Abstract
Structural abnormalities in striated muscle have been observed in numerous transcription factor gain- and loss-of-function phenotypes in animal and cell culture model systems, indicating that transcription is important in regulating the cytoarchitecture. While most characterized cytoarchitectural defects are largely indistinguishable by histological and ultrastructural criteria, analysis of dysregulated gene expression in each mutant phenotype has yielded valuable information regarding specific structural gene programs that may be uniquely controlled by each of these transcription factors. Linking the formation and maintenance of each subcellular structure or subset of proteins within a cytoskeletal compartment to an overlapping but distinct transcription factor cohort may enable striated muscle to control cytoarchitectural function in an efficient and specific manner. Here we summarize the available evidence that connects transcription factors, those with established roles in striated muscle such as MEF2 and SRF, as well as other non-muscle transcription factors, to the regulation of a defined cytoskeletal structure. The notion that genes encoding proteins localized to the same subcellular compartment are coordinately transcriptionally regulated may prompt rationally designed approaches that target specific transcription factor pathways to correct structural defects in muscle disease.
Keywords: Transcription factor, MEF2, Striated muscle, Costamere, Sarcomere, Myofibril
Introduction
High-resolution electron micrographs of striated muscle reveal the remarkable organization of the contractile apparatus. These images bring to mind the elaborate mechanisms that have been established to assemble and maintain this and other morphologically distinct cytoskeletal structures, which consist of an array of structurally and functionally diverse proteins. Intrinsic to the process of building the cytoarchitecture is the coordinated temporal expression of thin and thick filaments, and myofilament-associated proteins of the contractile apparatus [1–3]. Following their primary induction, continued expression is required to maintain levels necessary to preserve the integrity and function of the cytoarchitecture [4]. The importance of maintaining proper expression of structural proteins is exemplified by the numerous cardio- and skeletal myopathies that are either caused by a deficiency of a cytoskeletal protein, or are associated with altered expression of structural proteins resulting from the remodeling that occurs in response to a primary insult [5, 6]. Given the deleterious consequences caused by improperly synthesized structural proteins, striated muscle must be able to tightly regulate their expression.
Muscle structural defects have been observed in gain- and loss-of-function analyses of a number of transcription factors thought to function in muscle. Consistent with the mutant phenotypes, most of these abnormalities are associated with dysregulated expression of genes encoding structural proteins. These observations reflect the importance of transcription in the regulation of the cytoarchitecture. Not only are absolute expression levels finely controlled, but because of its structural complexity, striated muscle has likely evolved sophisticated mechanisms to coordinately regulate the synthesis of proteins localized within distinct compartments. Placing the synthesis of the entire repertoire of cytoarchitectural proteins under the control of one or the same cohort of transcription factors would presumably leave this intricate framework vulnerable to dysfunction, especially if there were a significant change in expression or activity of one of these transcription factors. Rather, it seems logical to place the assorted cytoskeletal structures under the control of overlapping but distinct sets of transcription factors. In this manner, the regulation of the various cytoarchitectural complexes can be coupled with specific transcriptional pathways throughout the differentiation process and in mechanotransduction, in both physiological and pathological conditions.
The purpose of this review is to highlight the role of transcription in the regulation of the cytoarchitecture in striated muscle. We begin this review by briefly describing the basic organization of the contractile apparatus and membrane-associated cytoskeletal complexes. We then summarize those transcription factors that regulate genes whose encoded proteins share a common subcellular localization, or whose misexpression results in a well-defined structural phenotype in striated muscle. We will not discuss transcription factors that regulate contractile gene expression in a fiber type-specific manner or those that have been identified as candidate regulatory factors via analysis of a single structural gene promoter in vitro. Instead, we will focus on transcription factors that seem to function in the global regulation of a distinct, macromolecular structure in striated muscle.
Striated muscle cytoskeletal structures
The striated muscle cytoarchitecture consists of several discrete structures that play specific roles in contraction, anchoring, and signaling [4]. The major cytoskeletal structure for contraction is the myofibril, which harbors the actin (thin) and myosin (thick) filaments and is organized into distinct domains as defined by their appearance in electron micrographs. A myofibril consists of numerous sarcomeres, the main contractile components, connected in tandem with each segment flanked by the Z-disc [7, 8] (Fig. 1). The Z-disc is a macromolecular complex consisting of numerous scaffolding proteins and functions as the primary anchoring point for thin filaments, where they interact with the actin cross-linking protein α-actinin [9, 10]. It also serves as the anchoring region for the giant protein titin, which extends a half sarcomere in length and is considered to dictate not only the length of the sarcomere but also assembly and alignment of the filament system [11, 12]. The Z-disc may also function as a signaling center for muscle contraction because of its unique molecular composition [13, 14]. Additional regions within myofibrils are designated the M-line, and I- and A-bands, each consisting of a distinct combination of filament and accessory proteins necessary for their specific role in muscle contraction [4].
Fig. 1.
Schematic representation of striated muscle cytoarchitecture. Sarcomeres are the most basic unit of contraction and are linked in tandem at the Z-disc to make up myofibrils, the major cytoskeletal structure for contraction [4, 7, 8]. The sarcomere is composed of distinct regions harboring actin (thin filament), myosin (thick filament), and several accessory proteins. The A-band encompasses the entire length of the thick filament, including segments overlapping thin filaments, whereas the I-band region only contains thin filaments. The M-line is composed of proteins that hold thick filaments together. The outermost myofibrils are connected to the sarcolemma through their association with the costamere, a macromolecular protein structure harboring dystrophin and integrin complexes, which registers in line with the Z-disc [15–18]. Cardiomyocytes also contain intercalated discs composed of gap junctions, adherens junctions, and desmosomes, and serve as an anchoring point for myofibrils in addition to enabling myocytes to communicate with each other [23, 24]
Myofibrils are anchored to the sarcolemma (muscle cell membrane) through elaborate protein networks called costameres (Fig. 1). Costameres connect the outermost myofibrils to the sarcolemma at each Z-disc [15–18]. These specialized structures enable myofibrils to transmit the force of each contraction throughout the entire myocyte. Because of their unique localization in striated muscle, costameres are positioned to sense changes in force and modulate contraction thereby enabling striated muscle to adapt to exercise, stress, and other conditions that affect muscle physiology [17]. Three types of multi-protein complexes can be found within the costamere: the dystrophin-glycoprotein complex (DGC), integrins, and spectrin-ankyrin cytoskeleton, which perform specialized functions consistent with their distinct protein composition within the costamere [4, 19, 20]. The costamere is undoubtedly a critical cytoarchitectural structure given that mutations in proteins localized to this region, particularly the DGC, can destabilize the complex and result in a spectrum of debilitating muscle diseases [21, 22].
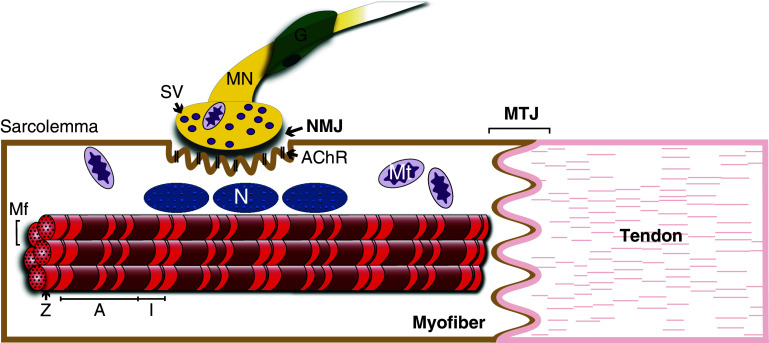
Other subsarcolemmal complexes, such as the intercalated discs in cardiac muscle and myotendinous junction in skeletal muscle, attach the ends of myofibrils to their respective striated muscle cell membrane. The intercalated discs of cardiomyocytes consist of gap junctions (connexins), adherens junctions (cadherins), and desmosomes (plakoglobin, desmoplakin), which not only function as anchoring points for myofibrils but also enable communication between neighboring myocytes [23, 24] (Fig. 1). Like the intercalated discs, the myotendinous junction is also a myofibrillar anchoring structure but in this instance functions as an attachment region between skeletal myofibers and tendons [25] (Fig. 2). Given their role in linking the ends of myofibrils to the sarcolemma, both of these complexes display many molecular similarities to costameres.
Fig. 2.
Schematic representation of a skeletal muscle cell connected to a tendon. The myotendinous junction (MTJ) is a cytoskeletal complex that not only connects muscle cells to tendons, but also links myofibrils to the sarcolemma [25]. The neuromuscular junction (NMJ) is a specialized synapse that potentiates signaling between skeletal muscle and motor neurons (MN) and is composed of three distinct cell types: myofiber, motor neuron, and glial cells (G). The sarcolemma contains highly specialized regions characterized by membrane folding at the NMJ which are enriched with acetylcholine receptors [26, 27]. A cluster of nuclei are situated just below the NMJ, which receive signals from the motor neuron in an activity-dependent manner to express genes encoding proteins that function at the NMJ. N nucleus, Mt mitochondria, Mf Myofibril, Z Z-disc, A A-band, I I-band, SV synaptic vesicle, AChR acetylcholine receptor
The neuromuscular junction (NMJ) functions as an important signaling interface between skeletal muscle and motor neurons. The NMJ constitutes a discrete macromolecular structure with specialized membrane features and has a vastly different molecular make-up than other regions of the sarcolemma. The NMJ consists of a cluster of neurotransmitter receptors and accessory transmembrane and subsarcolemmal proteins that are required for the activity of this specialized synapse (Fig. 2). Although the neuromuscular junction is not considered a cytoarchitectural element per se, components of this structure in skeletal muscle are closely associated with cytoskeletal proteins [26, 27].
Transcriptional regulation of distinct cytoarchitectural compartments
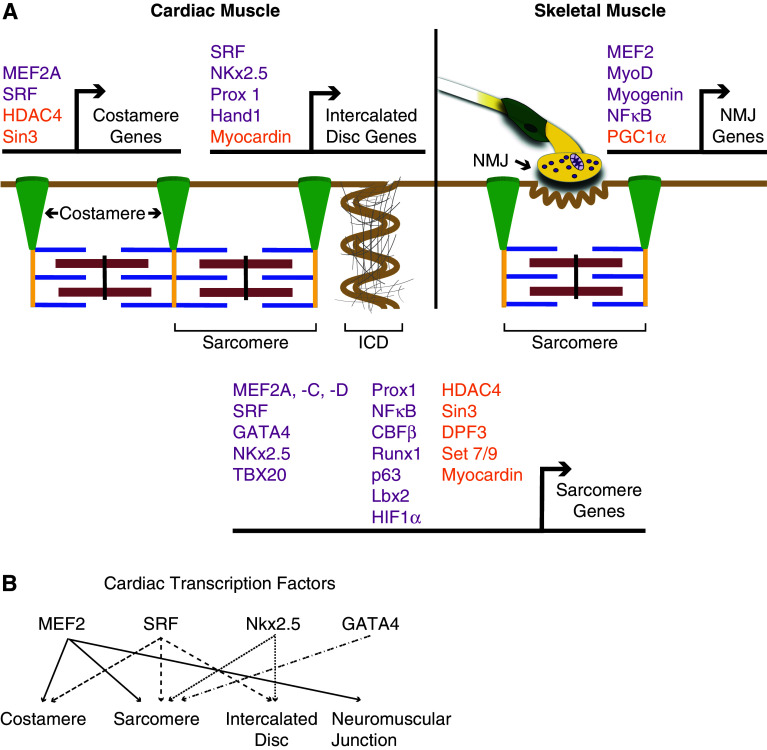
The contractile apparatus and the various macromolecular complexes of striated muscle have been studied nearly exclusively using proteomic approaches with considerable emphasis on the multitude of protein–protein interactions. Analyzing these structures at this level is not surprising given the extensive array of structurally diverse proteins, their post-translational modifications, and their association with signaling pathways. By contrast, despite the extensive investigations into dissecting the transcriptional mechanisms of individual muscle structural genes in vitro, much less is known regarding the role that transcription factors play in the coordinate regulation of a gene cohort whose encoded proteins comprise a molecularly defined structure in muscle. The notion that multiple components within a cytoarchitectural structure may be under the control of a particular transcriptional program is a distinct possibility. Given the regulation of the actin cytoskeleton network by the myocardin related transcription factor (MRTF)-serum response factor (SRF) pathway [28], it is not unreasonable to suggest that a designated transcription factor, including SRF, or specific transcription factor/co-factor combinations function in the differential regulation of morphologically distinct complexes in striated muscle (Fig. 3; Table 1).
Fig. 3.
Transcriptional networks regulating cytoarchitectural structures in striated muscle. a Striated muscle cytoarchitecture is tightly controlled by a distinct but overlapping set of transcription factors (purple) and co-factors (orange). While skeletal and cardiac muscles share many structural similarities, intercalated discs (ICD) are unique to cardiac cells and neuromuscular junctions (NMJ) are restricted to skeletal muscle. b The cardiac transcription factors MEF2, SRF, Nkx2.5, and GATA4 are evolutionarily conserved regulators of cardiac development, and are essential mediators in muscle disease pathways. All four transcription factors have also been shown to be important in regulating cytoarchitectural gene transcription. Arrows between the transcription factors and cytoskeletal structures illustrate the overlapping but distinct structural gene programs dependent on each factor in striated muscle
Table 1.
Regulatory factors involved in transcriptional regulation of striated muscle cytoarchitecture
| Transcription factor | Model system | Structure | Muscle type | Reference |
|---|---|---|---|---|
| MEF2 | ||||
| D-mef2 | Fly | Sarcomere, NMJ | Skeletal | [34, 54] |
| MEF2A | Mouse, NRVMa | Costamere | Cardiac | [35, 37–39] |
| Zebrafish | Sarcomere | Cardiac | [59] | |
| MEF2C | Mouse, Zebrafish | Sarcomere | Skeletal | [36, 60] |
| MEF2D | Zebrafish | Sarcomere | Skeletal | [60] |
| MEF2e | C2C12b, Fly | NMJ | Skeletal | [103, 104] |
| SRF | Mouse | Costamere, sarcomere, ICD | Cardiac | [46–48] |
| GATA4 | NRVMa | Sarcomere | Cardiac | [63, 64] |
| NKx2.5/ | Mouse | Sarcomere, ICD | Cardiac | [71, 113] |
| Tinman | Fly | Sarcomere | Cardiac | [70] |
| Tbx20/ | Mouse | Sarcomere | Cardiac | [74] |
| Neuromancer | Fly | Sarcomere | Cardiac | [73] |
| NF-ĸB | Mouse, C2C12b | Sarcomere, NMJ | Skeletal | [80, 110] |
| Prox1 | Mouse | Sarcomere, ICD | Cardiac | [78] |
| JunB–CBFβ | Zebrafish | Sarcomere | Cardiac, Skeletal | [84] |
| Runx1 | Mouse | Sarcomere | Skeletal | [85] |
| Lbx2 | Fly, Zebrafish | Sarcomere | Skeletal | [89] |
| HIF1α | Mouse | Sarcomere | Cardiac | [92] |
| p63 | Mouse, ESc | Sarcomere | Cardiac | [94] |
| Hand1 | Mouse | ICD | Cardiac | [116] |
| MyoD/Myogenin | Mouse, C2C12b, RMTd | NMJ | Skeletal | [105–109] |
| Chromatin factor | ||||
| HDAC4 | Mouse, C2C12b | Costamere, sarcomere | Skeletal | [50] |
| Sin3 | Mouse, C2C12b | Costamere, sarcomere | Skeletal | [53] |
| DPF3 | C2C12b, Zebrafish | Sarcomere | Cardiac, Skeletal | [100] |
| Set 7/9 | Zebrafish | Sarcomere | Skeletal | [102] |
| Co-activators | ||||
| Myocardin | Mouse, Xenopus | Sarcomere, ICD | Cardiac | [97, 98] |
| PCG-1α | Mouse, C2C12b | NMJ | Skeletal | [111] |
aNeonatal rat ventricular myocytes, a widely used primary cardiac muscle cell culture model
bSkeletal muscle cell line with differentiation potential
cEmbryonic stem cells
dRat myoblast-like tetracycline cell line (ref. [108])
eNo distinction between MEF2 isoforms has been made
Costamere
Transcription factors
MEF2
MEF2 is an evolutionarily conserved transcription factor [29–31] and seminal studies in Drosophila have shown that this regulatory protein is required for the terminal differentiation of cardiac, skeletal, and smooth muscle [32, 33]. MEF2 mutant fly embryos exhibit multiple defects in muscle along with reduced expression of several myosin genes [34], demonstrating that in vivo structural gene expression is dependent on MEF2 activity. Similar to the findings in Drosophila, characterization of the cardiac and skeletal muscle abnormalities in MEF2A and MEF2C mutant mice, respectively, have revealed that integrity of the cytoarchitecture and structural gene expression is dependent on MEF2 in vivo [35, 36]. However, as described below, it appears that vertebrates have evolved distinct cytoarchitectural gene programs that are differentially regulated by MEF2 family members.
MEF2A knockout mice die perinatally with severe cardiac defects including widespread myofibrillar fragmentation [35]. To dissect the molecular mechanism of the cardiac phenotype, microarray analysis was performed using MEF2A knockout hearts. This analysis led to the identification of numerous, novel dysregulated genes, one of which was myospryn, also known as cardiomyopathy gene 5 (CMYA5) [37]. The discovery of myospryn subsequently led to the identification of another downregulated CMYA gene, CMYA3 (myomaxin/Xirp2), in MEF2A knockout hearts [38]. Detailed characterization of myospryn and myomaxin/Xirp2 revealed a costamere localization of these proteins in cardiac muscle. The finding that two genes encoding costamere-localized proteins are sensitive to MEF2A raised the possibility that a defect in this cytoskeletal complex caused the myofibrillar disorganization in MEF2A knockout hearts.
A candidate approach was undertaken to examine the expression of genes encoding costamere localized proteins in MEF2A-deficient cardiomyocytes. A subset of these genes was found to be particularly sensitive to MEF2A deficiency in perinatal mouse hearts and neonatal rat ventricular myocytes [39]. This requirement, however, does not appear to extend to skeletal muscle as costamere gene expression and the cytoarchitecture are largely unaffected in MEF2A knockout skeletal muscle (unpublished data). Additionally, this particular cohort was significantly more dependent on MEF2A than either MEF2C or MEF2D, suggesting that the promoters of these genes are able to discriminate among the MEF2 transcription factor family either through differences in the MEF2 DNA binding sequence or via specific, cooperative interactions with other transcription factors.
Although slight differences in the MEF2 DNA binding consensus sequence have been identified in many MEF2-dependent genes, studies to date have failed to reveal any preference towards specific family members. On the other hand, it is firmly established that MEF2 proteins interact with cofactors to regulate target genes [40]. Related to this notion, the above report described a computational analysis of overrepresented transcription factor binding sites in costamere gene promoters compared to the background genome and found a number of significantly enriched DNA binding motifs located in these promoters [39]. These findings raise the possibility that MEF2A collaborates with those transcription factors bound to the enriched DNA binding motifs to regulate costamere gene expression. Furthermore, there is evidence to suggest that co-activators interact with MEF2 in an isoform-dependent manner. MAML1 (mastermind-like 1), an effector in the Notch signaling pathway, has been shown to function as a specific co-activator of MEF2C but not MEF2A or MEF2D [41]. In the future it will be important to determine the role of putative transcriptional co-regulators in costamere assembly or integrity, and whether they cooperate with MEF2A in an isoform-specific manner in costamere gene regulation.
SRF
Serum response factor is a broadly expressed transcription factor with an essential function in the differentiation of mesoderm derived tissues, such as muscle [42]. The most well studied aspect of SRF pertains to its role in the regulation of the actin cytoskeleton. However, we will not focus on this particular function of SRF as this is the subject of several other comprehensive and excellent reviews [28, 43, 44]. Instead, we will highlight the function of SRF in the regulation of striated muscle cytoarchitectural elements.
Because of the early embryonic lethality of global SRF knockout mice, a floxed allele of SRF was generated to address the role of this transcription factor in muscle development [45]. Cre-mediated deletion of SRF in the cardiovascular system in mice resulted in abnormal cardiac development consisting of cardiomyocytes with disorganized myofibrils and dysregulation of selected structural genes [46, 47]. Taking a similar conditional deletion approach, SRF inactivation in primary neonatal cardiomyocytes from floxed SRF mice resulted in the dysregulation of genes encoding costamere, sarcomere, and numerous other cytoskeletal proteins [48]. Included among the cohort of downregulated cytoskeletal genes were dystrophin, α-dystrobrevin, integrin β 1 (ITGB1), melusin (ITGB1BP2), and β-sarcoglycan, all of which encode proteins that reside within the DGC and integrin complexes. Although costamere structure in SRF-deficient cardiomyocytes was not analyzed in any of these studies, these results suggest that in addition to its established role in actin dynamics, SRF is also involved in a costamere gene regulatory program in cardiac muscle.
Chromatin factors linked to the costamere
HDAC4
It is well established that MEF2 transcriptional function is modulated via its interaction with class II HDACs [40]. Class II HDACs, such as HDAC4, are important modulators of myogenic differentiation [49]. Moreover, HDAC4 has been shown to control structural gene expression in skeletal muscle in vitro and in vivo. Overexpression of a constitutively nuclear form of HDAC4 in C2C12 myotubes and in mouse hindlimb muscle resulted in the repression of numerous muscle structural genes, many of which encode proteins localized to the costamere [50]. Among the downregulated genes, are those that encode dystrophin and β- and γ-sarcoglycan, components of the DGC, which resides in the costamere. Not surprisingly, the repressive effects of HDAC4 on structural gene expression were demonstrated to be mediated through MEF2. Dystrophin and β- and γ-sarcoglycan are also downregulated in MEF2A-deficient cardiomyocytes [39] suggesting that a class II HDAC-MEF2 circuit may operate in the control of costamere gene regulation in both cardiac and skeletal muscle.
Sin3
Mammalian Sin3, encoded by Sin3A and Sin3B, is a component of a multi-subunit chromatin modifying complex that contains histone deacetylases and is recruited to promoter regions to both positively and negatively regulate genes [51]. ChIP-on-chip (chromatin immunoprecipitation followed by DNA microarray) studies in C2C12 myoblasts suggested a role for Sin3 in myogenic differentiation by its ability to bind to numerous muscle regulatory genes, which prompted additional analysis of the role of Sin3 in muscle development [52]. Compound skeletal muscle-specific Sin3A and 3B knockout mice resulted in severe myofibrillar abnormalities in mutant skeletal muscle tissue and primary myotubes demonstrating an essential function for this chromatin factor in the muscle cytoarchitecture [53]. To identify the specific pathways dependent on Sin3 in muscle differentiation, ChIP-sequencing was performed using differentiated C2C12 myotubes. Subsequent pathway analysis of these Sin3 bound DNA sequences revealed an enrichment of genes encoding costamere proteins. A number of these, such as integrin α7 (Itga7) and β1 (Itgb1), dystroglycan 1 (dag1), and α- and β-syntrophins (snta1, sntb1), encode components of the DGC and integrin complexes. Taken together, these findings suggest that costamere gene regulation is dependent on Sin3 function.
Interestingly, the majority of the genes affected in both the Sin3 and SRF studies were not significantly dysregulated in MEF2A-deficient cardiomyocytes suggesting that different transcriptional sub-programs exist to regulate costamere gene expression. Moreover, the vast majority of transcription factor knockout models with cardiac and/or skeletal muscle cytoarchitectural defects have not specifically examined the costamere. There are likely additional transcriptional pathways that will be found to function in the regulation of the costamere due to its essential role in myofibrillar integrity and muscle contraction.
Sarcomere
Transcription factors
MEF2
To define the gene regulatory programs controlled by MEF2 in developing muscle in greater detail, genome-wide MEF2 binding along with gene expression profiling of MEF2 mutants was performed in Drosophila [54]. This study revealed that MEF2 regulates a diverse array of genes encoding proteins belonging to many different pathways, many which were previously demonstrated to be dependent on MEF2, such as actin57B [55], Mlp84B [56], tropomyosin I [57], and myosin light chain 2 [34]. Additionally, prior studies have shown that the troponin I gene, which encodes an essential contractile protein, is a direct target of MEF2 in flies [58]. Characterization of the dorsal vessel (heart) phenotype of a Mef2 hypomorphic allele in Drosophila, which results in severely reduced MEF2 protein levels, revealed reduced expression of myosin and three myosin subunit genes [34]. Taken together, these results support a role for MEF2 in the regulation of sarcomere integrity in invertebrate striated muscle.
An essential role for vertebrate MEF2 proteins in sarcomere stability has been demonstrated in the zebrafish model system. Morpholino knockdown of MEF2A in zebrafish resulted in cardiac contractility dysfunction, a defect that may be caused by the extensive myofibrillar disorganization in cardiomyocytes and downregulation of sarcomere genes [59]. Similarly, combined knockdown of MEF2C and MEF2D in zebrafish resulted in myofibrillar defects but this phenotype was restricted to skeletal myofibers [60]. Interestingly, the specific class of genes downregulated in MEF2C/2D morphants encodes proteins comprising the thick filament system, including fast myosin heavy (myhz1) and light (mylz2) chain genes, as well as the thick filament gene smybpc1. Thus, in zebrafish, it appears that sarcomere integrity in cardiac muscle is dependent on MEF2A, whereas skeletal muscle myofibrils are more dependent on MEF2C and MEF2D.
To investigate the role of MEF2C in mammalian myogenesis, a skeletal muscle-specific deletion of MEF2C in mice was generated in order to circumvent the early embryonic lethality associated with the global knockout. Skeletal muscle-specific knockout of MEF2C did not adversely affect muscle differentiation but resulted in myofiber irregularities [36]. Ultrastructural analysis of skeletal muscle revealed widespread fragmentation of the contractile apparatus, particularly at the M-line region. Consistent with the phenotype, the genes encoding the M-line proteins myomesin 1 and myomesin 2 (M protein) were downregulated in MEF2C-deficient skeletal muscle and subsequently shown to be direct targets of this factor. Additional sarcomere genes such as calsarcin 2 (myozenin 1) and 1 (myozenin 2), skeletal α-actin (Acta1), myosin (Myh7), and myotilin were downregulated in MEF2C conditional knockout skeletal muscle. These results demonstrate that, like zebrafish, the skeletal muscle sarcomere is dependent on MEF2C. Furthermore, the expression data suggest that this factor regulates M-line and Z-disc gene sub-programs within the sarcomere.
SRF
Three independent reports have described a role for SRF in the regulation of sarcomere formation and integrity through the inactivation of SRF in a cardiac-specific manner in mice [46–48]. In one of these studies, Cre-mediated deletion of SRF in primary cardiomyocytes isolated from floxed SRF neonatal mouse hearts resulted in severely disorganized contractile apparatus [48]. Subsequent genome-wide expression profiling revealed numerous dysregulated cytoarchitectural genes, most of which encode proteins localized to the sarcomere. Supporting these observations, ultrastructural examination of SRF-deficient hearts in mice revealed cardiomyocytes with highly disorganized myofibrils, and expression of genes encoding sarcomeric proteins was significantly downregulated [47]. Given the similarity in myofibrillar defects in the SRF knockout studies, it was not surprising when expression analysis revealed a substantial overlap in the dysregulated sarcomere gene set, which included myomesin 1, myosin light chain 1v (MLC1v), MYL9, and smooth muscle α-actin (ACTA2). The embryonic and perinatal sarcomere defects indicate that SRF has an essential role in the formation and maintenance of myofibrillar structure.
GATA4
The GATA4 transcription factor is required for proper cardiac morphogenesis and mediates hypertrophic signaling pathways in cardiomyocytes [61, 62]. Apart from its important role in cardiac development and disease, GATA4 function appears to be required for sarcomere assembly in differentiated cardiac muscle. Antisense-mediated inhibition of GATA4 expression in neonatal rat ventricular myocytes (NRVMs) treated with endothelin or phenylephrine, agonists which promote hypertrophy, attenuated sarcomere reorganization [63]. Conversely, overexpression of GATA4 in NRVMs, in the absence of hypertrophic agonists, was sufficient to induce sarcomere assembly and resulted in the upregulation of contractile protein gene expression. GATA4 is also required for sarcomere integrity downstream of doxorubicin, a chemotherapeutic drug which results in cardiotoxicity and breakdown of the contractile apparatus [64]. Overexpression of GATA4 was able to blunt the doxorubicin-induced myofibrillar disarray. The ability of GATA4 to regulate sarcomere integrity appears to be mediated through its direct activation of CARP (cardiac ankyrin repeat protein/Ankrd1), a member of the muscle ankyrin repeat protein family that is localized to the nucleus and the sarcomere Z-disc.
Nkx2.5
Murine Nkx2.5 and the fly ortholog, tinman, have established functions in cardiac specification and morphogenesis [65–67]. Tinman homozygous null flies fail to develop a heart, but heterozygotes are viable and have been used to reveal a requirement of this factor for cytoarchitectural integrity. Compound heterozygotes of tinman and the Cdc42 RhoGTPase, a small GTPase family member involved in the cell cycle, actin cytoskeleton, and in cardiac hypertrophy in mice [68, 69], resulted in functional and structural deficits in the heart. Examination of the adult heart from these double heterozygote mutant flies revealed a disruption of myofibrillar alignment [70]. Surprisingly, apart from α-actinin immunohistochemistry to detect myofibrillar organization, structural gene expression was not examined in these mutants. It is worth noting that in the same study, contrary to the observations in flies, compound heterozygote mutations of Nkx2.5 and Cdc42 in mice did not perturb sarcomere integrity in cardiac muscle.
Overexpression of a DNA binding-defective Nkx2.5 mutant during early (driven by the βMHC promoter) or late (αMHC promoter) cardiac development in transgenic mice has been shown to result in sarcomere abnormalities along with conduction defects [71]. Myofibrillar disorganization was observed in transgenic cardiomyocytes but detailed expression analysis for genes encoding sarcomere proteins was not performed. Therefore, it is unknown whether Nkx2.5 directly regulates a sarcomere gene program in cardiac muscle and warrants further investigation. Nevertheless, it seems that myofibrillar integrity in cardiac muscle is sensitive to Nkx2.5 dosage based on the observations in mutant flies and transgenic mice.
Tbx20
Mutations in the human TBX20 gene are associated with cardiac developmental abnormalities and cardiomyopathy [72]. The Drosophila homologue of Tbx20, neuromancer, is required for structural integrity in striated muscle. Alpha-actinin staining of neuromancer null mutant cardiac muscle revealed disorganization of Z-discs with associated irregular heartbeats [73]. Like tinman/Cdc42 heterozygote mutant flies, compound heterozygotes of neuromancer and tinman also displayed myofibrillar disorganization. Curiously, global expression analysis of structural genes was not performed in neuromancer null mutants, but expression of the gene encoding the costamere protein dystrophin was reduced in null and double mutant flies. These results suggest that, while neuromancer is clearly required for myofibrillar stability, the primary deficiency may be at the costamere. Because only one gene was examined it remains to be determined whether Tbx20 actually regulates a broad costamere gene program in the fly heart.
As global Tbx20 knockout mice are embryonic lethal, Tbx20 was ablated in a cardiomyocyte-specific manner in mice. Given that these adult mutant mice develop cardiomyopathy with conduction defects, the authors of this study analyzed candidate structural genes and reported the dysregulation of cypher and desmin [74]. Subsequently, ChIP sequencing of adult transgenic hearts overexpressing epitope-tagged Tbx20 revealed significantly enriched binding of Tbx20 to genes that function in sarcomere organization, contraction, and myofibril assembly. As analysis of the cytoarchitecture was not described, it remains to be determined whether myofibrillar integrity is disrupted in Tbx20 mutant hearts. Intriguingly, computational analysis of transcription factor binding motifs identified MEF2A as one of the overrepresented transcription factors associated with Tbx20 bound genes.
Additional nuclear regulatory factors linked to the sarcomere
Apart from those transcription factors with established functions in cardiac and skeletal muscle, a number of knockdown and/or knockout studies of other transcription factors, co-factors, and chromatin-regulatory proteins have also been shown to result in myofibrillar defects and/or dysregulated sarcomere gene expression. These factors will be discussed in the following section.
Transcription factors
Prox1
The homeobox transcription factor Prospero related homeobox 1 (Prox1) functions in a variety of tissues, particularly the hematopoietic and lymphatic systems [75, 76]. Based on the documented expression of Prox1 in the developing heart, the role of Prox1 was examined in this tissue in mice [77]. Cardiac-specific deletion of Prox1 resulted in defective embryonic heart formation with cardiomyocytes displaying extensive sarcomere disarray. Several genes encoding constituents of the sarcomere were shown to be dysregulated in Prox1-deficient cardiomyocytes [78]. For example, cardiac α-actin, βMHC, and sarcomeric α-actinin were downregulated, and myosin binding protein-C (MyBP-C) was upregulated. Additionally, titin and desmin were sensitive to the loss of Prox1. These results suggest a role for Prox1 in the assembly of the sarcomere in the developing heart.
NF-ĸB
The NF-ĸB transcription factor is a recognized central mediator downstream of signals involved in cell differentiation and survival, the immune system, and numerous diseases including cancer [79]. Inhibition of NF-ĸB transcriptional activity using the super repressor IκBα in proliferating C2C12 myoblasts resulted in the upregulation of a number of myofibrillar genes such as the troponins, myosin light and heavy chains, and α-actin [80]. These results suggest that NF-κB functions as a transcriptional repressor of muscle structural genes. However, this effect was found to be indirect since DNA binding supershift assays failed to reveal direct binding of NF-κB subunits on candidate NF-κB sites in the troponin 2 promoter. The repression of these genes by NF-κB was subsequently found to be mediated through the NF-κB-mediated regulation of transcription factor YY1, which is known to function as a transcriptional repressor. Consistent with the above findings, another report showed that the NF-κB subunit p65 directly repressed cardiac and smooth muscle reporters [81]. However, in this instance, repression by p65 required a CArG box (the SRF DNA binding motif) and was shown to directly interact with and inhibit myocardin activity.
CBFβ
Core-binding factor subunit β (CBFβ) along with the Runx transcription factors are part of a transcription factor complex involved in hematopoietic and bone development and leukemia [82]. Although ubiquitously expressed, CBFβ is highly enriched in cardiac and skeletal muscle [83]. Consistent with its high expression in cardiac and skeletal muscle, morpholino-mediated knockdown of CBFβ in zebrafish resulted in severe abnormalities in these tissues [84]. Cardiomyocytes and skeletal myofibers in these mutants consist of thin sarcomeres and diffuse Z-discs, as well as misaligned intercalated discs. Interestingly, CBFβ is localized to the nucleus and sarcomeric Z-disc so it is unknown whether the myofibrillar defects arise from the lack of CBFβ in the Z-disc, its transcriptional function, or both. This study also demonstrated that knockdown of the JunB transcription factor, previously shown to regulate the CBFβ gene, results in reduced CBFβ expression and cardiac contractility defects similar to CBFβ morphant zebrafish embryos. These result suggest that JunB and CBFβ function in the same genetic pathway to control sarcomere Z-disc integrity.
Runx1
Conditional Runx1 knockout mice were generated to study the role of this transcription factor in denervated skeletal muscle, as it had previously been shown to be upregulated in this model of muscle atrophy. Skeletal muscle denervation in these mutant mice resulted in misaligned and irregularly spaced Z-discs including a deficiency in A- and I-bands [85]. In addition, it appeared that thick filaments were absent in denervated Runx1 mutant skeletal muscle. Gene expression profiling experiments revealed several structural genes that were misregulated in denervated Runx1 mutant skeletal muscle. The sarcomeric myosin genes, myh2 and myh3, as well as the keratin genes Krt1-18 and Krt2-8, whose encoded proteins link the Z-disc to the costamere, were not appropriately expressed in Runx1-deficient denervated muscle. Since keratin connects the myofibrils to the costamere it is tempting to speculate that Runx1 co-regulates sarcomere and costamere gene programs.
Lbx2
Lbx1 and Lbx2 are vertebrate homeobox transcription factors related to the Drosophila ladybird transcription factor [86]. Drosophila ladybird is required for cell fate specification in the mesodermal lineage [87]. Conditional RNAi knockdown of ladybird in Drosophila leg muscle has been shown to cause irregular myofilament appearance [88]. While Lbx1 functions in muscle migration in vertebrates, the role of Lbx2 in muscle development is less clear. Zebrafish express Lbx2 in a subset of skeletal muscle precursors. Morpholino knockdown of lbx2 in zebrafish resulted in abnormally shaped slow muscle fibers, and myofibrils were thinner compared to wild type animals [89]. It was found that expression of genes encoding the thick and thin filament system, including skeletal α-actin, several troponin isoforms, and myosin heavy and light chains, was significantly reduced in lbx2 morphant embryos. Given that Lbx2 primarily functions as a transcriptional repressor, it is likely that sarcomere filament gene expression is regulated indirectly by this transcription factor.
HIF-1α
HIF-1α (hypoxia-inducible factor 1α) is a transcription factor involved in responding and adapting to changes in cellular oxygenation in normal physiology and in disease [90]. Global deficiency of HIF-1α in mice results in embryonic lethality with widespread developmental defects, including abnormalities in the cardiovascular system [91]. Cardiac-specific ablation of HIF-1α in mice also results in embryonic lethality and defective cardiac development with the mutant hearts displaying abnormal contractility [92]. Immunohistochemical analysis using α-actinin and myomesin, two sarcomeric proteins, revealed disorganized myofibrils and incomplete sarcomere assembly. In addition, these mice had reduced titin expression, a major protein of the sarcomere. These results suggest that the contractile dysfunction is due to sarcomere abnormalities and appear to link HIF-1α to the regulation of this cytoarchitectural structure.
p63
The p63 transcription factor, a member of the p53 family of transcription factors, is a key regulator of epidermal differentiation in mice [93]. Intriguingly, ChIP-seq studies using epidermal cells identified a number of p63 bound genes that function in cardiac development. This finding led investigators to examine p63 knockout mice for potential cardiac abnormalities. Newborn p63 knockout mice display embryonic cardiac defects with myofibrillar fragmentation and disorganization [94]. Additionally, Z-disc structures were not readily apparent. Supporting these results, siRNA-mediated knockdown of p63 in ES cell derived cardiomyocytes show reduced expression of MLC2v, α-actinin, and troponin-T suggesting that p63 regulates a sarcomere gene program.
Co-factors linked to the sarcomere
Myocardin
Myocardin is a potent transcriptional activator of SRF-dependent gene expression [95, 96]. While considerable emphasis has been placed on myocardin function in smooth muscle, several in vivo studies have shown that myocardin regulates myofibrillar gene expression in striated muscle. In Xenopus, overexpression of myocardin induced the expression of the contractile proteins cardiac α-actin, αMHC, and cardiac troponin I [97]. Conversely, morpholino knockdown of myocardin resulted in reduced expression of the myosin genes, αMHC and MLC2v. In mice, cardiac-specific deletion of myocardin resulted in dilated cardiomyopathy in the adult heart. Widespread loss of myofibrillar striations and reduced expression of cardiac a-actin, MLC2v, and α-tropomyosin genes were reported in this study [98]. Additionally, in the same report, inducible, cardiac-specific knockout of myocardin in adult mice resulted in similar structural defects in cardiomyocytes and a reduction in cardiac α-actin, αMHC, MLC2v, tropomyosin, α-actinin, and desmin gene expression. These results reinforce the notion that an SRF-myocardin pathway is essential for sarcomere assembly and integrity.
Chromatin factors linked to the sarcomere
HDAC4
In addition to regulating costamere genes, a MEF2-HDAC pathway is required for the proper expression of genes encoding sarcomere proteins. Microarray analysis performed using C2C12 myoblasts expressing constitutively nuclear HDAC4 revealed downregulation of a number of genes encoding sarcomere proteins such as, myotilin, myomesin-2, myozenin-1, β myosin heavy chain, and titin [50]. In a set of complementary experiments, skeletal muscle denervation, which promotes the downregulation of sarcomere genes, in mice with reduced HDAC4 levels resulted in derepression of these genes. HDAC-mediated regulation of the sarcomere in skeletal muscle was also demonstrated to be dependent on MEF2.
Sin3
As discussed in the previous section, Sin3 deficiency in mice resulted in severe myofibrillar abnormalities, including thin myofibrils and reduction in Z-disc structures. Subsequent ChIP-seq analysis revealed enriched binding of Sin3 to genes encoding titin, troponin C1, skeletal α-actin, desmin, and tropomyosins 2 and 4 [53]. Ablation of Sin3 in C2C12 and primary myotubes caused reduced expression of several of the aforementioned sarcomere genes demonstrating that Sin3 functions primarily as an activator of sarcomere genes.
DPF3
The chromatin remodeling factor DPF3 (double PHD finger 3) is involved in the regulation of the muscle cytoarchitecture. DPF3 has been shown to be upregulated in the congenital cardiac disorder Tetralogy of Fallot in humans, which is characterized by ventricular chamber and cardiac vessel malformations [99]. Because of its dysregulation in this cardiac disease, the function of DPF3 in striated muscle was examined in greater details using morpholino knockdown in zebrafish and siRNA knockdown in C2C12 myoblasts. Dpf3 depletion in zebrafish resulted in sarcomere disarray and fewer myofibrils in cardiac and skeletal muscle [100]. Gene expression profiling using whole Dpf3 morphant embryos revealed a number of dysregulated structural genes. Knocking down DPF3 in C2C12 myoblasts caused a similar structural phenotype. In addition, genome-wide analysis of DPF3 binding using C2C12 myoblasts identified an enrichment of numerous cytoskeletal genes encoding proteins localized to the sarcomere.
Set7/9
Histone methyltransferases have been shown to play a role in myogenic differentiation [101]. To further understand the role of histone methylation in myogenesis the histone methyltransferase Set7/9 was knocked down in C2C12 myoblasts and zebrafish. Whereas Set7 knockdown impaired C2C12 myogenic differentiation, Set7/9 depletion in zebrafish did not disrupt myogenesis in vivo. Instead, Set7/9 depletion caused severe myofibrillar defects, as determined by a decrease in MHC staining in these morphants [102]. Ultrastructural analysis of skeletal muscle from these Set7/9 morphants revealed narrower myofibrils, fewer Z-bands, and A-band irregularities. It is worth noting that Mef2a expression is affected in this model suggesting that MEF2A may function in the same genetic pathways as Set7/9 to modulate myofibrillar formation and/or integrity in skeletal muscle.
Neuromuscular junction
Transcription factors
MEF2
As previously discussed, a genome-wide analysis of MEF2 target genes in Drosophila revealed a wide array of cellular pathways potentially regulated by this factor in myogenesis [54]. One of the pathways that showed a significant enrichment of MEF2 binding consists of genes encoding components of the neuromuscular junction. Along these lines, neuromuscular junction defects have been reported in somatic muscle in Mef2 mutant flies [103]. In mammalian muscle, MEF2 binding was enriched for genes encoding neuromuscular junction proteins in C2C12 myoblasts [104]. Collectively, these results indicate a role for MEF2 in the regulation of the neuromuscular junction, but the specific isoform(s) that function in this capacity have not been elucidated. Along these lines, examining the various MEF2 knockout mice for potential NMJ defects is likely to be an exciting and fruitful area of investigation in the future.
MyoD, myogenin
Genome-wide ChIP-on-chip analysis using C2C12 myoblasts showed significant enrichment of the myogenic basic helix-loop-helix (bHLH) transcription factors MyoD and myogenin on genes encoding proteins localized to the neuromuscular junction [105]. Supporting these observations, MyoD-deficient skeletal muscle shows abnormal neuromuscular junctions [106, 107]. Acute myogenin depletion in the conditionally immortalized muscle cell line (RMT) results in a reduction in acetylcholine receptor clustering in differentiated myotubes [108]. It is interesting to note that skeletal myofibers in neonatal myogenin knockout mice have normal sarcomere organization but showed reduced expression of the α- and γ-acetylcholine receptor subunit genes [109]. Taken together, these results indicate a central role for the myogenic bHLH factors in neuromuscular junction formation and maintenance.
NF-κB
In addition to affecting myofibrillar gene expression, inhibition of NF-κB expression or activity in C2C12 myoblasts results in a perturbation in acetylcholine receptor clustering (AChR). One of the primary targets appears to be rapsyn, a molecule that promotes clustering at the NMJ. Supporting these observations, skeletal muscle-specific deletion of an NF-κB subunit, RelA/p65, caused a decrease in AChR density at the NMJ in early postnatal mice [110]. Therefore, NF-κB also appears to be required for NMJ integrity in skeletal muscle.
Co-factors linked to the NMJ
PGC-1α
The peroxisome proliferator-activated receptor gamma co-activator 1α (PGC-1α), an established coactivator of nuclear hormone receptors, was found to regulate a cohort of genes encoding proteins in the neuromuscular junction in skeletal muscle [111]. Overexpression of PGC1α in C2C12 myoblasts and skeletal muscle of transgenic mice resulted in increased neuromuscular junction gene expression. Conversely, expression of NMJ genes was downregulated in PGC1α knockout skeletal muscle, and acetylcholine receptor clustering, an indicator of NMJ formation, was dependent on PGC1α activity. Finally, a number of NMJ promoters were found to be cooperatively activated by PGC1α and GABP.
Intercalated discs
Transcription factors
Nkx2.5
In addition to disrupting sarcomere integrity, cardiac overexpression of a DNA binding-defective Nkx2.5 mutant in transgenic mice also resulted in severe conduction abnormalities. These mice exhibited dysregulation of connexin 43 gene expression and perturbations in their contractile apparatus [71, 112]. Connexins are the major constituents of gap junctions, which reside within intercalated discs, and function to chemically couple cardiomyocytes. Related to these findings, conditional deletion of Nkx2.5 resulted in perinatal lethality with conduction defects and reduced expression of the gap junction protein connexin 40 [113]. Available evidence suggests that the myofibrillar defects in these mice arise from abnormalities of the intercalated discs. Thus, one of the principal roles of Nkx2.5 in the mature heart may be to regulate intercalated disc and conduction gene programs.
SRF
The downregulation of the genes encoding the intercalated disc proteins Xin (CMYA1), plakophilin 2 (PKP2), and tight junction protein 1 (ZO-1) has been reported in SRF-deficient cardiac myocytes, which display myofibrillar defects [48]. Although an intercalated disc phenotype was not described in SRF knockout cardiomyocytes, it is possible that SRF is also involved in the regulation of this macromolecular structure given the dysregulation of these genes and the myofibrillar defects.
Consistent with its role as an SRF coactivator, myocardin also results in intercalated disc abnormalities. Myocardin mutant cardiomyocytes had a drastic reduction in expression of the gap junction protein connexin 43 [98]. Moreover, malformations in desmosomes and adherens junctions, two complexes within the intercalated disc, were observed.
Prox1
The downregulation of N-RAP (nebulin-related anchoring protein) and zyxin, encoding proteins localized to the intercalated disc, has been demonstrated in cardiomyocyte-specific Prox1 mutant hearts [83, 114]. Both of these genes were also identified as targets of Prox1 in global ChIP studies. Although expression of these genes was significantly downregulated, defects in this structure were not reported in Prox1 mutant cardiomyocytes. Nevertheless, the possibility remains that Prox1 is part of a transcriptional program in the regulation of intercalated discs in cardiac myocytes.
Hand1
The bHLH transcription factor Hand1 is required for the differentiation of myocardial and non-myocardial cell types in the developing heart [115]. While Hand1 function has been analyzed primarily in early cardiogenic events, cardiac overexpression of Hand1 in adult transgenic mice results in cardiac arrhythmias in adult mice [116]. Detailed examination of these hearts showed reduced expression of connexin 43 and increased β-catenin expression. Although both of these proteins are localized to the intercalated discs there did not appear to be major structural perturbations of this structure. Nevertheless, the conduction abnormalities along with reduced connexin expression suggest a role for Hand1 in intercalated disc function.
Concluding remarks
Although numerous muscle structural genes have been characterized both as a means to dissect the fundamental mechanisms of tissue-specific gene regulation and to identify upstream regulatory pathways in cellular differentiation, the notion that expression of these and other genes whose encoded proteins reside within a defined compartment of the muscle cytoarchitecture may be controlled by a particular transcription factor cohort has received little attention. Despite the advances in understanding the transcriptional control of differentiation and regeneration, there still remains a knowledge gap concerning the transcriptional regulatory programs involved in the formation of the various structural elements in striated muscle. Of the many aspects required to assemble the diverse cytoskeletal structures, ensuring that appropriate protein levels are achieved and maintained for the proper function of a given structure is of critical importance. Furthermore, because of its intricate structural design, striated muscle has likely developed mechanisms to coordinate the synthesis and activity of the various subcellular structures with specific transcription factor pathways. Not only would this mode of regulation be important for differentiation and regeneration, but also for the adaptation to chronic alterations in contractile function in muscle disease.
We have reviewed evidence demonstrating that distinct structural complexes in striated muscle display differential sensitivity to certain transcription factors in both vertebrate and invertebrate model systems. Since many of the structural phenotypes associated with transcription factor mutations have not been thoroughly characterized at the genome level, one primary objective in the muscle field would be to delve deeper into their structural gene expression profiles in order to gain a better handle of the specific gene program(s) controlled by these factors. Conversely, performing a large-scale enhancer or binding motif analysis on a group of genes whose encoded proteins localize to a common subcellular structure may help in the identification of novel transcription factors and regulatory inputs controlling essential structural elements in striated muscle. Although the focus of this review has centered on the direct connection of transcription factors to cytoarchitectural gene expression, many of the highlighted studies lack evidence of direct regulation of these genes and a number of these pathways may utilize post-transcriptional mechanisms such as microRNAs to control levels of transcripts encoding structural genes.
Acknowledgments
We thank members of the Naya laboratory for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health HL73304 to FJN and a diversity supplement (HL73304-S1) to NLE.
References
- 1.Lin Z, Lu MH, Schultheiss T, Choi J, Holtzer S, DiLullo C, Fischman DA, Holtzer H. Sequential appearance of muscle-specific proteins in myoblasts as a function of time after cell division: evidence for a conserved myoblast differentiation program in skeletal muscle. Cell Motil Cytoskeleton. 1994;29(1):1–19. doi: 10.1002/cm.970290102. [DOI] [PubMed] [Google Scholar]
- 2.Sanger JW, Kang S, Siebrands CC, Freeman N, Du A, Wang J, Stout AL, Sanger JM. How to build a myofibril. J Muscle Res Cell Motil. 2005;26(6–8):343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- 3.Sparrow JC, Schöck F. The initial steps of myofibril assembly: integrins pave the way. Nat Rev Mol Cell Biol. 2009;10(4):293–298. doi: 10.1038/nrm2634. [DOI] [PubMed] [Google Scholar]
- 4.Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 5.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104(4):557–567. doi: 10.1016/S0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 6.Nicol RL, Frey N, Olson EN. From the sarcomere to the nucleus: role of genetics and signaling in structural heart disease. Annu Rev Genomics Hum Genet. 2000;1:179–223. doi: 10.1146/annurev.genom.1.1.179. [DOI] [PubMed] [Google Scholar]
- 7.Boateng SY, Goldspink PH. Assembly and maintenance of the sarcomere night and day. Cardiovasc Res. 2008;77(4):667–675. doi: 10.1093/cvr/cvm048. [DOI] [PubMed] [Google Scholar]
- 8.Ehler E, Gautel M. The sarcomere and sarcomerogenesis. Adv Exp Med Biol. 2008;642:1–14. doi: 10.1007/978-0-387-84847-1_1. [DOI] [PubMed] [Google Scholar]
- 9.Pyle WG, Solaro RJ. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ Res. 2004;94(3):296–305. doi: 10.1161/01.RES.0000116143.74830.A9. [DOI] [PubMed] [Google Scholar]
- 10.Frank D, Frey N. Cardiac Z-disc signaling network. J Biol Chem. 2011;286(12):9897–9904. doi: 10.1074/jbc.R110.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MK, Granzier H, Ehler E, Gregorio CC. The sensitive giant: the role of titin-based stretch sensing complexes in the heart. Trends Cell Biol. 2004;14(3):119–126. doi: 10.1016/j.tcb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Gautel M. The sarcomeric cytoskeleton: who picks up the strain? Curr Opin Cell Biol. 2011;23(1):39–46. doi: 10.1016/j.ceb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Lange S, Ehler E, Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16(1):11–18. doi: 10.1016/j.tcb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Sanger JM, Sanger JW. The dynamic Z bands of striated muscle cells. Sci Signal. 2008;1(32):pe37. doi: 10.1126/scisignal.132pe37. [DOI] [PubMed] [Google Scholar]
- 15.Danowski BA, Imanaka-Yoshida K, Sanger JM, Sanger JW. Costameres are sites of force transmission to the substratum in adult rat cardiomyocytes. J Cell Biol. 1992;118(6):1411–1420. doi: 10.1083/jcb.118.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ervasti JM. Costameres: the Achilles’ heel of Herculean muscle. J Biol Chem. 2003;278(16):13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 17.Samarel AM. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am J Physiol Heart Circ Physiol. 2005;289(6):H2291–H2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- 18.Hoshijima M. Mechanical stress-strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol Heart Circ Physiol. 2006;290(4):H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rando TA. The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve. 2001;24(12):1575–1594. doi: 10.1002/mus.1192. [DOI] [PubMed] [Google Scholar]
- 20.Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta. 2007;1772(2):108–117. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Davies KE, Nowak KJ. Molecular mechanisms of muscular dystrophies: old and new players. Nat Rev Mol Cell Biol. 2006;7(10):762–773. doi: 10.1038/nrm2024. [DOI] [PubMed] [Google Scholar]
- 22.Heydemann A, McNally EM. Consequences of disrupting the dystrophin-sarcoglycan complex in cardiac and skeletal myopathy. Trends Cardiovasc Med. 2007;17(2):55–59. doi: 10.1016/j.tcm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Severs NJ. The cardiac gap junction and intercalated disc. Int J Cardiol. 1990;26(2):137–173. doi: 10.1016/0167-5273(90)90030-9. [DOI] [PubMed] [Google Scholar]
- 24.Bennett PM. From myofibril to membrane; the transitional junction at the intercalated disc. Front Biosci (Landmark Ed) 2012;17:1035–1050. doi: 10.2741/3972. [DOI] [PubMed] [Google Scholar]
- 25.Charvet B, Ruggiero F, Le Guellec D. The development of the myotendinous junction. A review. Muscles Ligaments Tendons J. 2012;2(2):53–63. [PMC free article] [PubMed] [Google Scholar]
- 26.Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci. 2001;2(11):791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
- 27.Burden SJ. Building the vertebrate neuromuscular synapse. J Neurobiol. 2002;53(4):501–511. doi: 10.1002/neu.10137. [DOI] [PubMed] [Google Scholar]
- 28.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11(5):353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 30.Naya FJ, Olson E. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol. 1999;11(6):683–688. doi: 10.1016/S0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- 31.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134(23):4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 32.Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9(6):730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 33.Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267(5198):688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 34.Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171(1):169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- 35.Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8(11):1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 36.Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol. 2007;27(23):8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durham JT, Brand OM, Arnold M, Reynolds JG, Muthukumar L, Weiler H, Richardson JA, Naya FJ. Myospryn is a direct transcriptional target for MEF2A that encodes a striated muscle, alpha-actinin-interacting, costamere-localized protein. J Biol Chem. 2006;281(10):6841–6849. doi: 10.1074/jbc.M510499200. [DOI] [PubMed] [Google Scholar]
- 38.Huang HT, Brand OM, Mathew M, Ignatiou C, Ewen EP, McCalmon SA, Naya FJ. Myomaxin is a novel transcriptional target of MEF2A that encodes a Xin-related alpha-actinin interacting protein. J Biol Chem. 2006;281(51):39370–39379. doi: 10.1074/jbc.M603244200. [DOI] [PubMed] [Google Scholar]
- 39.Ewen EP, Snyder CM, Wilson M, Desjardins D, Naya FJ. The Mef2A transcription factor coordinately regulates a costamere gene program in cardiac muscle. J Biol Chem. 2011;286(34):29644–29653. doi: 10.1074/jbc.M111.268094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27(1):40–47. doi: 10.1016/S0968-0004(01)02031-X. [DOI] [PubMed] [Google Scholar]
- 41.Shen H, McElhinny AS, Cao Y, Gao P, Liu J, Bronson R, Griffin JD, Wu L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20(6):675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miano JM. Role of serum response factor in the pathogenesis of disease. Lab Invest. 2010;90(9):1274–1284. doi: 10.1038/labinvest.2010.104. [DOI] [PubMed] [Google Scholar]
- 43.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16(11):588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292(1):C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 45.Arsenian S, Weinhold B, Oelgeschläger M, Rüther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17(21):6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miano JM, Ramanan N, Georger MA, de Mesy Bentley KL, Emerson RL, Balza RO, Jr, Xiao Q, Weiler H, Ginty DD, Misra RP. Restricted inactivation of serum response factor to the cardiovascular system. Proc Natl Acad Sci USA. 2004;101(49):17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu Z, Iyer D, Conway SJ, Martin JF, Ivey K, Srivastava D, Nordheim A, Schwartz RJ. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc Natl Acad Sci USA. 2008;105(46):17824–17829. doi: 10.1073/pnas.0805491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balza RO, Jr, Misra RP. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem. 2006;281(10):6498–6510. doi: 10.1074/jbc.M509487200. [DOI] [PubMed] [Google Scholar]
- 49.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6(2):233–244. doi: 10.1016/S1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 50.Cohen TJ, Barrientos T, Hartman ZC, Garvey SM, Cox GA, Yao TP. The deacetylase HDAC4 controls myocyte enhancing factor-2-dependent structural gene expression in response to neural activity. FASEB J. 2009;23(1):99–106. doi: 10.1096/fj.08-115931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47(1):1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 52.van Oevelen C, Wang J, Asp P, Yan Q, Kaelin WG, Jr, Kluger Y, Dynlacht BD. A role for mammalian Sin3 in permanent gene silencing. Mol Cell. 2008;32(3):359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Oevelen C, Bowman C, Pellegrino J, Asp P, Cheng J, Parisi F, Micsinai M, Kluger Y, Chu A, Blais A, David G, Dynlacht BD. The mammalian Sin3 proteins are required for muscle development and sarcomere specification. Mol Cell Biol. 2010;30(24):5686–5697. doi: 10.1128/MCB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10(6):797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Kelly KK, Meadows SM, Cripps RM. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech Dev. 2002;110(1–2):39–50. doi: 10.1016/S0925-4773(01)00586-X. [DOI] [PubMed] [Google Scholar]
- 56.Stronach BE, Renfranz PJ, Lilly B, Beckerle MC. Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis. Mol Biol Cell. 1999;10(7):2329–2342. doi: 10.1091/mbc.10.7.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin MH, Nguyen HT, Dybala C, Storti RV. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc Natl Acad Sci USA. 1996;93(10):4623–4628. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marín MC, Rodríguez JR, Ferrús A. Transcription of Drosophila troponin I gene is regulated by two conserved, functionally identical, synergistic elements. Mol Biol Cell. 2004;15(3):1185–1196. doi: 10.1091/mbc.E03-09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang YX, Qian LX, Yu Z, Jiang Q, Dong YX, Liu XF, Yang XY, Zhong TP, Song HY. Requirements of myocyte-specific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett. 2005;579(21):4843–4850. doi: 10.1016/j.febslet.2005.07.068. [DOI] [PubMed] [Google Scholar]
- 60.Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development. 2007;134(13):2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou P, He A, Pu WT. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol. 2012;100:143–169. doi: 10.1016/B978-0-12-387786-4.00005-1. [DOI] [PubMed] [Google Scholar]
- 62.Oka T, Xu J, Molkentin JD. Re-employment of developmental transcription factors in adult heart disease. Semin Cell Dev Biol. 2007;18(1):117–131. doi: 10.1016/j.semcdb.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charron F, Tsimiklis G, Arcand M, Robitaille L, Liang Q, Molkentin JD, Meloche S, Nemer M. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev. 2001;15(20):2702–2719. doi: 10.1101/gad.915701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen B, Zhong L, Roush SF, Pentassuglia L, Peng X, Samaras S, Davidson JM, Sawyer DB, Lim CC. Disruption of a GATA4/Ankrd1 signaling axis in cardiomyocytes leads to sarcomere disarray: implications for anthracycline cardiomyopathy. PLoS ONE. 2012;7(4):e35743. doi: 10.1371/journal.pone.0035743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118(3):719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 66.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nk2-5. Genes Dev. 1995;9(13):1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126(6):1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 68.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 69.Maillet M, Lynch JM, Sanna B, York AJ, Zheng Y, Molkentin JD. Cdc42 is an antihypertrophic molecular switch in the mouse heart. J Clin Invest. 2009;119(10):3079–3088. doi: 10.1172/JCI37694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qian L, Wythe JD, Liu J, Cartry J, Vogler G, Mohapatra B, Otway RT, Huang Y, King IN, Maillet M, Zheng Y, Crawley T, Taghli-Lamallem O, Semsarian C, Dunwoodie S, Winlaw D, Harvey RP, Fatkin D, Towbin JA, Molkentin JD, Srivastava D, Ocorr K, Bruneau BG, Bodmer R. Tinman/Nk2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J Cell Biol. 2011;193(7):1181–1196. doi: 10.1083/jcb.201006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kasahara H, Ueyama T, Wakimoto H, Liu MK, Maguire CT, Converso KL, Kang PM, Manning WJ, Lawitts J, Paul DL, Berul CI, Izumo S. Nkx2.5 homeoprotein regulates expression of gap junction protein connexin 43 and sarcomere organization in postnatal cardiomyocytes. J Mol Cell Cardiol. 2003;35(3):243–256. doi: 10.1016/S0022-2828(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 72.Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, Mackay JP, Waddell LB, Cole AD, Hayward C, Keogh A, Macdonald P, Griffiths L, Fatkin D, Sholler GF, Zorn AM, Feneley MP, Winlaw DS, Harvey RP. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81(2):280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qian L, Mohapatra B, Akasaka T, Liu J, Ocorr K, Towbin JA, Bodmer R. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc Natl Acad Sci USA. 2008;105(50):19833–19838. doi: 10.1073/pnas.0808705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, Boogerd CJ, Cai CL, Peterson K, Chen J, Nobrega MA, Evans SM. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121(12):4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Francois M, Harvey NL, Hogan BM. The transcriptional control of lymphatic vascular development. Physiology (Bethesda) 2011;26(3):146–155. doi: 10.1152/physiol.00053.2010. [DOI] [PubMed] [Google Scholar]
- 76.Hope KJ, Sauvageau G. Roles for MSI2 and PROX1 in hematopoietic stem cell activity. Curr Opin Hematol. 2011;18(4):203–207. doi: 10.1097/MOH.0b013e328347888a. [DOI] [PubMed] [Google Scholar]
- 77.Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44(1):3–16. doi: 10.1016/0925-4773(93)90012-M. [DOI] [PubMed] [Google Scholar]
- 78.Risebro CA, Searles RG, Melville AA, Ehler E, Jina N, Shah S, Pallas J, Hubank M, Dillard M, Harvey NL, Schwartz RJ, Chien KR, Oliver G, Riley PR. Prox1 maintains muscle structure and growth in the developing heart. Development. 2009;136(3):495–505. doi: 10.1242/dev.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 80.Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, Carathers M, Davuluri R, Guttridge DC. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol. 2007;27(12):4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang RH, Zheng XL, Callis TE, Stansfield WE, He J, Baldwin AS, Wang DZ, Selzman CH. Myocardin inhibits cellular proliferation by inhibiting NF-kappaB (p65)-dependent cell cycle progression. Proc Natl Acad Sci USA. 2008;105(9):3362–3367. doi: 10.1073/pnas.0705842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 83.Chiba N, Watanabe T, Nomura S, Tanaka Y, Minowa M, Niki M, Kanamaru R, Satake M. Differentiation-dependent expression and distinct subcellular localization of the protooncogene product, PEBP2beta/CBFbeta, in muscle development. Oncogene. 1997;14(21):2543–2552. doi: 10.1038/sj.onc.1201109. [DOI] [PubMed] [Google Scholar]
- 84.Meder B, Just S, Vogel B, Rudloff J, Gärtner L, Dahme T, Huttner I, Zankl A, Katus HA, Rottbauer W. JunB-CBFbeta signaling is essential to maintain sarcomeric Z-disc structure and when defective leads to heart failure. J Cell Sci. 2010;123(Pt 15):2613–2620. doi: 10.1242/jcs.067967. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, Littman DR, Burden SJ. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2005;19(14):1715–1722. doi: 10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buckingham M. Skeletal muscle formation in vertebrates. Curr Opin Genet Dev. 2001;11(4):440–448. doi: 10.1016/S0959-437X(00)00215-X. [DOI] [PubMed] [Google Scholar]
- 87.Jagla K, Bellard M, Frasch M. A cluster of Drosophila homeobox genes involved in mesoderm differentiation programs. BioEssays. 2001;23(2):125–133. doi: 10.1002/1521-1878(200102)23:2<125::AID-BIES1019>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 88.Maqbool T, Soler C, Jagla T, Daczewska M, Lodha N, Palliyil S, VijayRaghavan K, Jagla K. Shaping leg muscles in Drosophila: role of ladybird, a conserved regulator of appendicular myogenesis. PLoS ONE. 2006;1:e122. doi: 10.1371/journal.pone.0000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ochi H, Westerfield M. Lbx2 regulates formation of myofibrils. BMC Dev Biol. 2009;9:13. doi: 10.1186/1471-213X-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12(2):149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krishnan J, Ahuja P, Bodenmann S, Knapik D, Perriard E, Krek W, Perriard JC. Essential role of developmentally activated hypoxia-inducible factor 1alpha for cardiac morphogenesis and function. Circ Res. 2008;103(10):1139–1146. doi: 10.1161/01.RES.0000338613.89841.c1. [DOI] [PubMed] [Google Scholar]
- 93.Koster MI, Roop DR. The role of p63 in development and differentiation of the epidermis. J Dermatol Sci. 2004;34(1):3–9. doi: 10.1016/j.jdermsci.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 94.Rouleau M, Medawar A, Hamon L, Shivtiel S, Wolchinsky Z, Zhou H, De Rosa L, Candi E, de la Forest Divonne S, Mikkola ML, van Bokhoven H, Missero C, Melino G, Pucéat M, Aberdam D. TAp63 is important for cardiac differentiation of embryonic stem cells and heart development. Stem Cells. 2011;29(11):1672–1683. doi: 10.1002/stem.723. [DOI] [PubMed] [Google Scholar]
- 95.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20(12):1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 96.Parmacek MS. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circ Res. 2007;100(5):633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 97.Small EM, Warkman AS, Wang DZ, Sutherland LB, Olson EN, Krieg PA. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development. 2005;132(5):987–997. doi: 10.1242/dev.01684. [DOI] [PubMed] [Google Scholar]
- 98.Huang J, Min Lu M, Cheng L, Yuan LJ, Zhu X, Stout AL, Chen M, Li J, Parmacek MS. Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc Natl Acad Sci USA. 2009;106(44):18734–18739. doi: 10.1073/pnas.0910749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaynak B, von Heydebreck A, Mebus S, Seelow D, Hennig S, Vogel J, Sperling HP, Pregla R, Alexi-Meskishvili V, Hetzer R, Lange PE, Vingron M, Lehrach H, Sperling S. Genome-wide array analysis of normal and malformed human hearts. Circulation. 2003;107(19):2467–2474. doi: 10.1161/01.CIR.0000066694.21510.E2. [DOI] [PubMed] [Google Scholar]
- 100.Lange M, Kaynak B, Forster UB, Tönjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, Mebus S, Lehrach H, Lurz R, Gobom J, Rottbauer W, Abdelilah-Seyfried S, Sperling S. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22(17):2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25(14):3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R, Zhang Y, Jin SW, Wang DZ. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol. 2011;194(4):551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prokop A, Landgraf M, Rushton E, Broadie K, Bate M. Presynaptic development at the Drosophila neuromuscular junction: assembly and localization of presynaptic active zones. Neuron. 1996;17(4):617–626. doi: 10.1016/S0896-6273(00)80195-6. [DOI] [PubMed] [Google Scholar]
- 104.Paris J, Virtanen C, Lu Z, Takahashi M. Identification of MEF2-regulated genes during muscle differentiation. Physiol Genomics. 2004;20(1):143–151. doi: 10.1152/physiolgenomics.00149.2004. [DOI] [PubMed] [Google Scholar]
- 105.Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19(5):553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang ZZ, Washabaugh CH, Yao Y, Wang JM, Zhang L, Ontell MP, Watkins SC, Rudnicki MA, Ontell M. Aberrant development of motor axons and neuromuscular synapses in MyoD-null mice. J Neurosci. 2003;23(12):5161–5169. doi: 10.1523/JNEUROSCI.23-12-05161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Macharia R, Otto A, Valasek P, Patel K. Neuromuscular junction morphology, fiber-type proportions, and satellite-cell proliferation rates are altered in MyoD(−/−) mice. Muscle Nerve. 2010;42(1):38–52. doi: 10.1002/mus.21637. [DOI] [PubMed] [Google Scholar]
- 108.Macpherson PC, Cieslak D, Goldman D. Myogenin-dependent nAChR clustering in aneural myotubes. Mol Cell Neurosci. 2006;31(4):649–660. doi: 10.1016/j.mcn.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 109.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364(6437):501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 110.Wang J, Fu XQ, Lei WL, Wang T, Sheng AL, Luo ZG. Nuclear factor kappaB controls acetylcholine receptor clustering at the neuromuscular junction. J Neurosci. 2010;30(33):11104–11113. doi: 10.1523/JNEUROSCI.2118-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Handschin C, Kobayashi YM, Chin S, Seale P, Campbell KP, Spiegelman BM. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007;21(7):770–783. doi: 10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kasahara H, Wakimoto H, Liu M, Maguire CT, Converso KL, Shioi T, Huang WY, Manning WJ, Paul D, Lawitts J, Berul CI, Izumo S. Progressive atrioventricular conduction defects and heart failure in mice expressing a mutant Csx/Nkx2.5 homeoprotein. J Clin Invest. 2001;108(2):189–201. doi: 10.1172/JCI12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Briggs LE, Takeda M, Cuadra AE, Wakimoto H, Marks MH, Walker AJ, Seki T, Oh SP, Lu JT, Sumners C, Raizada MK, Horikoshi N, Weinberg EO, Yasui K, Ikeda Y, Chien KR, Kasahara H. Perinatal loss of Nk2-5 results in rapid conduction and contraction defects. Circ Res. 2008;103(6):580–590. doi: 10.1161/CIRCRESAHA.108.171835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crawford GL, Horowits R. Scaffolds and chaperones in myofibril assembly: putting the striations in striated muscle. Biophys Rev. 2011;3(1):25–32. doi: 10.1007/s12551-011-0043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vincentz JW, Barnes RM, Firulli AB. Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011;91(6):485–494. doi: 10.1002/bdra.20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Breckenridge RA, Zuberi Z, Gomes J, Orford R, Dupays L, Felkin LE, Clark JE, Magee AI, Ehler E, Birks EJ, Barton PJ, Tinker A, Mohun TJ. Overexpression of the transcription factor Hand1 causes predisposition towards arrhythmia in mice. J Mol Cell Cardiol. 2009;47(1):133–141. doi: 10.1016/j.yjmcc.2009.04.007. [DOI] [PubMed] [Google Scholar]