Abstract
Factors contributing to the variability of serum 25-hydroxyvitamin D [25(OH)D] in response to a given dose of vitamin D supplementation are largely unknown. We examined whether DNA methylation levels of Cytochrome P450 (CYP) enzymes (CYP2R1, CYP24A1, CYP27A1, and CYP27B1) are potential biomarkers predicting vitamin D response variation. We randomized 446 white postmenopausal women to a calcium and vitamin D (1100 IU/day) intervention for at least 12 months. From these subjects, 18 with the highest 12-month increase in serum 25(OH)D were selected as “responders.” Another 18 with the lowest 12-month increase in serum 25(OH)D were selected as “non-responders.” DNA methylation levels between the groups were compared. To validate findings in the first study, association between DNA methylation levels and vitamin D response variation was studied in another 145 extended independent white postmenopausal women. In the first study, compared to non-responders, responders had significantly lower baseline DNA methylation levels in the promoter region of CYP2R1 (8% in the responders vs 30% in the non-responders, P=0.004), and CYP24A1 (13% in the responders vs 32% in the non-responders, P=0.001). In the validation study, for CYP2R1, baseline DNA methylation levels at eight CpG sites were negatively associated with 12-month increases in serum 25(OH)D (P<0.05). For CYP24A1, baseline DNA methylation levels at two CpG sites were also negatively associated with vitamin D response variation (r=−0.151, P=0.011; r=−0.131, P=0.025). These negative associations were consistent with the first study’s results. Our findings indicate that baseline DNA methylation levels of CYP2R1 and CYP24A1 may predict vitamin D response variation.
Keywords: CYP2R1, CYP24A1, Vitamin D response variation, Methylation
1. Introduction
Vitamin D deficiency is related to many diseases, such as rickets and fractures due to osteoporosis [1–4]. Serum 25-hydroxyvitamin D [25(OH)D] is the functional indicator of vitamin D status. Vitamin D supplementation is a simple, safe, and inexpensive approach for achieving adequate serum 25(OH)D levels. However, high variability of serum 25(OH)D in response to a given dose of vitamin D supplementation is widely observed [5–8]. The wide vitamin D dose-response variation is a daunting challenge for vitamin D deficiency treatment.
Previous studies have indicated that the ratio of serum 24,25-dihydroxyvitamin D to 25-hydroxyvitamin D [9], polymorphisms in vitamin D binding protein [10], or obesity status [11] contribute to the response variation. Those factors generally explain 8–16% of the response variation [9–11], leaving factors contributing to the wide response variation largely unknown.
There is a significant need to identify factors contributing to the vitamin D response variation. Rises in serum 25(OH)D occur gradually after beginning supplementation, reaching a plateau at about three months [12, 13]. This is a long wait to discover whether a dose is adequate, especially for some vulnerable individuals in whom the deficiency should be urgently treated (e.g., infants and pregnant women). Further, since there are concerns about vitamin D toxicity [14], high-dose supplements cannot be recommended for all. Therefore, it is vital to find biomarkers that can effectively identify individuals with high or low efficacy of vitamin D usage.
Cytochrome P450 (CYP) enzymes play key roles in vitamin D metabolism. Carried in the bloodstream to the liver, vitamin D from various sources is converted to circulating 25(OH)D by CYP2R1 and CYP27A1. Then, in the kidneys and other tissues, CYP27B1 converts 25(OH)D to the active form, 1,25-dihydroxyvitamin D [1,25(OH)2D]. Both 25(OH)D and 1,25(OH)2D can be inactivated after being hydroxylated at C24 by CYP24A1.
DNA methylation is one of the most common epigenetic mechanisms for change in gene expression. High methylation levels in a promoter region of a CYP gene may cause gene silencing, and thereby may affect vitamin D response variation. Previous studies indicated that DNA methylation levels in some CYP family genes were associated with vitamin D status [15], and methylation levels of the CYP family of genes can be modulated by drugs [16]. It is unknown whether DNA methylation levels of the CYP family can be used to predict vitamin D response variation. The primary aim of this study was to test whether the DNA methylation status of the CYP family of genes (CYP2R1, CYP27A1, CYP24A1, and CYP27B1) at baseline is associated with the serum 25(OH)D levels in response to vitamin D supplementation.
2. Methods and Materials
2.1. Subjects
Subjects for the study came from a completed NIH project (R01AG014683, (Calcium and Vitamin D Malnutrition in Elderly Women Study, CaMEWS). This was a population-based, four-year calcium and vitamin D intervention clinical trial. The participants were non-Hispanic white postmenopausal women aged ≥ 55 years who were randomly selected from a rural area of Nebraska (approximately 41.4 degrees N). To be eligible for the study, participants needed to be generally healthy with no history of renal calculi or Paget’s disease. Women with a history of cancer were included only if they had been cancer-free for 10 years or more. The study began in 2000 and was completed in 2005. In total, 1,179 non-Hispanic white postmenopausal women were enrolled, and 446 of them were assigned to receive supplemental calcium (1400–1500 mg/day) plus 1100 IU/day of vitamin D (Ca+D group). Details of the study have been reported previously [17, 18].
For this analysis, we selected subjects from the group randomized to the Ca+D group. We selected subjects who met the following criteria: 1) Body mass index (BMI), age, serum 25(OH)D, and vitamin D intake levels were available at both baseline and at the 12-month visit; 2) At least 500 μl frozen serum were stored at −80°C at both the baseline and 12-month visit; 3) DNA were successfully extracted from sera at both the baseline and 12-month visits. This analysis was approved by the Creighton University Institutional Review Board.
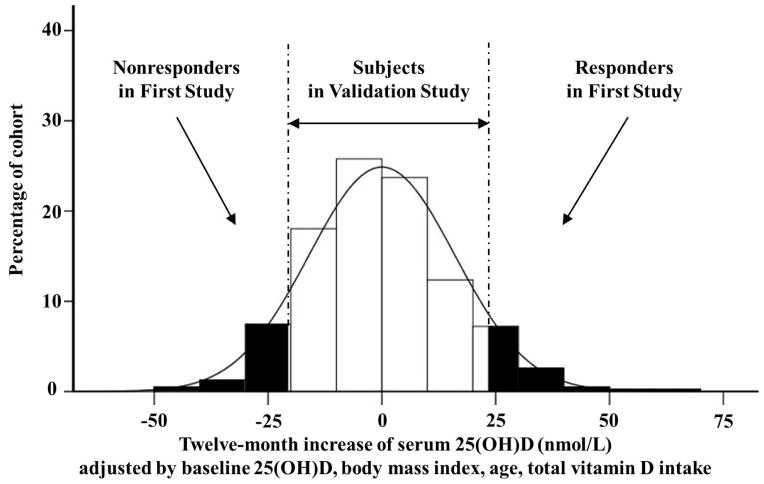
With these samples, we conducted two studies, a first study and a validation study. The first study used extreme samples selected from the 446 samples. After adjusting the 12-month serum 25(OH)D increments using the baseline 25(OH)D concentration, age, BMI, and total vitamin D intake, 18 responders and 18 non-responders were selected out of the two extreme tails in the distribution of the 12-month increase in serum 25(OH)D (Figure 1). Responders were subjects who had the highest adjusted 12-month increase in serum 25(OH)D, and non-responders had the lowest adjusted 12-month increase in serum 25(OH)D. In order to identify factors responsible for the vitamin D response variation in the population with modest vitamin D, all responders and non-responders should have baseline serum 25(OH)D levels of <75 nmol/L, which is considered an optimal vitamin D status in some publications [19, 20]. The Institute of Medicine recommends a 25(OH)D target of 50 nmol/L [21]. Considering that the average serum 25(OH)D level in our samples was ~71 nmol/L, we used 75 nmol/L as the cutoff line to select responders and non-responders. The characteristics of the responders and non-responders are presented in Table 1.
Figure 1. Subject frequency distribution of the adjusted 12-month increase of serum 25(OH)D after 1,100 IU/day vitamin D supplementation.
Eighteen responders and 18 non-responders were selected out of the extreme tails. Responders were defined as subjects who had the highest adjusted 12-month increase in serum 25(OH)D levels after vitamin D supplementation. Non-responders were subjects who had the lowest increase in adjusted serum 25(OH)D after vitamin D supplementation. Subjects in the validation study came from the remainder of the 446 subjects in the Calcium and Vitamin D Malnutrition in Elderly Women Study (CaMEWS).
Table 1.
Characteristics of subjects used in the first study and the validation study1
| First Study | Validation Study | |||
|---|---|---|---|---|
| Responders (n 18) | Non-responders (n=18) | Subjects for CYP2R1 (n=145) | Subjects for CYP24A1 (n=117) | |
|
| ||||
| Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | |
| Age (years) | 68.84 (7.68) | 70.05 (6.46) | 67.71 (8.45) | 67.72 (8.67) |
|
| ||||
| Height (m) | 1.64 (0.07) | 1.64 (0.07) | 1.62 (0.06) | 1.62 (0.06) |
|
| ||||
| Weight (kg) | 74.77 (12.02) | 79.39 (17.15) | 75.46 (15.22) | 76.55 (15.63) |
|
| ||||
| Body mass index (kg/m2) | 27.87 (4.61) | 29.48 (6.10) | 28.89 (5.57) | 29.26 (5.67) |
|
| ||||
| Baseline serum 25(OH)D (nmol/L) | 67.59 (11.30) | 58.55 (12.73) | 71.27 (20.30) | 71.80 (20.07) |
|
| ||||
| Serum 25(OH)D (nmol/L) after 12-month vitamin D supplementation2 | 109.70 (7.22)2 | 59.87 (10.79)2 | 92.91 (22.76) | 92.97 (21.75) |
Note:
Raw data are presented.
Serum 25(OH)D levels after 12-month vitamin D supplementation were significantly different between responders and non-responders, by t-test.
The validation study also used serum samples from the N=446 who met our selection criteria, but excluded the 36 subjects used in the first study. Due to the limitation in the amount of serum in the samples and minute amounts of DNA available in a serum sample, we had 145 subjects who had serum DNA at both baseline and 12-month visits for the CYP2R1 gene data analyses, and 117 subjects for the CYP24A1 gene data analyses. The characteristics of the subjects for the validated study are presented in Table 1.
2.2. Clinical measurements
Serum 25(OH)D in each participant was measured by radioimmunoassay using the IDS kit (Fountain Hills, AZ) at baseline and at the 12-month visit in the Osteoporosis Research Center (ORC) at Creighton University. The ORC laboratory participates in the international quality assessment process for vitamin D assays (DEQAS) for 25(OH)D. Over the course of the study, the findings from test samples were always close to the international mean. The coefficient of variation (CV) for intra-assay was 5.1% and inter-assay was 7.9%.
Other factors that may influence variation in serum 25(OH)D were collected, such as age, height, weight, BMI, blood collection date, serum 25(OH)D collection date, serum calcium concentration, daily dietary vitamin D intake, physical activity, cigarette-smoking status, adherence to the trial vitamin D supplementation, and self-selected vitamin D supplementation outside of the study. All the related factors were included in the data analyses.
In the clinical trial, adherence to supplementation was determined by weighing each bottle of vitamin D tablets before distributing the bottle and again when it was returned by the participant. Vitamin D treatment compliance rate for the clinical study averaged 85.7% [17].
2.3. DNA extraction from frozen serum
Variable amounts of circulating genomic DNA exists in serum in healthy humans. We extracted serum genomic DNA according to the protocol of the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA), with some minor revision [22]. In order to achieve high DNA yield, we began with 400 μl frozen serum, instead of 200 μl, which was suggested by the manufacturer’s protocol. Reagent amounts were adjusted to meet the need for increased initial serum volume.
2.4. DNA methylation measurement
2.4.1. Methylation specific PCR (MSP) for the first study
For the first study, we used MSP to measure DNA methylation. Bisulfite treatment was carried out using 400 ng of serum DNA with the EZ DNA Methylation-Direct kit (Zymo Research, Orange, CA). This process deaminates unmethylated cytosine residues to uracil, leaving methylated cytosine residues unchanged [23].
In order to increase the amount of DNA for DNA methylation measurement, bisulfite converted gene specific primers (BS) were designed. PCR reactions were conducted with 5 ng of bisulfite-modified DNA in a total volume of 25 μl for 35 cycles using Roche Diagnostic Corporation (Indianapolis, IN) FastStart Taq DNA Polymerase (1.0 U), MgCl2 solution (3.5 mM), dNTPs (0.2 mM), sense primer and antisense primer (0.3 μM) (Supplementary Table S1), with denaturation at 95°C for 30 seconds, annealing at a temperature reported in Supplementary Table S1 for 45 seconds, and extension at 72°C for 1 minute.
To determine DNA methylation levels in the promoter region of the CYP family of genes, methylation-specific primers were designed (Supplementary Table S1). MSP for each gene was carried out as described above using primers representing methylated and unmethylated CpGs and 2.5 ng of purified (Qiagen, Valencia, CA) BS PCR product as template, fully CpG methylated DNA as a positive control, and bisulfite-treated Roche Human Genomic DNA and no-template control DNA as negative controls. After MSP, PCR products underwent electrophoresis on 0.8% agarose gel and were stained with ethidium bromide and visualized under a Bio-Rad Laboratories (Hercules, CA) Gel-Doc UV illuminator. All the bands were read using a Bio-Rad Gel Imager (Hercules, CA). The methylation level was calculated as:
2.4.2. Pyrosequencing for validation study
In order to identify specific CpG sites contributing to vitamin D response variation, we used pyrosequencing for DNA methylation measurement in the validation study. First, 400 ng of serum DNA was treated with sodium bisulfite using the EZ DNA Methylation-Direct kit (Zymo Research, Orange, CA). To perform PCR reactions, 42 ng of bisulfite-modified DNA was used as a template. The PCR reactions were performed in a total volume of 50 μl for 40 cycles using Roche Diagnostic Corporation (Indianapolis, IN) FastStart Taq DNA Polymerase (1.0 U), MgCl2 solution (3.5 mM), dNTPs (0.2 mM), sense primer (0.24 μM), antisense primer (0.18 μM) (Supplementary Table S2), with denaturation at 95°C for 30 seconds, annealing temperature for 45 seconds, indicated in Supplementary Table 2, and extension at 72°C for 1 minute. Roche Diagnostic Corporation (Indianapolis, IN) methylated human genomic DNA was used as a positive control. Sodium bisulfite-treated Roche human genomic DNA (1 μg) served as negative control. All PCR products underwent electrophoresis on 0.8% agarose gel and were stained with ethidium bromide and visualized for appropriate and pure product before proceeding with all analyses using a Bio-Rad Laboratories (Hercules, CA) Gel-Doc UV illuminator. Methylation percentage of each CpG was determined using a Qiagen (Valencia, CA) Pyromark Q24 pyrosequencer; sequencing primers are indicated in Supplementary Table S2, and were used according to the manufacturer’s manual.
2.5. Statistical Analyses
Statistical analyses were performed with SPSS version 15.0. Unless otherwise stated, all tests were two-tailed, and P<0.05 was considered statistically significant.
In the first study, descriptive analyses were performed for the serum 25(OH)D status and clinical information. We calculated vitamin D response variation using the 12-month increases in serum 25(OH)D levels, which reduced the confounding effect of the season. Potential confounding factors (e.g., BMI, season) were first assessed for their importance to the 12-month increase in serum 25(OH)D using the stepwise linear regression model. Then the increases in serum 25(OH)D levels were adjusted using significant factors, such as baseline serum 25(OH)D concentration, age, BMI, and total amount of vitamin D intake. Repeated measures ANOVA tests were conducted to compare the differences in DNA methylation levels between responders and non-responders, and between baseline and after a 12-month vitamin D intervention.
In the validation study, paired t-tests were applied to analyze the change in DNA methylation levels after a 12-month vitamin D intervention. Bonferroni corrections were performed after the t-tests. Bivariate correlations were used to examine the correlation between DNA methylation levels and adjusted 12-month increase in serum 25(OH)D. The adjusted factors included baseline serum 25(OH)D concentration, age, BMI, and total amount of vitamin D intake. The contribution of CpG sites to vitamin D response variation was determined using a nonlinear regression model.
3. Results
In order to investigate the association between DNA methylation status and vitamin D response variation, we conducted two studies, the first a study in extreme samples from CaMEWS and the second a validation study using independent samples from CaMEWS.
3.1. First study in extreme samples
3.1.1. Basic characteristics of responders and non-responders
The 18 responders and 18 non-responders for the first study were selected according to their vitamin D response value. Figure 1 depicts the samples used for the studies. They were selected from the extreme tails of the distribution of the 12-month increase in serum 25(OH)D. The responder and non-responder groups had no statistically significant differences in age, height, weight, BMI, and baseline serum 25(OH)D concentration (Table 1). After 12 months of vitamin D supplementation, as expected, responders had significantly higher serum 25(OH)D levels compared to non-responders.
3.1.2. Comparing DNA methylation levels between responders and non-responders at baseline
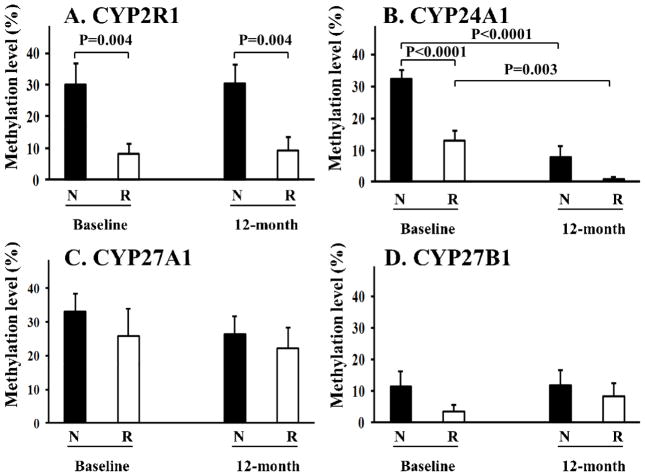
In order to identify genes predicting vitamin D response variation, we used MSP to measure the general DNA methylation levels of the promoter regions of CYP27A1, CYP2R1, CYP27B1, and CYP24A1. Figure 2 presents the MSP results of the 18 responders and 18 non-responders at baseline and after a 12-month vitamin D supplementation.
Figure 2. Methylation levels in responders and non-responders at baseline and after a 12-month vitamin D supplementation.
For the CYP2R1 gene, non-responders had significantly higher DNA methylation levels in the promoter region, compared to that in the responders at baseline and 12-month visit (A). DNA methylation level of the CYP24A1 gene was significantly higher in the non-responders compared to responders at baseline, but not at 12-month visit (B). DNA methylation level of the CYP24A1 gene was significantly reduced in both responders and non-responders at 12-month visit (B). For CYP27A1 and CY27B1, no significant differences were identified in DNA methylation levels between responders and non-responders at baseline or 12-month visit (C and D). “R” denotes responders (n=18) and “N” denotes non-responders (n=18). Data are expressed as means ± SE.
At baseline, for the CYP2R1 gene, which converts vitamin D to 25(OH)D, non-responders had significantly higher DNA methylation levels in the promoter region compared to that in the responders [30±27% (mean±SD) in the non-responders vs 8±13% (mean±SD) in responders, P=0.004] (Figure 2A). Similarly, the baseline DNA methylation level of the CYP24A1 gene was significantly higher in non-responders compared to responders [32±12% (mean±SD) in non-responders, vs. 13±13% (mean±SD) in responders, P=0.001] (Figure 2B). For CYP27A1 and CY27B1, we did not identify significant differences in DNA methylation levels between responders and non-responders at baseline (Figure 2C and 2D).
3.1.3. The effect of vitamin D supplementation on DNA methylation levels in responders and non-responders
To determine whether vitamin D would affect DNA methylation levels in vivo in humans, we compared MSP results for the four genes (CYP27A1, CYP2R1, CYP27B1, and CYP24A1) at baseline and after a 12-month vitamin D supplementation. For CYP2R1, CYP27A1, and CYP27B1, no statistically significant difference was identified between DNA methylation levels at the two time points (Figure 2A, 2C, 2D). For the CYP24A1 gene, we found a significant reduction in the DNA methylation levels in both responders [13±13% (mean±SD) at baseline, and 1±3% (mean±SD) at 12-months, P=0.001] and non-responders [32±12% (mean±SD) at baseline, and 8±14% (mean±SD) at 12-months, P=0.003] (Figure 2B). Among the four tested genes (CYP2R1, CYP24A1, CYP27A1, and CYP27B1), the time by treatment was significant (P=0.028) only for CYP24A1.
3.2. Validation study in extended independent samples
In the first study, non-responders had higher DNA methylation levels of the CYP2R1 and CYP24A1 genes at baseline. Based on the first results, we hypothesized that DNA methylation levels of the CYP2R1 and CYP24A1 genes at baseline are negatively associated with a 12-month increase in serum 25(OH)D levels. In order to test the hypothesis, we conducted a validation study in extended independent samples from CaMEWs.
3.2.1. Basic characteristics of the samples
The characteristics of the subjects for the validation study are presented in Table 1. The average baseline serum 25(OH)D for the participants was about 71 nmol/L. Baseline serum 25(OH)D levels were not associated with methylation levels of CYP2R1 and CYP24A1 (data not shown). After a 12-month 1,100 IU/day vitamin D supplementation, the subjects raised their serum 25(OH)D from ~71 nmol/L to ~92 nmol/L.
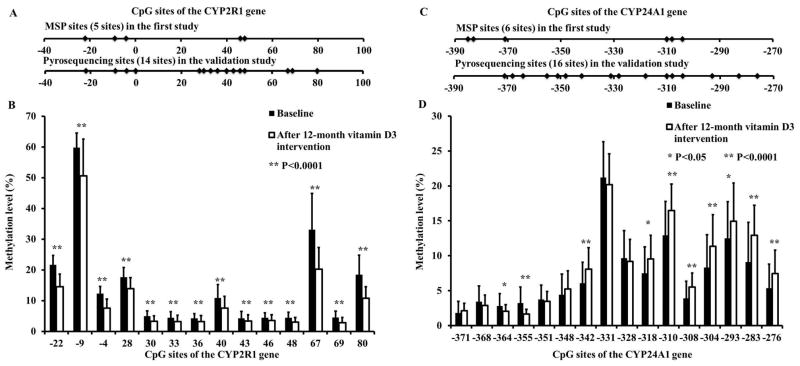
For the CYP2R1 gene, 14 CpG sites of the CYP2R1 gene were scanned. Among them, five CpG sites were shared in both the pyrosequencing analysis in the validation study and the MSP analysis in the first study (Figure 3A). Sixteen CpG sites were scanned in the CYP24A1 gene, and four of them were shared in both the validation study and first study (Figure 3C).
Figure 3. Methylation levels of CYP2R1 and CYP24A1 at baseline and after a 12-month vitamin D supplementation.
For the CYP2R1 gene, DNA methylation levels of each tested CpG site were significantly lower at the 12-month visit compared to the baseline visit (A and B). For the CYP24A1 gene, CpG sites reacted differently to the vitamin D intervention. Among the 16 tested CpG sites, the DNA methylation levels significantly decreased in two CpG sites, and significantly increased in another eight sites; six were unchanged (C and D). Data are expressed as means ± SD.
3.2.2. Association between baseline DNA methylation levels and vitamin D response variation
For CYP2R1, baseline DNA methylation levels at eight CpG sites (−4C, +28C, +30C, +33C, +40C, +43C, +69C, +80C) were negatively associated with the 12-month increase in serum 25(OH)D (P<0.05 for each site, Table 2). Baseline DNA methylation levels at +30C and +40C were still negatively associated with the 12-month increase in serum 25(OH)D, even after the conservative Bonferroni correction. The average of the 14 CpG sites of CYP2R1 was also negatively associated with the 12-month increase in serum 25(OH)D (R=−0.182, P=0.028).
Table 2.
Negative correlations between baseline DNA methylation levels of CpG sites in the CYP2R1 and CYP24A1 genes and adjusted vitamin D response variation
| Significant CpG sites | r value | P value1 |
|---|---|---|
|
| ||
| CYP2R1 (14 CpG sites examined in 145 subjects) | ||
| −4C | −0.133 | 0.025 |
| +28C | −0.134 | 0.022 |
| +30C | −0.188 | 0.002 |
| +33C | −0.122 | 0.046 |
| +40C | −0.174 | 0.003 |
| +43C | −0.147 | 0.014 |
| +69C | −0.128 | 0.033 |
| +80C | −0.140 | 0.015 |
|
| ||
| CYP24A1 (16 CpG sites examined in 117 subjects) | ||
|
| ||
| −342C | −0.105 | 0.011 |
| −293C | −0.131 | 0.025 |
Note:
P values were generated before Bonferroni corrected, by bivariate correlations. After Bonferroni correction, +30C and +40C sites of CYP2R1 remain significant.
For CYP24A1, baseline DNA methylation levels at −342C and −293C were also negatively associated with vitamin D response variation (r=−0.151, P=0.011; r =−0.131, P=0.025, respectively) (Table 2).
These negative associations were consistent with our hypothesis. In total, the detected 10 CpG sites of the two genes explained 6.4% of the variation in the 12-month increase of serum 25(OH)D.
3.2.3. The effect of vitamin D supplementation on DNA methylation in the validation study
For the CYP2R1 gene, DNA methylation levels of each tested CpG site were significantly lower at the 12-month visit compared to the baseline visit (P=0.001 for each site, Figure 3B). That is, vitamin D supplementation significantly decreased DNA methylation levels in the CYP2R1 gene in all the 14 CpG sites [average DNA methylation level: 15±3% (mean±SD) at baseline vs. 11±3% (mean±SD) at 12-month visit, P=0.001] (Figure 3B). For the CYP24A1 gene, CpG sites reacted differently to the vitamin D intervention. Among the 16 tested CpG sites, the DNA methylation levels significantly decreased in two CpG sites, and significantly increased in another eight sites; six were unchanged (Figure 3D).
4. Discussion
To the best of our knowledge, this is the first study to test whether the DNA methylation status of the CYP family of genes (CYP2R1, CYP27A1, CYP24A1, and CYP27B1) is associated with vitamin D response variation, and whether vitamin D supplementation will affect DNA methylation of the CYP family of genes in vivo. Our study indicates that the baseline DNA methylation levels of CYP2R1 and CYP24A1 genes are associated with serum 25(OH)D levels in response to vitamin D supplementation.
In our first study, we found that the baseline DNA methylation levels of the CYP2R1 gene significantly differed between responders and non-responders. The results were further verified in the validation study. However, for another 25-hydroxlase, CYP27A1, we did not find this difference. This finding is consistent with the recent finding that CYP2R1 is a major vitamin D 25-hydroxylase [24]. CYP2R1 showed higher affinity and specificity for vitamin D than CYP27A1 [24, 25]. A genetic mutation of CYP2R1 will cause vitamin D deficiency [26], while no genetic mutation was associated with vitamin D deficiency for the CYP27A1 gene. Recent large-scale genome-wide association studies have also emphasized the important CYP2R1 association with baseline 25(OH)D levels [27, 28], but the association with CYP27A1 was not found.
In our first study, non-responders had higher baseline DNA methylation levels in the CYP2R1 gene compared to responders. High DNA methylation levels in the promoter region of the gene may lead to lower levels of serum 25(OH)D. On the other hand, low DNA methylation levels in responders allow normal expression of the CYP2R1 gene and conversion of vitamin D to serum 25(OH)D. Consistent with the CYP2R1 gene, DNA methylation levels of the CYP24A1 gene were significantly higher in non-responders than in responders at the baseline (Figure 2B). However, due to the significant reduction of CYP24A1 gene in response to vitamin D supplementation in both groups, there is no difference in the gene’s DNA methylation levels between responders and non-responders after the 12-month vitamin D intervention.
The different patterns of the DNA methylation levels of the CYP2R1 gene and CYP24A1 gene may explain the diverse vitamin D response between responders and non-responders. CYP2R1 and CYP24A1 are key enzymes responsible for the synthesis and breakdown of serum 25(OH)D, respectively. The distinct function of the two enzymes allows us to propose a hypothesis (Figure 4) of how the DNA methylations of the two genes may influence vitamin D response variation between responders and non-responders. As shown in Figure 4, we assumed there were two phases of serum 25(OH)D levels: intermediate phase and final phase. The “intermediate” phase serum 25(OH)D was synthesized by CYP2R1, and the “final” phase serum 25(OH)D was the level after degradation by CYP24A1. At baseline, compared to non-responders, relatively low methylation levels of CYP2R1 and CYP24A1 led to high “intermediate” and similar “final” serum 25(OH)D levels in the responders (Figure 4A and Figure 2A, 2B). That explains why we didn’t find a significant difference in the baseline serum 25(OH)D levels between responders and non-responders (Figure 4A). During the 12-month vitamin D intervention, the amount of vitamin D intake was dramatically increased for both responders and non-responders. Since DNA methylation level of CYP2R1 was relatively lower in responders (Figure 2A), responders were expected to have higher “intermediate” 25(OH)D levels than non-responders (Figure 4B). Meanwhile, the DNA methylation levels of CYP24A1 were reduced significantly in both responders and non-responders after vitamin D supplementation (Figure 2B). Higher “intermediate” 25(OH)D and similar degradation rate of 25(OH)D caused greater increase in “final” 25(OH)D level in responders than that in the non-responder (Figure 4B). All of this may explain why responders have higher serum 25(OH)D levels at the 12-month visit than non-responders (Figure 4B).
Figure 4. The hypothesized mechanism for DNA methylation levels influence on vitamin D response variation in the first study.
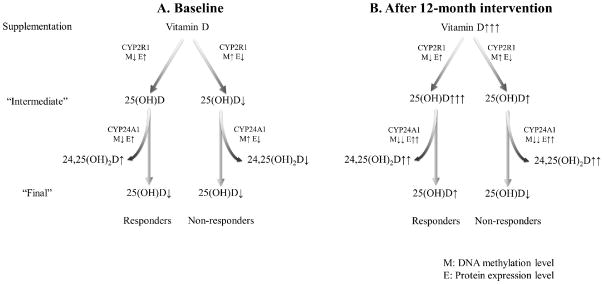
The “Intermediate” phase denotes the 25(OH)D levels converted by CYP2R1. The “Final” phase indicates the 25(OH)D levels after degradation by CYP24A1. The number of arrows indicates the levels of DNA methylation, enzyme expression or 25(OH)D concentration. “M” denotes methylation level and “E” denotes expression level.
In our validation study, we also found the inverse correlation between vitamin D response and baseline methylation levels of CYP2R1 and CYP24A1. The results confirmed our hypothesis deduced from the first study. The contribution of the methylation levels of the two genes to the vitamin D response variation are moderate (R2=6.4%), similar to other known factors, such as genotype of vitamin D binding protein (R2=8.5%) [10], and body fat mass (R2=8.4%) [11]. Our results indicate that vitamin D supplementation may change the DNA methylation levels in the promoter regions of some of the CYP family of genes in vivo, although the specific results are different between our first study and the validation study. In the first study, DNA methylation levels of the CYP2R1 gene were not different between the baseline and 12-month visit in either responders or non-responders (Figure 2A), while the methylation level of CYP24A1 decreased significantly (Figure 2B). However, in the validation study, we found that DNA methylation for all CpG sites of the CYP2R1 gene were significantly lower after vitamin D intervention (Figure 3B), while the CpG sites of CYP24A1 showed various responses. Several factors may contribute to the inconsistent results: 1) Sample difference. The first study focused on extreme and small samples, but the validation study focused on a general population. 2) Approach difference. The MSP approach in the first study offers general DNA methylations for all examined CpG sites (five CpG sites for CYP2R1 and six sites for CYP24A1). The pyrosequencing approach provides specific methylation level for each CpG site (14 CpG sites for CYP2R1 and 16 CpG sites for CYP24A1).
This study had several limitations. First, the nature of the parent study, CaMEWS, limits the sample sizes and available tissue for this study. For example, we didn’t archive patients’ tissue other than their serum. Second, using genomic DNA from archived serum has advantage and disadvantages. On one hand, serum is easy to obtain from a human subject. Our experience with >200 samples in another study [29] indicated that serum DNA exists and can be isolated from fresh serum/plasma. On the other hand, the source of serum DNA makes the explanation of our results complicated. DNA methylation is tissue-specific. In healthy subjects, serum DNA derives from apoptosis of lymphocytes [30]. Therefore, the DNA methylation detected in the study is an integrated reflection of the DNA methylation levels in these cells. Third, we didn’t measure the serum 24,25(OH)2D levels, and no serum samples were collected between baseline and the 12-month visit. The lack these data may make using our results difficult when explaining the relationship between methylation patterns and enzymatic activity. Fourth, we only investigated the association between vitamin D dose responses with methylation levels of the key enzymes in vitamin D metabolism. Vitamin D dose may affect methylation levels of other genes which also contribute to the response status of vitamin D level.
In summary, our findings suggest that DNA methylation levels of CYP2R1 and CYP24A1 at baseline may be potential biomarkers in predicting vitamin D response variation. That is, subjects with high DNA methylation levels in the two genes may need high vitamin D supplementation to reach optimal serum 25(OH)D levels. Meanwhile, our study indicated that vitamin D supplementation may change the DNA methylation levels of the CYP family of genes in vivo. Studies with large sample sizes are needed to verify the results from the study. In addition, this study was limited to non-Hispanic white postmenopausal women. Studies in males and other ethnicities are needed to test the population generality of our results.
Supplementary Material
Highlights.
We examined the methylation levels of CYPs family in patients who had vitamin D supplementation.
Baseline methylation level of CYP2R1 is negatively associated with vitamin D response variation.
Baseline methylation level of CYP24A1 is negatively associated with vitamin D response variation.
Baseline DNA methylation levels of CYP2R1 and CYP24A1 may predict vitamin D response variation.
Acknowledgments
The authors’ responsibilities were as follows: LJZ, YZ, RRR, and JM designed the study; DTG supervised and conducted the subject recruitment; YZ, XX, AY, BZ, and HWW conduct the experiment and performed the statistical data analysis; YZ and LJZ prepared the first draft of the manuscript; LJZ coordinated and supervised the study; RRR, JMP, HWD and LLM contributed to the data interpretation and critical review; and all authors contributed to reading and approval of the final content of the manuscript.
Abbreviations used
- 1,25(OH)2D
1,25-dihydroxyvitamin D
- 25(OH)D
25-hydroxyvitamin D
- BMI
Body mass index
- BS
Bisulfite converted gene specific primers
- CaMEWS
Calcium and Vitamin D Malnutrition in Elderly Women Study
- CV
Coefficient of variation
- CYP
Cytochrome P450
- MSP
Methylation-specific PCR
- ORC
Osteoporosis Research Center
Footnotes
This work was partially supported by grants from Cancer and Smoking Disease Research Bone Biology Program, the Nebraska Tobacco Settlement Biomedical Research Development Award, an NIH grant (3R01CA129488-01A2S2), and a grant from State of Nebraska Cancer and Smoking Disease Research Program (LB595). Yu Zhou, Xiaojing Xu, An Ye and Lan-Juan Zhao were partially supported by Dr. Zhao’s start up fund from Tulane University. Dr. Hong-Wen Deng is partially supported by grants from National Institute of Health (R01AG026564, R01AR050496, R01AR057049, and R03TW008221), a SCOR (Specialized Center of Research) grant (P50 AR055081) supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) and the Office of Research on Women’s Health (ORWH), and Edward G. Schlieder Endowment from Tulane University.
Author Disclosure: Y. Zhou, LJ Zhao, X Xu, A. Ye, D. Travers-Gustafson, B. Zhou, HW Wang, W Zhang, LL Hamm, HW Deng, RR Recker, and JM Lappe have no conflicts of interest.
This clinical trial entitled Calcium and Vitamin D Malnutrition in Elderly Women Study (CaMEWS) was registered at clinicaltrials.gov as NCT00352170.
Conference presentation: Abstracts of the first study and the validation study were submitted to the American Society for Bone and Mineral Research (ASBMR) 2010 and 2011 Annual Meetings, respectively. Oral presentations were delivered at the meetings.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wick JY. Spontaneous fracture: multiple causes. Consult Pharm. 2009;24(2):100–102. 105–108, 110–102. doi: 10.4140/tcp.n.2009.100. [DOI] [PubMed] [Google Scholar]
- 2.Grant AM, Avenell A, Campbell MK, McDonald AM, MacLennan GS, McPherson GC, Anderson FH, Cooper C, Francis RM, Donaldson C, Gillespie WJ, Robinson CM, Torgerson DJ, Wallace WA. Oral vitamin D3 and calcium for secondary prevention of low-trauma fractures in elderly people (Randomised Evaluation of Calcium Or vitamin D, RECORD): a randomised placebo-controlled trial. Lancet. 2005;365(9471):1621–1628. doi: 10.1016/S0140-6736(05)63013-9. [DOI] [PubMed] [Google Scholar]
- 3.Garland CF, Gorham ED, Mohr SB, Garland FC. Vitamin D for cancer prevention: global perspective. Ann Epidemiol. 2009;19(7):468–483. doi: 10.1016/j.annepidem.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 4.Haddow JE. Vitamin D and rickets: much has been accomplished, but there is room for improvement. J Med Screen. 2011;18(2):58–59. doi: 10.1258/jms.2011.011059. [DOI] [PubMed] [Google Scholar]
- 5.Cashman KD, Hill TR, Lucey AJ, Taylor N, Seamans KM, Muldowney S, Fitzgerald AP, Flynn A, Barnes MS, Horigan G, Bonham MP, Duffy EM, Strain JJ, Wallace JM, Kiely M. Estimation of the dietary requirement for vitamin D in healthy adults. Am J Clin Nutr. 2008;88(6):1535–1542. doi: 10.3945/ajcn.2008.26594. [DOI] [PubMed] [Google Scholar]
- 6.Cashman KD, Wallace JM, Horigan G, Hill TR, Barnes MS, Lucey AJ, Bonham MP, Taylor N, Duffy EM, Seamans K, Muldowney S, Fitzgerald AP, Flynn A, Strain JJ, Kiely M. Estimation of the dietary requirement for vitamin D in free-living adults >=64 y of age. Am J Clin Nutr. 2009;89(5):1366–1374. doi: 10.3945/ajcn.2008.27334. [DOI] [PubMed] [Google Scholar]
- 7.Aloia JF, Patel M, Dimaano R, Li-Ng M, Talwar SA, Mikhail M, Pollack S, Yeh JK. Vitamin D intake to attain a desired serum 25-hydroxyvitamin D concentration. Am J Clin Nutr. 2008;87(6):1952–1958. doi: 10.1093/ajcn/87.6.1952. [DOI] [PubMed] [Google Scholar]
- 8.Genaro PS, Pereira GA, Pinheiro MM, Szejnfeld VL, Martini LA. Relationship between nutrient intake and vitamin D status in osteoporotic women. Int J Vitam Nutr Res. 2007;77(6):376–381. doi: 10.1024/0300-9831.77.6.376. [DOI] [PubMed] [Google Scholar]
- 9.Wagner D, Hanwell HE, Schnabl K, Yazdanpanah M, Kimball S, Fu L, Sidhom G, Rousseau D, Cole DE, Vieth R. The ratio of serum 24,25-dihydroxyvitamin D(3) to 25-hydroxyvitamin D(3) is predictive of 25-hydroxyvitamin D(3) response to vitamin D(3) supplementation. J Steroid Biochem Mol Biol. 2011;126(3–5):72–77. doi: 10.1016/j.jsbmb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Fu L, Yun F, Oczak M, Wong BY, Vieth R, Cole DE. Common genetic variants of the vitamin D binding protein (DBP) predict differences in response of serum 25-hydroxyvitamin D [25(OH)D] to vitamin D supplementation. Clin Biochem. 2009;42(10–11):1174–1177. doi: 10.1016/j.clinbiochem.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Blum M, Dallal GE, Dawson-Hughes B. Body size and serum 25 hydroxy vitamin D response to oral supplements in healthy older adults. J Am Coll Nutr. 2008;27(2):274–279. doi: 10.1080/07315724.2008.10719700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieth R. Vitamin D supplementation, 25-hydroxyvitamin D concentrations, and safety. Am J Clin Nutr. 1999;69(5):842–856. doi: 10.1093/ajcn/69.5.842. [DOI] [PubMed] [Google Scholar]
- 13.Bacon CJ, Gamble GD, Horne AM, Scott MA, Reid IR. High-dose oral vitamin D3 supplementation in the elderly. Osteoporos Int. 2009;20(8):1407–1415. doi: 10.1007/s00198-008-0814-9. [DOI] [PubMed] [Google Scholar]
- 14.Binkley N, Gemar D, Engelke J, Gangnon R, Ramamurthy R, Krueger D, Drezner MK. Evaluation of ergocalciferol or cholecalciferol dosing, 1,600 IU daily or 50,000 IU monthly in older adults. J Clin Endocrinol Metab. 2011;96(4):981–988. doi: 10.1210/jc.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu H, Wang X, Shi H, Su S, Harshfield GA, Gutin B, Snieder H, Dong Y. A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. The Journal of pediatrics. 2013;162(5):1004–1009. e1001. doi: 10.1016/j.jpeds.2012.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escher G, Hoang A, Georges S, Tchoua U, El-Osta A, Krozowski Z, Sviridov D. Demethylation using the epigenetic modifier, 5-azacytidine, increases the efficiency of transient transfection of macrophages. J Lipid Res. 2005;46(2):356–365. doi: 10.1194/jlr.D400014-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Lappe JM, Davies KM, Travers-Gustafson D, Heaney RP. Vitamin D status in a rural postmenopausal female population. J Am Coll Nutr. 2006;25(5):395–402. doi: 10.1080/07315724.2006.10719551. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Zhao LJ, Watson P, Zhang Q, Lappe JM. The effect of calcium and vitamin D supplementation on obesity in postmenopausal women: secondary analysis for a large-scale, placebo controlled, double-blind, 4-year longitudinal clinical trial. Nutr Metab (Lond) 2010;7:62. doi: 10.1186/1743-7075-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson-Hughes B, Heaney RP, Holick MF, Lips P, Meunier PJ, Vieth R. Estimates of optimal vitamin D status. Osteoporos Int. 2005;16(7):713–716. doi: 10.1007/s00198-005-1867-7. [DOI] [PubMed] [Google Scholar]
- 20.Holick MF, Binkley NC, Bischoff-Ferrari HA, Gordon CM, Hanley DA, Heaney RP, Murad MH, Weaver CM. Guidelines for preventing and treating vitamin D deficiency and insufficiency revisited. J Clin Endocrinol Metab. 2012;97(4):1153–1158. doi: 10.1210/jc.2011-2601. [DOI] [PubMed] [Google Scholar]
- 21.I.o. Medicine. Dietary reference intakes for calcium and vitamin D. Washington, DC: National Academies Press; 2011. [PubMed] [Google Scholar]
- 22.Chen XQ, Stroun M, Magnenat JL, Nicod LP, Kurt AM, Lyautey J, Lederrey C, Anker P. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996;2(9):1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng JB, Motola DL, Mangelsdorf DJ, Russell DW. De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxilase. J Biol Chem. 2003;278(39):38084–38093. doi: 10.1074/jbc.M307028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinkyo R, Sakaki T, Kamakura M, Ohta M, Inouye K. Metabolism of vitamin D by human microsomal CYP2R1. Biochem Biophys Res Commun. 2004;324(1):451–457. doi: 10.1016/j.bbrc.2004.09.073. [DOI] [PubMed] [Google Scholar]
- 26.Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci U S A. 2004;101(20):7711–7715. doi: 10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, Jacobs EJ, Ascherio A, Helzlsouer K, Jacobs KB, Li Q, Weinstein SJ, Purdue M, Virtamo J, Horst R, Wheeler W, Chanock S, Hunter DJ, Hayes RB, Kraft P, Albanes D. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19(13):2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, Peltonen L, Cooper JD, O’Reilly PF, Houston DK, Glazer NL, Vandenput L, Peacock M, Shi J, Rivadeneira F, McCarthy MI, Anneli P, de Boer IH, Mangino M, Kato B, Smyth DJ, Booth SL, Jacques PF, Burke GL, Goodarzi M, Cheung CL, Wolf M, Rice K, Goltzman D, Hidiroglou N, Ladouceur M, Wareham NJ, Hocking LJ, Hart D, Arden NK, Cooper C, Malik S, Fraser WD, Hartikainen AL, Zhai G, Macdonald HM, Forouhi NG, Loos RJ, Reid DM, Hakim A, Dennison E, Liu Y, Power C, Stevens HE, Jaana L, Vasan RS, Soranzo N, Bojunga J, Psaty BM, Lorentzon M, Foroud T, Harris TB, Hofman A, Jansson JO, Cauley JA, Uitterlinden AG, Gibson Q, Jarvelin MR, Karasik D, Siscovick DS, Econs MJ, Kritchevsky SB, Florez JC, Todd JA, Dupuis J, Hypponen E, Spector TD. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376(9736):180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bu FX, Armas L, Lappe J, Zhou Y, Gao G, Wang HW, Recker R, Zhao LJ. Comprehensive association analysis of nine candidate genes with serum 25-hydroxy vitamin D levels among healthy Caucasian subjects. Hum Genet. 2010;128(5):549–556. doi: 10.1007/s00439-010-0881-9. [DOI] [PubMed] [Google Scholar]
- 30.Takayama T, Yamada S, Watanabe Y, Hirata K, Nagai A, Nakamura I, Bunai Y, Ohya I. Origin of DNA in human serum and usefulness of serum as a material for DNA typing. Leg Med (Tokyo) 2001;3(2):109–113. doi: 10.1016/s1344-6223(01)00018-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.