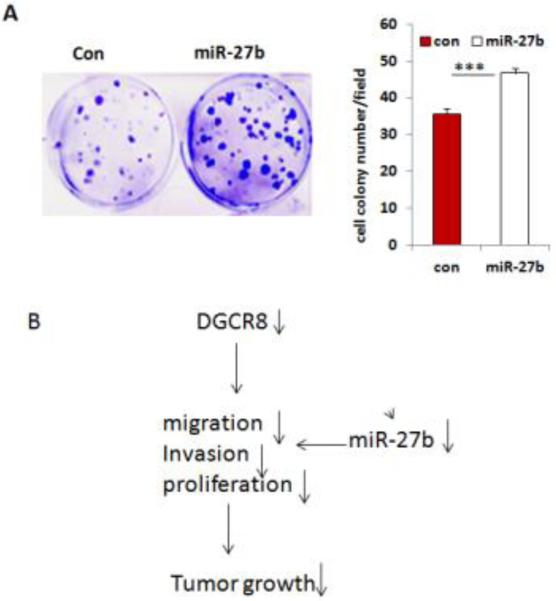
miR-27b contributes to oncogenic properties of DGCR8
To determine the contribution of these dysregulated miRNAs to DGCR8 function, we selected miR-27b for further investigation based on the finding that miR-27b is the most down-regulated miRNA upon DGCR8 knockdown. To determine the contribution of miR-27b to the oncogenic properties of DGCR8, we transduced SKOV3 cells using a lentiviral vector expressing miR-27b. We found that overexpression of miR-27b significantly promotes colony formation of SKOV3 cells (Fig.7A), suggesting that miR-27b may partially contributes to the properties of DGCR8 in SKOV3 cells (Fig.7B).
Figure 7. miR-27b promotes cell proliferation in ovarian cancer cells.
A. Colony formations were significantly increased in SKOV3 cells transduced with lentiviral miR-27b overexpression vector compared to controls. B. Molecular model for the oncogenic properties of DGCR8.
DISCUSSION
The correlation of DGCR8 and ovarian cancer
While DGCR8 is a double-stranded RNA binding protein and a key component of miRNA biogenesis, its role in ovarian cancer has not been examined. In this study, we revealed that the expression of DGCR8 in ovarian cancer was highly expressed compared to normal ovary tissue. Previous studies showed that DGCR8 was upregulated in skin epithelial cancer (13), cervical squamous cell carcinoma (14) and salivary gland pleomorphic adenoma (15), suggesting that DGCR8 may function as an oncogene in those cancers. Low Dicer and Drosha expression has been associated with advanced tumor stage, reduced survival and poor response to chemotherapy in ovarian cancer (10,11). However, Dicer was also found to be upregulated in ovarian serous carcinomas and associated with poor survival, which is similar to the findings we have observed on DGCR8 in ovarian cancer (12). Given that DGCR8 plays a critical role in miRNA maturation, DGCR8 may be an important biomarker for diagnosis and therapeutic target for ovarian cancer treatment.
The role of DGCR8 in ovarian cancer
The function of DGCR8 in ovarian cancer is unresolved. We found that knockdown of DGCR8 in ovarian cancer cells leads to the significant reduction in cell proliferation, migration, invasion, and sensitizes cells to apoptosis induced by chemotherapeutic drug cisplatin. Most importantly DGCR8 appears to play an important role in tumor growth in vivo, since DGCR8 knockdown markedly retards SKOV3 tumor growth. Moreover, silencing DGCR8 in ovarian cancer cells attenuates two cellular survival pathways ERK1/2 and PI3K/AKT, indicating that DGCR8 functions as an oncogene in part by promoting cell growth. A recent study also showed that silencing DGCR8 and Drosha in breast cancer cells inhibited colony formation (18).
miR-27b is an oncogenic miRNA in ovarian cancer
DGCR8 facilitates miRNA maturation by interacting with a RNase III enzyme Drosha. Silencing DGCR8 interferes miRNA maturation, and thus miRNA gene expression. By miRNA array profiling, we identified a specific subset of miRNAs regulated by DGCR8. miRNAs downregulated in DGCR8KD cells, include miR-10a, 221, 19a, 181a, and miR-27b, which have been found to oncogenic miRNAs in a variety of cancers. However their role in ovarian cancer has not been studied. For example, miR-10a is an oncomiRNA in cervical cancer (19), pancreatic cancer (20). miR-19a is an oncomiR in B-cell lymphoma (21) and breast cancer by targeting PTEN (22). miR-181a plays an oncogenic role in oral squamous cell carcinoma (23) and lymphoblastic leukemia (24). miR-221 functions as an oncomiRNA in melanoma (25), breast cancer (26) and prostate cancer (27). miR-27b promotes cell proliferation in breast cancer by targeting tumor suppressor of tumorigenicity 14 (ST14) (28). Silencing miR-27b in glioma was found to induce cell apoptosis and inhibit cell proliferation, suggesting that miR-27b functions as an oncomiRNA (29). In our current study, we showed that miR-27b is the most downregulated miRNA in DGCR8 KD cells as compared to controls, which leads to the hypothesis that miR-27b may function as an oncomiRNA and contributes to the oncogenic properties of DGCR8 in ovarian cancer. We showed a potential oncogenic role of miR-27b in ovarian cancer cells by overexpressing miR-27b using lentiviral vector, which resulted in significant increase of colony formation in miR-27b overexpressing cells. Therefore, we conclude that miR-27b at least in part contributes to the oncogenic properties of DGCR8 in ovarian cancer.
Supplementary Material
Acknowledgments
This work was supported by NIH awards HL095957, HD061420 and funding from UTHSC to J. Yue.
Footnotes
Authors’ Contributions
YG, PT, CY, ZL, ML, MS performed experiments. JY designed experiments and wrote the manuscript. LP provided reagents and wrote the manuscript. LL, ZZ and HL provided reagents.
Disclosure
None
References
- 1.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 2.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landthaler M, Yalcin A, Tuschl T. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Han L, Zhang A, Zhou X, Xu P, Wang GX, Pu PY, Kang CS. Int J Oncol. 2010;37:299–305. doi: 10.3892/ijo_00000678. [DOI] [PubMed] [Google Scholar]
- 5.Sand M, Gambichler T, Skrygan M, Sand D, Scola N, Altmeyer P, Bechara FG. Cancer Invest. 2010;28:649–653. doi: 10.3109/07357901003630918. [DOI] [PubMed] [Google Scholar]
- 6.Faber C, Horst D, Hlubek F, Kirchner T. Eur J Cancer. 2011;47:1414–1419. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Ma Z, Swede H, Cassarino D, Fleming E, Fire A, Dadras SS. PLoS One. 2011;6:e20494. doi: 10.1371/journal.pone.0020494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu JF, Shen W, Liu NZ, Zeng GL, Yang M, Zuo GQ, Gan XN, Ren H, Tang KF. Med Oncol. 2011;28:804–809. doi: 10.1007/s12032-010-9520-5. [DOI] [PubMed] [Google Scholar]
- 9.Shu GS, Yang ZL, Liu DC. Pathol Res Pract. 2012;208:392–397. doi: 10.1016/j.prp.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pampalakis G, Diamandis EP, Katsaros D, Sotiropoulou G. Clin Biochem. 2010;43:324–327. doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Flavin RJ, Smyth PC, Finn SP, Laios A, O'Toole SA, Barrett C, Ring M, Denning KM, Li J, Aherne ST, Aziz NA, Alhadi A, Sheppard BL, Loda M, Martin C, Sheils OM, O'Leary JJ. Mod Pathol. 2008;21:676–684. doi: 10.1038/modpathol.2008.33. [DOI] [PubMed] [Google Scholar]
- 13.Sand M, Skrygan M, Georgas D, Arenz C, Gambichler T, Sand D, Altmeyer P, Bechara FG. Mol Carcinog. 2011 doi: 10.1002/mc.20861. [DOI] [PubMed] [Google Scholar]
- 14.Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, Pett MR, Coleman N. J Pathol. 2007;212:368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Cairns M, Rose B, O'Brien C, Shannon K, Clark J, Gamble J, Tran N. Int J Cancer. 2009;124:2855–2863. doi: 10.1002/ijc.24298. [DOI] [PubMed] [Google Scholar]
- 16.Yue J, Sheng Y, Ren A, Penmatsa S. Biochem Biophys Res Commun. 2010;394:667–672. doi: 10.1016/j.bbrc.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Balazs L, Tigyi G, Yue J. Biochem Biophys Res Commun. 2011;408:369–374. doi: 10.1016/j.bbrc.2011.02.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peric D, Chvalova K, Rousselet G. Oncogene. 2012;31:2039–2048. doi: 10.1038/onc.2011.391. [DOI] [PubMed] [Google Scholar]
- 19.Long MJ, Wu FX, Li P, Liu M, Li X, Tang H. Cancer Lett. 2012;324:186–196. doi: 10.1016/j.canlet.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N, Toma H, Nakamura M, Nagai E, Hashizume M, Tanaka M. Ann Surg Oncol. 2012;19:2394–2402. doi: 10.1245/s10434-012-2252-3. [DOI] [PubMed] [Google Scholar]
- 21.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Z, Li Y, Huang K, Wagar N, Shim H. Pharm Res. 2011;28:3091–3100. doi: 10.1007/s11095-011-0570-y. [DOI] [PubMed] [Google Scholar]
- 23.Yang CC, Hung PS, Wang PW, Liu CJ, Chu TH, Cheng HW, Lin SC. J Oral Pathol Med. 2011;40:397–404. doi: 10.1111/j.1600-0714.2010.01003.x. [DOI] [PubMed] [Google Scholar]
- 24.Fragoso R, Mao T, Wang S, Schaffert S, Gong X, Yue S, Luong R, Min H, Yashiro-Ohtani Y, Davis M, Pear W, Chen CZ. PLoS Genet. 2012;8:e1002855. doi: 10.1371/journal.pgen.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini M, Colombo MP, Peschle C, Care A. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 26.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, Calin GA, Mukherjee P. Cancer Res. 2009;69:9090–9095. doi: 10.1158/0008-5472.CAN-09-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Wang L, Matyunina LV, Hill CG, McDonald JF. Gynecol Oncol. 2011;121:200–205. doi: 10.1016/j.ygyno.2010.12.339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.