Abstract
In this review, we compare and contrast current knowledge about in-vitro and in-vivo protein folding. Major advances in understanding fundamental principles underlying protein folding in optimized in-vitro conditions have yielded detailed physicochemical principles of folding landscapes for small, single domain proteins. In addition, there has been increased research focusing on the key features of protein folding in the cell that differentiate it from in-vitro folding, such as co-translational folding, chaperone-facilitated folding, and folding in crowded conditions with many weak interactions. Yet these two research areas have not been bridged effectively in research carried out to date. This review points to gaps between the two that are ripe for future research. Moreover, we emphasize the biological selection pressures that impact protein folding in-vivo and how fitness drives the evolution of protein sequences in ways that may place foldability in tension with other requirements on a given protein. We suggest that viewing the physicochemical process of protein folding through the lens of evolution will unveil new insights and pose novel challenges about in-cell folding landscapes.
Introduction
In the sixty-odd years since Anfinsen’s pioneering work showing the ability of RNaseA to re-fold from a reductively denatured state [1] the mechanism of protein folding and how an amino acid sequence encodes a folding reaction have been extensively studied. Increasingly powerful experimental and computational methods have been focused on the intellectually seductive in-vitro ‘protein folding problem’. As a consequence, we know a great deal about protein folding, but our knowledge is largely confined to how a protein folds at high dilution in conditions that are optimized for folding success.
In parallel with progress in understanding in-vitro folding, the chaperone concept has emerged, and chaperones have been recognized as essential players that facilitate protein folding in vivo [2]. More recently, researchers have begun to pay attention to the cellular complexities of folding during ribosomal synthesis of a polypeptide (co-translational folding), the critical need for proteins to be degraded in an ongoing way, and the impact of the highly concentrated cellular milieu with concomitant macromolecular crowding, spatial organization, and weak intermolecular interactions (reviewed in [3]).
Thus, it is clear that both in vitro and in vivo folding have seen major progress over the past decades. Ironically, there remain major gaps between these two perspectives on folding, and more crucially between most research in protein folding and the biological driving forces that exert selective pressures on protein folding in vivo. The irony of this gap is made all the greater by the realization that misfolding is a very dangerous problem for an organism—one that must be prevented and reckoned with efficiently when it occurs. Both the loss of a protein’s function and the potential toxicity of misfolded and aggregated states must be avoided [4]. Only very recently has attention begun to be paid to the evolutionary context that underlies selection of protein sequences and their folding landscapes [5]. This last point is crucial because present-day protein sequences are the result of a complex fabric of selective pressures, and evolutionary selection may not produce generalized solutions to the protein folding problem.
In this short review, we first give a brief reminder of issues affecting folding that are different in vivo and in vitro; we then highlight examples from the last two years of in-vitro folding research that may be relevant to in-vivo folding; we next describe recent work on in-vivo folding; and we end by describing recent research that should provoke the reader to think about protein folding in an evolutionary context and how folding relates to the fitness of an organism. We want to issue an apology up front: The brevity of this review and restricted number of papers that can be cited unavoidably led us to leave out many excellent and informative papers. We apologize to the authors!
Folding In vitro versus Folding in the Cell: What’s Different?
Proteins initially fold in vivo upon their biosynthesis. Hence, the first environment they are subjected to is created by the ribosome and ribosome-associated enzymes and chaperones. In addition, chains may fold co-translationally before the entire chain has been made. In contrast, folding of proteins in vitro generally is initiated from an unfolded ensemble in which a population of full-length chains (or in the case of single molecule experiments, one polypeptide) is subjected to folding conditions. Thus, the possibility of co-translational folding constitutes a major difference between the de novo folding reaction in the test tube and in a living organism.
Second, in vitro proteins sample their unfolded state in a dynamic equilibrium governed by their thermodynamic stability. Whether proteins spend much time unfolded in vivo is unclear. Many factors may disfavor accumulation of any significant population in the unfolded state, including chaperone binding, ongoing degradation, and kinetic barriers. Nonetheless, there may be lessons to be drawn from in vitro studies of unfolded states.
Third, protein-folding experiments in vitro are done at high dilution. In vivo, macromolecule concentrations range from 200 to 400 mg/ml, and surfaces present all around a folding chain are highly interactive. Thus, the impact of crowding and the influence of protein-protein interactions, including weak “quinary” interactions [6], must be taken into account.
Fourth, while proteins fold on their own in vitro, a significant fraction have ‘helpers’ in vivo [2]. It remains unclear to what extent and how chaperones alter the fundamental folding energy landscapes of proteins.
Fifth, proteins are vulnerable to competing intermolecular aggregation reactions to an extent that depends quite straightforwardly in vitro on the concentration of aggregation-prone species. Aggregation also competes with folding in vivo, but translating the parameters and mechanistic insights from aggregation studies in vitro to the in vivo context must be done with caution.
Lastly, folding reactions in vivo are spatially organized such that some interactions will be preferred over others. In vitro it is very difficult to mimic a spatially organized, inhomogeneous environment. This point is absolutely central to the folding of membrane proteins, which, despite their importance, will not be a focus of this review due to space constraints. Similarly, protein folding in organelles, in particular the endoplasmic reticulum and the secretory pathway, depend critically on the compartmentalization and sub-cellular organization (for a recent review, see [7]).
Recent in-vitro protein folding research advances with potential relevance to in-vivo folding
Small fast-folding domains have been the subject of extensive in-depth study in vitro because they are amenable to detailed physico-chemical analysis. For multiple reasons, it might be expected that the intrinsic folding behavior of these domains will determine their in-vivo folding properties. They fold on time scales that are much faster (e.g., microseconds to msec) than co-translational events (average polypeptide synthesis rate in eukaryotes 5 aa/sec, or 15 aa/sec in E. coli). Also, they generally do not populate long-lived intermediates and do not present extensive hydrophobic surfaces—both necessary to mediate binding with many chaperones. If such domains retain their intrinsic properties and their properties are not dominated by context, they may be viewed as the “building blocks” of well-folded proteins in the cell. This view would allow researchers to treat large proteins as composites of smaller domains (and, if parsed into even smaller units, ‘foldons’ [8]). Thus, the insights on folding of small domains provided by ever more powerful experimental methods and impressive new computational capabilities may translate to folding in vivo. For example, the description of transition path times using single-molecule fluorescence resonant energy transfer (FRET) sheds light on timescales of fundamental folding events [9], and the promising simulations of folding at realistic timescales have afforded the opportunity to compare experiment and theory directly [10,11]. Analysis of the folding trajectories computed by Anton, the supercomputer designed for protein folding simulations, offered a unifying mechanism for a dozen proteins and suggested that native-like contacts are formed in the unfolded state, with successive stabilization of key contacts driving the folding reaction. Progress in the simulation of folding reactions has also been reported by the Pande group, who showed the utility of Markov state modeling, a method that enables the generation of kinetically relevant folding trajectories from molecular dynamics simulations over time scales that would otherwise be insufficient to sample folding steps [12]. These researchers found evidence of glass-like kinetics using Markov state modeling of folding [13]. Computational studies such as these may provide a possible bridge from theory to experiment.
However, small single domain proteins are quite rare; for example, they represent less than 15% of the E. coli proteome [14]. Moreover, recent work from several groups suggests that, even though large proteins can generally be broken down into smaller units by domain dissection, the folding of these component domains may not be independent, and thus what is seen for free-standing small-domain proteins may not be applicable to the universe of larger proteins in the proteomes of all organisms. Specifically, the domains of repeat proteins have been found to display context-dependent folding [15,16]. In addition, the coupling of domains of large proteins is often a key part of the function of the large protein [17].
Thus it will be necessary to push the envelope of in vitro approaches and tackle larger proteins. Some recent research has taken on this challenge, and results show how new complexities in folding landscapes will emerge when larger proteins are examined: Pirchi et al. deployed single molecule FRET coupled to hidden Markov analysis to uncover six metastable states and multiple folding routes along the folding landscape of adenylate kinase, a three-domain 23.5 kDa protein [18]. The Reif laboratory used optical tweezer pulling experiments and hidden Markov analysis to study the folding of the two-domain, 17-kDa protein, calmodulin, and observed four on-pathway intermediates along with two off-pathway intermediates that compete with the productive folding reaction [19]. Dahiya and Chaudhuri examined the folding of the 82-kDa multi-domain protein, malate synthase G, and concluded that weak interdomain cooperativity may add complexity to a folding pathway, including the possibility of a functional intermediate [20].
Another research topic in in-vitro folding that has seen impressive progress recently is the nature of the denatured or unfolded state ensemble [21,22] and under what conditions the chains collapse [23]. A subject of long standing debate is how collapsed the unfolded state ensemble is under differing denaturant concentrations, and a recent study shows that the apparent results depend on the method of observation [24]. In any case, it remains unclear whether and when a polypeptide freely explores the unfolded state in vivo, apart from intrinsically disordered proteins, which need to maintain some degree of flexibility in order to participate in diverse interactions. Domains may transiently unfold or populate non-native states as they interact with chaperones (see below), and molecular machines that facilitate either translocation across membranes or degradation likely actively unfold proteins [25,26]. Thus, the connections between non-native, unfolded states in vitro and in vivo should continue to be explored.
Recent advances in in-vivo folding
In vivo, proteins must fold and be stable in a heterogeneous environment as concentrated as 400 g/L. Recent work by Pielak and colleagues reveals that the influence of the crowded in-vivo environment may be dominated by the prevalence of weak interactions, rather than the effects of excluded volume from macromolecular crowding, as previously believed [27,28]. These researchers found that protein crowding agents (bovine serum albumin, lysozyme) destabilized a test protein, CI2, in contrast to the stabilization expected from excluded volume effects [28]. Such effects are expected to be protein- and context-dependent, and indeed Guo et al. used a novel rapid laser temperature stepping method capable of measuring complete thermal melts and kinetic traces in vivo to deduce that phosphoglycerate kinase was more stable in mammalian cells than in vitro [29]. The seemingly contrasting results may differ because the experiments were performed at different temperatures, and the entropic component of crowding is temperature-dependent [30]. In addition, Dixit and Maslov have argued compellingly that protein-protein interaction networks will stabilize proteins in vivo relative to in vitro [31]. In a recent commentary, Gruebele and colleagues underlined the importance of the panoply of weak interactions influencing a protein in vivo, both specific and non-specific: terming them ‘quinary structure’ [32], as originally suggested by McConkey [6] and re-introduced in an earlier review of ours [33].
How co-translational folding modulates the folding landscape of proteins has been examined in a number of recent experimental and computational studies. O’Brien et al. introduced a computational approach to explore the impact of factors such as translation rate on folding [34]. Their findings suggest that mutations in mRNA that lead to altered translation rates may markedly alter folding outcomes. In a subsequent study, this group compared folding of ribosome nascent chain complexes that are arrested with those that are actively translating and concluded that at in-vivo translation rates, one-third of E. coli proteins would fold co-translationally. Krobath et al. also applied computational methods and found major differences between co-translational folding of arrested chains and freely folding (untethered) chain fragments [35]. They observed that the ribosome enhanced the population of low energy conformations dominated by local interactions. The interrelatedness of translation rate and folding points to a level of selective pressure acting at the RNA level. Experiments with synonomous codons [36], as well as a computational analysis correlating codon usage with protein structural motifs [37], and ribosome display [38] indeed point to the encoding of RNA-level information that might be woven together with the sequence code for folding in vivo. The ribosome itself has been shown to affect folding. Using single molecule force experiments, Kaiser et al. found that electrostatic interactions between the ribosome and their test protein (T4 lysozyme) retarded premature folding and allowed the nascent chain to remain in a folding-competent state [39]. Knight et al. examined the dynamics of a model nascent chain (a disordered protein) with varying charge and concluded that the ribosome surface electrostatically influenced the behavior of the chain, causing nascent protein variants carrying more negative charge to be more mobile [40].
Viewing co-translational folding in terms of a naked nascent chain exploring conformational space is, however, greatly oversimplified. A whole host of chaperones and quality control mechanisms lie in wait to greet the emerging polypeptide chain and assist its folding. The nature of this ribosome-associated greeting committee in E. coli is reviewed by Bukau and co-authors in this issue of COSB [41]. Their studies and others have revealed the order of events upon ‘birth’ of a nascent polypeptide, beginning with N-terminal processing, and followed by chaperone interactions with trigger factor, the chaperone that has privileged access to nascent chains of cytoplasmic proteins. These authors have provided compelling arguments for an unfolding role of trigger factor [42]. Single-molecule pulling experiments on maltose binding protein by Mashaghi et al. make a strong argument that trigger factor promotes productive folding by protecting partially folded states from misinteractions with neighboring molecules [43]. The emerging role of trigger factor in nascent chain folding is supported by computational work from Dobson and colleagues [44], which posits that trigger factor interacts with emerging chains and retards folding in addition to shielding the polypeptide from unfavorable interactions. Moving to eukaryotes, the Frydman lab has recently examined the co-translational roles of Hsp70 in yeast through a global analysis of ribosome-associated nascent chains [45]. They found that Hsp70 interacted preferentially with large multidomain proteins of complex topology that were unlikely to be able to fold co-translationally, consistent with the function of Hsp70 in maintaining the nascent chain in a folding-competent state.
Once a newly synthesized chain is away from the ribosome, it is further assisted by chaperones to ensure its successful folding and minimize competing aggregation processes. While data have been rapidly accumulating on the client repertoire of various chaperones in vivo, much less is known about how chaperone interactions affect protein folding reactions. For example, recent studies have asked how many and which proteins in E. coli are facilitated by the major chaperone systems GroEL/GroES and DnaK/DnaJ/GrpE: The Hsp70 system interacts with 700 cytoplasmic proteins, with particularly strong interaction with a subset of 180 that are aggregation-prone [46]. GroEL/ES was found in a proteomic study to support the folding of 250 proteins, with 84 of these obligately dependent on the chaperonin for folding [47]; a recent revisiting of this question concluded that there were fewer true GroEL substrates [48], but the two studies agreed on the nature of the obligate substrates: small enough to fit in the cavity, and enriched in metabolic enzymes and TIM barrels. Interestingly, Taguchi and coworkers found using a cell-free system that the major E. coli chaperone systems GroEL/GroES and DnaK/DnaJ/GrpE improved the solubility of 66% of their test group of 800 marginally soluble E. coli proteins [48].
These studies have provided insight into the cellular dependence on chaperones for productive folding. Yet how do chaperone-substrate interactions sculpt folding landscapes? Single particle cryo-electron microscopy has provided glimpses of substrates encapsulated in the GroEL chaperonin cavity, suggesting that they are quite collapsed [49,50]. Using in vivo experiments and monitoring growth as a criterion for fitness when mutant versions of the essential protein dihydrofolate reductase were expressed in the presence of differing amounts of GroEL/ES or the major protease Lon, Bershtein et al. concluded that both the chaperonin and the protease act on the molten globule intermediate [51]. These studies are consistent with current models in which GroEL smooths the folding landscape of poor folders, while DnaK largely acts to unfold its substrates, or to maintain folding-competent or unfolded states [2,52]. There have been numerous efforts to determine the clients of Hsp90 chaperones [53], and several labs have applied biophysical methods to deduce the nature of the binding interaction and likely impact on substrate folding, but many questions remain for this chaperone as well [54]. Data suggest that Hsp90 substrates are folding intermediates that have dynamic character. As an example, p53 was observed to adopt a molten-globule state upon interaction with Hsp90, and the model substrate staphylococcal nuclease has been proposed to bind Hsp90 in an unfolded state via a local structural element [55]. The elegant recent single molecule study of trigger factor-substrate interactions described above demonstrated directly an unfolding activity [43]. The interaction of small heat shock proteins with their clients has been recently reviewed [56], but here also we lack mechanistic understanding about how these chaperones affect folding. Similarly, the periplasmic chaperone HdeA binds molten globular substrates at low pH [57,58], but the consequent effects on their folding are as yet unexplored The eukaryotic chaperonin, TRiC, has to deal with larger proteins than encountered in E. coli, and a recent study concludes that it binds partially folded intermediates at domain boundaries, which helps explain how it may act on multidomain substrates but does not reveal details of its impact on their folding [59]. All told, current understanding of the impact of chaperone interactions on the folding landscapes of proteins remains incomplete, and the confluence of data and ideas from both in vitro and in vivo experiments will be needed to shed further light on this key question.
Chaperones work in teams and in partnership with degradation enzymes to facilitate folding in vivo and maintain protein homeostasis. A recent thrust is focused on admitting the complexity of integrated chaperone networks to elucidate the impact on folding of a substrate. A computational model for the flux of protein through the E. coli protein homeostasis network (including chaperones, degradation enzymes, disaggregases), beginning with a translated nascent chain, has been developed jointly by Powers’ and our labs [60]. This model enables generation of hypotheses about the involvement of the proteostasis machinery and the folding outcome of a polypeptide given its folding parameters. Also, by implementing in vivo FRET on fluorescently labeled chaperones Kumar and Sourjik were able to capture some of the interplay between the chaperone systems in E. coli, thus showing that the quality control systems are not isolated, but rather synergistic [61]. The authors show how DnaK (or more generally Hsp70) seems to be a central player in the de novo and re-folding branches of the proteostasis system.
Evolutionary perspectives: balancing function, evolvability, and successful folding
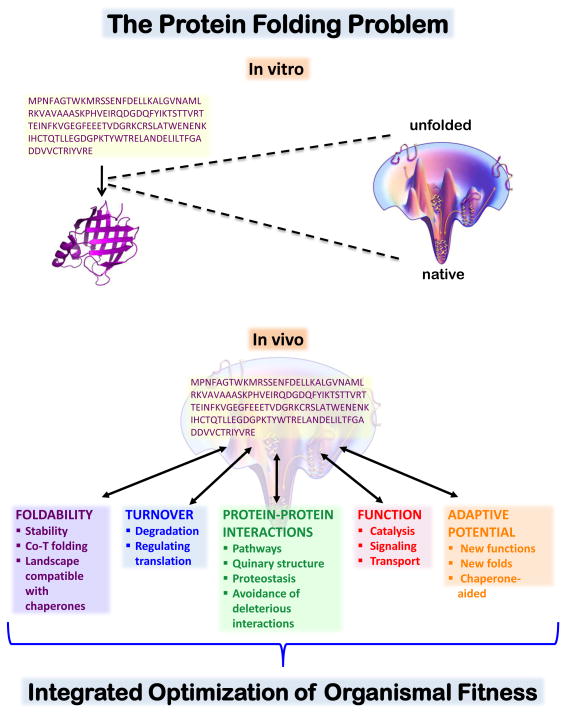
The canonical definition of ‘the protein folding problem’ viz., how is the information for a protein folding landscape encoded in a given sequence vastly oversimplifies the many selective pressures and stochastic events that have led to the existence of that sequence in a particular organism. Figure 1 depicts the panoply of protein properties that likely contributed to the evolution of current protein sequences. Adding to this complexity is the fact that there are also many pressures acting on nucleic acid sequences, for example pressures to adjust translation rate and to enable regulatory processes to occur in transcription and translation [62]. The importance of understanding the impact of evolutionary selection on protein sequences and consequently, their folding is a rapidly developing area of research.
Figure 1.
This figure draws a contrast between the in-vitro and in-vivo protein folding problems.
Top panel: In vitro, the protein folding problem is conceived as the challenge of understanding the physical chemical basis for an energy landscape. From this energy landscape, one can deduce the nature of the ensemble of folding polypeptides at different points in a protein folding reactions. Such landscapes have been elaborated with increasing detail as experimental and computational methods have advanced. However, the protein folding reaction depicted is one that occurs under high dilution, optimized conditions. Moreover, most of our knowledge about folding landscapes comes from studies of small, single domain proteins.
Bottom panel: In vivo, the protein folding problem comprises a complex set of interdependent responses to selective pressures integrated to optimize the fitness of an organism. Protein sequences and their behaviors must be viewed in light of all of the pressures acting simultaneously, and in some cases, orthogonally. For example, a protein must be able to fold under cellular conditions well enough to perform its function, be cleared with a physiologically necessary half-life, avoid pathological interactions, and be a favorable subject for evolution of new folds and functions (innovable). Relating these selective pressures to the folding landscapes pictured in the top panel is a challenging goal.
Protein stability naturally appears to be a property that would be selected for during evolution [63]. Using a theory-based and simulations approach, Shakhnovich and colleagues make a strong case that destabilizing mutations are selected against in highly abundant proteins, thus explaining their slow evolutionary rate. Yet, proteins designed in a laboratory generally display significantly higher stability than naturally occurring proteins [64]. This observation suggests that stability is not a dominant driving force for sequence selection [65]. Interestingly, a recent study demonstrates that experimental measures of fitness may underestimate the effects of mutations on protein function, which do not affect stability, unless dependence on expression level is taken into consideration [66]. In toto, a protein must possess a number of properties that are related to its folding, beyond stability, to survive a selection for organismal fitness.
Perhaps the most obvious evolutionary pressure that impacts folding properties is the requirement for function. Many have noted that the selection for folding and function frequently leads to a trade-off [67,68]. Tawfik recently noted that some folds, like TIM barrels, may possess a property, which he terms polarization, that enables them to adapt to new functions (innovability) while maintaining foldability and stability [68]. Additionally, deep mutational scanning by Fields and colleagues demonstrates the capacity of mutations that stabilize the native state to increase the tolerance to additional secondary mutations [69]. Mechanistic impacts of the folding-function tradeoff were also postulated for interleukin-1β (IL-1β). Capraro et al. observed that a functionally important structural feature, a β-bulge, acts to shape the IL-1β functional landscape so that only one folding route is followed, whereas variants in which this bulge was mutated follow multiple routes [70].
Tawfik’s term ‘innovability’ may also apply to the probability that a protein evolutionary path will lead to new folds. In a very thought provoking study, He et al. [71] experimentally identified ‘mutational tipping points’ that enabled proteins to switch folds and evolve new functions. On the other hand, a study of ancestral thioredoxin proteins by the Gavira group points out that although the ancestral protein differed considerably in sequence from the present version and was more thermo-stable, it folds into the same conformation as extant thioredoxins [72]. This highlights the robustness of a protein sequence to tolerate destabilizing mutations yet fold to carry out its function. It may well be an evolutionary advantage to retain this sequence nimbleness–the ability to absorb mutations that may cause a change in fold or function, which may improve organismal fitness, and in turn will have an impact on the ‘winning’ sequences we see in current proteomes.
The fine-tuning of protein sequences under selection integrates agnostically over all protein properties that contribute to fitness of an organism. The idea that chaperones can buffer destabilizing mutations that directly improve their function, or serve as stepping-stones to increase the rate of protein adaptation, has been experimentally supported [73,74]. Mapa and colleagues performed experiments on a set of model substrates that populated kinetic intermediates and demonstrated that each selectively bound its cognate chaperone from the whole spectrum of E. coli chaperones present in lysate [75]. They postulated that chaperone preferences co-evolve with foldability of protein sequences. This notion was recently emphasized in a provocative review on the origins of proteostasis [76]. Furthermore, the authors of this review, among others, have pointed out that protein evolution under the aegis of proteostasis is also environment dependent, and that integration of all factors operating on an organism leads to proteomic diversity [76,77].
Another factor that constrains sequence evolution is the requirement that proteins in vivo form productive interactions and avoid non-productive interactions [78]. A corollary of this selective pressure is the avoidance of pathological aggregation, which may be viewed as a non-productive interaction. As noted recently by Levy et al., the constraints on proteome evolution imposed by the need to form productive interactions and to avoid non-productive interactions is enhanced under the crowded conditions of the cell [79]. A computational study by Yang and co-workers postulates that avoiding deleterious interactions causes abundantly expressed proteins to evolve more slowly [80]. In addition, evolutionary trends also suggest that there has been a decrease in the fraction of hydrophobic residues and a tendency for increased disorder within the proteome over time [81]. Such changes may arise as a function of natural selection; however, they have consequences on folding and protein-protein interactions. Furthermore, interaction of proteins to form networks based on favored partners has recently been hypothesized to add to protein stability [31]. This concept is similar to that of chaperones being evolutionary buffers as discussed above, allowing proteins to accrue destabilizing mutations, yet fold and be better at their function [73].
Protein function also involves the formation of higher order protein structures such as quarternary and quinary structures [6], which require proteins to productively interact with each other. These higher levels of “folding” have long been implicated in metabolic functions, where the resulting organized pathways were termed ‘metabolons’ [82], and in signaling pathways [83]. Although such weakly associated complexes are difficult to study in situ, and would be difficult to isolate, recent efforts have led to new methods to interrogate them [84,85]. A recent study from the Teichmann and Robinson labs utilizes nano-electrospray ionization and gene fusion analysis to determine how several multimeric complexes are assembled and disassembled [86]. Through their analyses the authors find that the formation of quaternary structure and protein assembly pathways also appear to be under evolutionary pressure. The roles of selection and drift in protein-protein interactions is an emerging area of research, with theoretical work indicating that quaternary structure can be driven by stochastic forces [87].
Quo vadis?
The questions we have touched on in this review are extremely challenging to answer. Our hope for future clarity in understanding how protein landscapes in vivo relate to the deep and detailed descriptions we are privileged to be learning in vitro is buoyed by emergence of new methods to observe and simulate processes in intact cells. Work from the Xie lab quantifying the E. coli transcriptome and proteome [88], methods to observe translation at the resolution of a codon [89], advances in in-vivo NMR (e.g., [90] and [91]), and bold computational efforts from the Elcock [92] and Luthey-Schulten [93] groups, among others, should open doors in the future.
In parting, we cite words of Francisco Ayala written in an obituary for Theodosius Dobzhansky [94] (N.B. We have taken the liberty of substituting the words ‘protein sequence we examine today’ in place of the word ‘individual’ in the original quotation.): “the protein sequence we examine today is not the embodiment of some ideal type or norm, but rather a unique and unrepeatable realization in the field of quasi-infinite possible genetic combinations.” Thus, pity those of us who seek to elaborate general principles from what we see in protein behaviors in vivo! Nevertheless, physical chemistry abides….and its laws will surely reveal emergent principles.
HIGHLIGHTS.
This review discusses how folding in the test tube differs from folding in vivo.
New research is shedding light on the complex in vivo folding landscape.
Many selective pressures, in addition to foldability, shape protein sequence space.
Acknowledgments
Work on in-cell protein folding and chaperone mechanisms in the laboratory of L.M.G. has been supported by National Institutes of Health grants GM027616 and OD00945. K.S.H. is supported, in part, by a fellowship from the University of Massachusetts, Amherst, as a part of the Chemistry-Biology Interface Program (funded by NIH Grant T32 GM08515). The authors would like to thank Anne Gershenson, Eugenia Clerico, Peter Chien, and Dan Bolon for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 3.Gershenson A, Gierasch LM. Protein folding in the cell: challenges and progress. Curr Opin Struct Biol. 2011;21:32–41. doi: 10.1016/j.sbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 5.Grahnen JA, Nandakumar P, Kubelka J, Liberles DA. Biophysical and structural considerations for protein sequence evolution. BMC Evol Biol. 2011;11:361. doi: 10.1186/1471-2148-11-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McConkey EH. Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc Natl Acad Sci U S A. 1982;79:3236–3240. doi: 10.1073/pnas.79.10.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gidalevitz T, Stevens F, Argon Y. Orchestration of secretory protein folding by ER chaperones. Biochim Biophys Acta. 2013;1833:2410–2424. doi: 10.1016/j.bbamcr.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maity H, Maity M, Krishna MM, Mayne L, Englander SW. Protein folding: the stepwise assembly of foldon units. Proc Natl Acad Sci U S A. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung HS, McHale K, Louis JM, Eaton WA. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335:981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piana S, Lindorff-Larsen K, Shaw DE. Atomic-level description of ubiquitin folding. Proc Natl Acad Sci U S A. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How fast-folding proteins fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 12.Pande VS, Beauchamp K, Bowman GR. Everything you wanted to know about Markov State Models but were afraid to ask. Methods. 2010;52:99–105. doi: 10.1016/j.ymeth.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber JK, Jack RL, Pande VS. Emergence of glass-like behavior in Markov state models of protein folding dynamics. J Am Chem Soc. 2013;135:5501–5504. doi: 10.1021/ja4002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Braselmann E, Chaney JL, Clark PL. Folding the proteome. Trends Biochem Sci. 2013;38:337–344. doi: 10.1016/j.tibs.2013.05.001. This comprehensive review addresses the current state of the protein folding problem, emphasizing the limited coverage of the proteome in studies carried out to date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawyer N, Chen J, Regan L. All repeats are not equal: a module-based approach to guide repeat protein design. J Mol Biol. 2013;425:1826–1838. doi: 10.1016/j.jmb.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vieux EF, Barrick D. Deletion of internal structured repeats increases the stability of a leucine-rich repeat protein, YopM. Biophys Chem. 2011;159:152–161. doi: 10.1016/j.bpc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. On the role of frustration in the energy landscapes of allosteric proteins. Proc Natl Acad Sci U S A. 2011;108:3499–3503. doi: 10.1073/pnas.1018980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirchi M, Ziv G, Riven I, Cohen SS, Zohar N, Barak Y, Haran G. Single-molecule fluorescence spectroscopy maps the folding landscape of a large protein. Nat Commun. 2011;2:493. doi: 10.1038/ncomms1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Stigler J, Ziegler F, Gieseke A, Gebhardt JC, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. The authors tackle the folding of a multidomain protein, calmodulin, using a combination of single molecule force spectroscopy and hidden markov analysis, revealing a competition between cooperative and non-productive interactions. [DOI] [PubMed] [Google Scholar]
- 20.Dahiya V, Chaudhuri TK. Functional intermediate in the refolding pathway of a large and multidomain protein malate synthase G. Biochemistry. 2013;52:4517–4530. doi: 10.1021/bi400328a. [DOI] [PubMed] [Google Scholar]
- 21.Meng W, Lyle N, Luan B, Raleigh DP, Pappu RV. Experiments and simulations show how long-range contacts can form in expanded unfolded proteins with negligible secondary structure. Proc Natl Acad Sci U S A. 2013;110:2123–2128. doi: 10.1073/pnas.1216979110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng W, Luan B, Lyle N, Pappu RV, Raleigh DP. The denatured state ensemble contains significant local and long-range structure under native conditions: analysis of the N-terminal domain of ribosomal protein L9. Biochemistry. 2013;52:2662–2671. doi: 10.1021/bi301667u. [DOI] [PubMed] [Google Scholar]
- 23.Haran G. How, when and why proteins collapse: the relation to folding. Curr Opin Struct Biol. 2012;22:14–20. doi: 10.1016/j.sbi.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo TY, Meisburger SP, Hinshaw J, Pollack L, Haran G, Sosnick TR, Plaxco K. Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state. J Mol Biol. 2012;418:226–236. doi: 10.1016/j.jmb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomkiewicz D, Nouwen N, Driessen AJ. Pushing, pulling and trapping--modes of motor protein supported protein translocation. FEBS Lett. 2007;581:2820–2828. doi: 10.1016/j.febslet.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Sarkar M, Smith AE, Krois AS, Pielak GJ. Macromolecular crowding and protein stability. J Am Chem Soc. 2012;134:16614–16618. doi: 10.1021/ja305300m. [DOI] [PubMed] [Google Scholar]
- 28.Miklos AC, Li C, Sorrell CD, Lyon LA, Pielak GJ. An upper limit for macromolecular crowding effects. BMC Biophys. 2011;4:13. doi: 10.1186/2046-1682-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo M, Xu Y, Gruebele M. Temperature dependence of protein folding kinetics in living cells. Proc Natl Acad Sci U S A. 2012;109:17863–17867. doi: 10.1073/pnas.1201797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou HX. Influence of crowded cellular environments on protein folding, binding, and oligomerization: biological consequences and potentials of atomistic modeling. FEBS Lett. 2013;587:1053–1061. doi: 10.1016/j.febslet.2013.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Dixit PD, Maslov S. Evolutionary capacitance and control of protein stability in protein-protein interaction networks. PLoS Comput Biol. 2013;9:e1003023. doi: 10.1371/journal.pcbi.1003023. This paper uses a statistical mechanical approach to estimate how protein-protein interactions affect the stability of proteins inside cells. The authors hypothesize that there is a gain of stability, which would decrease the cellular concentration of unfolded species, and in turn may enhance organismal fitness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Wirth AJ, Gruebele M. Quinary protein structure and the consequences of crowding in living cells: Leaving the test-tube behind. Bioessays. 2013 doi: 10.1002/bies.201300080. This recent commentary discusses the effects of cellular crowding conditions on protein folding, with an emphasis on weak ‘quinary’ protein-protein interactions. [DOI] [PubMed] [Google Scholar]
- 33.Gierasch LM, Gershenson A. Post-reductionist protein science, or putting Humpty Dumpty back together again. Nat Chem Biol. 2009;5:774–777. doi: 10.1038/nchembio.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Ciryam P, Morimoto RI, Vendruscolo M, Dobson CM, O’Brien EP. In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc Natl Acad Sci U S A. 2013;110:E132–140. doi: 10.1073/pnas.1213624110. This computational study simulates the effects of translation rates and mRNA sequences on co-translational folding and concludes that a substantial fraction of the E. coli proteome is likely to fold co-translationally. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krobath H, Shakhnovich EI, Faisca PF. Structural and energetic determinants of co-translational folding. J Chem Phys. 2013;138:215101. doi: 10.1063/1.4808044. [DOI] [PubMed] [Google Scholar]
- 36.Shabalina SA, Spiridonov NA, Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013;41:2073–2094. doi: 10.1093/nar/gks1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. Genome-wide annotation and quantitation of translation by ribosome profiling. Curr Protoc Mol Biol. 2013;Chapter 4(Unit 4):18. doi: 10.1002/0471142727.mb0418s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaiser CM, Goldman DH, Chodera JD, Tinoco I, Jr, Bustamante C. The ribosome modulates nascent protein folding. Science. 2011;334:1723–1727. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knight AM, Culviner PH, Kurt-Yilmaz N, Zou T, Ozkan SB, Cavagnero S. Electrostatic Effect of the Ribosomal Surface on Nascent Polypeptide Dynamics. ACS Chem Biol. 2013 doi: 10.1021/cb400030n. e-pub ahead of print Apr 5. [DOI] [PubMed] [Google Scholar]
- 41.Bukau B. Ribosome-associated chaperones. Curr Opin Struct Biol. 2014 in press. [Google Scholar]
- 42.Hoffmann A, Becker AH, Zachmann-Brand B, Deuerling E, Bukau B, Kramer G. Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Mol Cell. 2012;48:63–74. doi: 10.1016/j.molcel.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 43•.Mashaghi A, Kramer G, Bechtluft P, Zachmann-Brand B, Driessen AJ, Bukau B, Tans SJ. Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature. 2013;500:98–101. doi: 10.1038/nature12293. This study applied powerful single molecule force experiments to examine the effect of trigger factor on the folding landscape of maltose-binding protein, and concluded that the chaperone reshaped the folding landscape, favoring and protecting partially folded intermediates that may then be productively folded. [DOI] [PubMed] [Google Scholar]
- 44.O’Brien EP, Christodoulou J, Vendruscolo M, Dobson CM. Trigger factor slows co-translational folding through kinetic trapping while sterically protecting the nascent chain from aberrant cytosolic interactions. J Am Chem Soc. 2012;134:10920–10932. doi: 10.1021/ja302305u. [DOI] [PubMed] [Google Scholar]
- 45.Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell. 2013;152:196–209. doi: 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calloni G, Chen T, Schermann SM, Chang HC, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 47.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 48•.Niwa T, Kanamori T, Ueda T, Taguchi H. Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc Natl Acad Sci U S A. 2012;109:8937–8942. doi: 10.1073/pnas.1201380109. Using a reconstituted cell-free system, these authors asked what the effects of the major E. coli chaperones were on the solubility (presumably, folding) of ~800 aggregation-prone E. coli proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen DH, Madan D, Weaver J, Lin Z, Schroder GF, Chiu W, Rye HS. Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell. 2013;153:1354–1365. doi: 10.1016/j.cell.2013.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR. Chaperonin complex with a newly folded protein encapsulated in the folding chamber. Nature. 2009;457:107–110. doi: 10.1038/nature07479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Bershtein S, Mu W, Serohijos AW, Zhou J, Shakhnovich EI. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol Cell. 2013;49:133–144. doi: 10.1016/j.molcel.2012.11.004. The authors explore using fitness criteria (growth rate) when a mutant protein (dihydrofolate reductase) is expressed what folding states are acted on in E. coli by the major components of the protein homeostasis network, GroEL/ES and Lon, the major protease. They compare their in vivo observations with in vitro experiments, and conclude that these protein homeostasis components act on the molten globule intermediate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013 doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohl A, Rohrberg J, Buchner J. The chaperone Hsp90: changing partners for demanding clients. Trends Biochem Sci. 2013;38:253–262. doi: 10.1016/j.tibs.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basha E, O’Neill H, Vierling E. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foit L, George JS, Zhang BW, Brooks CL, 3rd, Bardwell JC. Chaperone activation by unfolding. Proc Natl Acad Sci U S A. 2013;110:E1254–1262. doi: 10.1073/pnas.1222458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong W, Wu YE, Fu X, Chang Z. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 2012;20:328–335. doi: 10.1016/j.tim.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Russmann F, Stemp MJ, Monkemeyer L, Etchells SA, Bracher A, Hartl FU. Folding of large multidomain proteins by partial encapsulation in the chaperonin TRiC/CCT. Proc Natl Acad Sci U S A. 2012;109:21208–21215. doi: 10.1073/pnas.1218836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Powers ET, Powers DL, Gierasch LM. FoldEco: a model for proteostasis in E. coli. Cell Rep. 2012;1:265–276. doi: 10.1016/j.celrep.2012.02.011. The authors construct a computational model based on coupled differential equations and information mined from the literature to simulate the action of the E. coli protein homeostasis network of chaperones and degradation on a folding protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar M, Sourjik V. Physical map and dynamics of the chaperone network in Escherichia coli. Mol Microbiol. 2012;84:736–747. doi: 10.1111/j.1365-2958.2012.08054.x. [DOI] [PubMed] [Google Scholar]
- 62.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Serohijos AW, Rimas Z, Shakhnovich EI. Protein biophysics explains why highly abundant proteins evolve slowly. Cell Rep. 2012;2:249–256. doi: 10.1016/j.celrep.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. This paper describes and tests general principles that have emerged from the Baker lab regarding the design of proteins with high stability, and successful designs of sequences that take up five different topologies. The authors note that the design process is completely distinct from the evolutionary process that has given rise to protein folds and stabilities we observe today. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Reynolds KA, Russ WP, Socolich M, Ranganathan R. Evolution-based design of proteins. Methods Enzymol. 2013;523:213–235. doi: 10.1016/B978-0-12-394292-0.00010-2. This paper describes how information for protein folding and function is embedded in statistical correlations within natural protein sequences and how the understanding derived from experimental tests of evolution-based designed sequences may shed light on the relationship of fitness and natural proteins. [DOI] [PubMed] [Google Scholar]
- 66.Jiang L, Mishra P, Hietpas RT, Zeldovich KB, Bolon DN. Latent effects of Hsp90 mutants revealed at reduced expression levels. PLoS Genetics. 2013;9:e1003600. doi: 10.1371/journal.pgen.1003600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gosavi S. Understanding the folding-function tradeoff in proteins. PLoS One. 2013;8:e61222. doi: 10.1371/journal.pone.0061222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68••.Dellus-Gur E, Toth-Petroczy A, Elias M, Tawfik DS. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J Mol Biol. 2013;425:2609–2621. doi: 10.1016/j.jmb.2013.03.033. The authors explore properties of protein folds that enable evolution of new functions through mutation. The attribute ‘innovability’ to the polarization of structures such that high stability in one portion of the structure is compensatory to the destabilizing effects of mutations in another region of the protein. [DOI] [PubMed] [Google Scholar]
- 69.Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci U S A. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Capraro DT, Roy M, Onuchic JN, Gosavi S, Jennings PA. β-Bulge triggers route-switching on the functional landscape of interleukin-1β. Proc Natl Acad Sci U S A. 2012;109:1490–1493. doi: 10.1073/pnas.1114430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He Y, Chen Y, Alexander PA, Bryan PN, Orban J. Mutational tipping points for switching protein folds and functions. Structure. 2012;20:283–291. doi: 10.1016/j.str.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ingles-Prieto A, Ibarra-Molero B, Delgado-Delgado A, Perez-Jimenez R, Fernandez JM, Gaucher EA, Sanchez-Ruiz JM, Gavira JA. Conservation of protein structure over four billion years. Structure. 2013;21:1690–1697. doi: 10.1016/j.str.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 74.Wyganowski KT, Kaltenbach M, Tokuriki N. GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J Mol Biol. 2013;425:3403–3414. doi: 10.1016/j.jmb.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 75.Mapa K, Tiwari S, Kumar V, Jayaraj GG, Maiti S. Information encoded in non-native states drives substrate-chaperone pairing. Structure. 2012;20:1562–1573. doi: 10.1016/j.str.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 76.Powers ET, Balch WE. Diversity in the origins of proteostasis networks--a driver for protein function in evolution. Nat Rev Mol Cell Biol. 2013;14:237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bogumil D, Dagan T. Cumulative impact of chaperone-mediated folding on genome evolution. Biochemistry. 2012;51:9941–9953. doi: 10.1021/bi3013643. [DOI] [PubMed] [Google Scholar]
- 78.Pastore A, Temussi PA. The two faces of Janus: functional interactions and protein aggregation. Curr Opin Struct Biol. 2012;22:30–37. doi: 10.1016/j.sbi.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 79•.Levy ED, De S, Teichmann SA. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc Natl Acad Sci U S A. 2012;109:20461–20466. doi: 10.1073/pnas.1209312109. The authors analyze several proteins of known structure from E. coli, yeast and man to determine what properties enable proteins to be abundant and avoid unwanted interactions. They find that abundant proteins are less ‘sticky’, based on a definition of surface character that they develop, than less abundant ones, and posit that avoidance of nonfunctional interactions is a major constraint in evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang JR, Liao BY, Zhuang SM, Zhang J. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc Natl Acad Sci U S A. 2012;109:E831–840. doi: 10.1073/pnas.1117408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mannige RV, Brooks CL, Shakhnovich EI. A universal trend among proteomes indicates an oily last common ancestor. PLoS Comput Biol. 2012;8:e1002839. doi: 10.1371/journal.pcbi.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Velot C, Mixon MB, Teige M, Srere PA. Model of a quinary structure between Krebs TCA cycle enzymes: a model for the metabolon. Biochemistry. 1997;36:14271–14276. doi: 10.1021/bi972011j. [DOI] [PubMed] [Google Scholar]
- 83.Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, Panning B, Krogan NJ. Quantitative genetic-interaction mapping in mammalian cells. Nature Methods. 2013;10:432–437. doi: 10.1038/nmeth.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fraser JS, Gross JD, Krogan NJ. From systems to structure: bridging networks and mechanism. Mol Cell. 2013;49:222–231. doi: 10.1016/j.molcel.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lane LA, Nadeau OW, Carlson GM, Robinson CV. Mass spectrometry reveals differences in stability and subunit interactions between activated and nonactivated conformers of the (αβγδ)4 phosphorylase kinase complex. Mol Cell Proteomics. 2012;11:1768–1776. doi: 10.1074/mcp.M112.021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marsh JA, Hernandez H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell. 2013;153:461–470. doi: 10.1016/j.cell.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lynch M. Evolutionary diversification of the multimeric states of proteins. Proc Natl Acad Sci U S A. 2013;110:E2821–2828. doi: 10.1073/pnas.1310980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han Y, David A, Liu B, Magadan JG, Bennink JR, Yewdell JW, Qian SB. Monitoring cotranslational protein folding in mammalian cells at codon resolution. Proc Natl Acad Sci U S A. 2012;109:12467–12472. doi: 10.1073/pnas.1208138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tochio H. Watching protein structure at work in living cells using NMR spectroscopy. Curr Opin Chem Biol. 2012;16:609–613. doi: 10.1016/j.cbpa.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 91.Banci L, Barbieri L, Bertini I, Luchinat E, Secci E, Zhao Y, Aricescu AR. Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat Chem Biol. 2013;9:297–299. doi: 10.1038/nchembio.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frembgen-Kesner T, Elcock AH. Computer Simulations of the Bacterial Cytoplasm. Biophys Rev. 2013;5:109–119. doi: 10.1007/s12551-013-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roberts E, Magis A, Ortiz JO, Baumeister W, Luthey-Schulten Z. Noise contributions in an inducible genetic switch: a whole-cell simulation study. PLoS Comput Biol. 2011;7:e1002010. doi: 10.1371/journal.pcbi.1002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ayala FJ. Theodosius Dobzhansky: the man and the scientist. Annu Rev Genet. 1976;10:1–6. doi: 10.1146/annurev.ge.10.120176.000245. [DOI] [PubMed] [Google Scholar]