Abstract
An actinomycete was isolated from mangrove soil collected from Nellore region of Andhra Pradesh, India, and screened for its ability to produce bioactive compounds. The cultural, morphological, and biochemical characters and 16S rRNA sequencing suggest that the isolated strain is Nocardiopsis alba. The bioactive compounds produced by this strain were purified by column chromatography. The in vitro antioxidant capacity of the isolated compounds (fractions) was estimated and fraction F2 showed very near values to the standard ascorbic acid. The potential fraction obtained by column chromatography was subjected to HPLC for further purification, then this purified fraction F2 was examined by FTIR, NMR, and mass spectroscopy to elucidate its chemical structure. By spectral data, the structure of the isolated compound was predicted as “(Z)-1-((1-hydroxypenta-2,4-dien-1-yl)oxy)anthracene-9,10-dione.”
1. Introduction
Because of their useful biological activities, microbial secondary metabolites have received considerable attention especially in the beneficial effects of human health. Biosynthesis of these secondary metabolites through metabolic engineering and industrial biotechnology offers significant advantage over conventional methods for extraction from biomass. Among the microorganisms, marine bacteria produce unique and novel secondary metabolites and these organisms display interesting biological activities. Along with the different types of marine bacteria, actinomycetes also play an extensive role in the pharmaceutical and medical industry for their capacity to produce secondary metabolites with diverse chemical structures and biological activities. Thousands of bioactive compounds have been isolated and characterized, many of which have been developed into drugs for treatment of wide range of diseases in human, veterinary, and agriculture sectors [1–3]. Hence, the actinomycetes are considered to be the most potent source for the production of secondary metabolites, antibiotics, and other bioactive compounds. It is well established that each actinomycete strain has probably genetic potential ability to produce 10–20 secondary metabolites [4, 5]. A large body of evidences stated that actinomycetes are noteworthy as antibiotic producers, making 75% of all known products, and the Streptomyces has special role in antibiotic production [6, 7]. Streptomyces yielded many therapeutic agents which include antibacterial such as tetracyclines, antifungal such as amphotericin, and anticancer drugs exemplified by Adriamycin and the immunosuppressant tacrolimus [8]. Streptomyces has been reported to contribute nearly 70% of metabolites described under actinobacteria [9]. Streptomycetes and related actinomycetes continue to be useful sources of novel secondary metabolites with a range of biological activities that may ultimately find applications as anti-infectives, anticancer agents, or other pharmaceutically useful compounds [10].
Therefore, screening, isolation, and characterization of promising strains of actinomycetes producing potential antibiotics and other therapeutics have been a major part of research [11, 12]. Recent studies are focusing on the response of antioxidant system of bacteria, which is important in terms of biotechnology, such as Streptomyces growth in various oxidative stress conditions [13]. Nocardiopsis species is one of the actinomycetes which may produce different types of pharmacological compounds with antioxidant, antitumor, anti-inflammatory, antibacterial, and antioxidant properties. Searching for unique actinomycete that metabolized an essential component in natural product-based drug is becoming more and more interesting and meaningful. Antioxidants play an important role in inhibiting and scavenging free radicals, thus providing protection to humans against various infections and degenerative diseases [14]. Either increased free radicals or decreased antioxidant can lead to oxidative stress, which signifies the identification of natural antioxidative agents. There are certain naturally occurring antioxidants that can give protection against oxidative stress induced damage in human cells. Modern research is now directed towards natural antioxidants from plants and microorganisms which serves as safe therapeutics [15]. Therefore, the main objective of this work was the production and characterization of novel bioactive compound from marine actinomycetes and to screen their antioxidant properties.
2. Materials and Methods
2.1. Collection of Soil
Mangrove soil was collected from a field in Kandaleru creek, near Konamala, Gudur, Nellore (dist.), Andhra Pradesh, India. The soil sample was collected from 2 inches below the soil surface and soil samples were transferred to lab in sterilized polythene bags.
2.2. Isolation of Actinomycetes from Mangrove Soil
Starch casein agar (SCA) medium with 50% seawater was used for isolation of actinomycetes as described by El-Nakeeb [16] and Küster and Williams [17].
2.3. Screening for Bioactive Compound Producing Actinomycetes
The isolated strains were screened for the production of bioactive compounds. The isolated actinomycetes strains were inoculated in ISP2 medium with 50% seawater. The inoculated flask was kept for incubation at room temperature for a period of 5 days on rotary shaker (120 rpm) at 28°C. After incubation the broth was filtered and the filtrate was used to test antimicrobial activity. The overnight cultures like E. coli (ATCC 9837), Staphylococcus aureus (ATCC 6538), Bacillus subtilis (ATCC 9856), and Pseudomonas aeruginosa (ATCC 9027) were used as test organisms. The test organisms were spread uniformly on agar plate and wells were bored on the agar surface; then the wells were filled with the culture filtrate. Then plates were incubated at 37°C for 24 to 48 hrs.
2.4. Morphological and Taxonomical Identification of Isolated Strain
Based on the results of screening, one potential strain was selected for further investigations. The potential isolate (GN2) was observed for aerial spore, mycelia color, spore chain morphology, and other microscopic characters. The taxonomic identification of actinomycetes sp. was based on Nonomura's key and Bergey's Manual [18, 19]. Finally the strain was identified by 16s DNA sequencing.
2.5. Production and Extraction of Bioactive Compounds
All the media used in this study were prepared in 50 mL filtered seawater and 50 mL distilled water. Hence the growth was optimum at that proportion. Spores of potential actinomycete strain were scrapped from starch casein agar and inoculated into 50 mL of inoculation medium in 250 mL conical flask and kept in rotary shaker at 120 rpm for 48 hours at 28°C. Then 10% of inoculum was transferred into 100 mL of production medium and kept in rotary shaker at 120 rpm for 7 days at 28°C. After fermentation, mycelium and supernatant were separated first by filtration and finally by centrifugation at 10,000 rpm for 30 minutes at 4°C. The extracellular compounds from culture supernatant were extracted by liquid-liquid extraction method using equal amount of ethyl acetate and concentrated by Rota evaporation.
The crude extract (2 g) was subjected to silica gel column (15 × 2.5 cm with 200 to 300 mesh size) using different solvent systems. The separation of the crude extract was conducted via gradient elution with hexane: ethyl acetate. The solvent fractions F1, F2, F3, and F4 were collected at the concentration of 8 : 2 ratio; the purity of the compound was found by TLC at 2 : 8 ratio of mobile phase (hexane : ethyl acetate).
2.6. Determination of Biological Activities of the Bioactive Compounds
2.6.1. Determination of the Total Phenolics
The total phenolic content of four fractions (bioactive compounds) was determined spectrophotometrically with Folin-Ciocalteu reagent, using a slightly modified method by Junaid et al. [20]. The extract was mixed with Folin-Ciocalteu reagent (1 : 1) and 4 mL of sodium carbonate (1 M) was added and allowed to stand for 15 minutes. The absorbance was read spectrophotometrically at 765 nm. A standard curve was plotted using different concentrations of gallic acid (standard, 0–1000 μg/mL) and the total phenolic content of extract was estimated as μg gallic acid equivalents (GAE)/mg of extract. The reaction was conducted in triplicate, and the results were averaged.
2.6.2. Total Antioxidant Activity
Total antioxidant activity of the fractions was determined according to the method of Prieto et al. [21]. 0.3 mL of each fraction was mixed with 3.0 mL of reagent solution (0.6 M sulphuric acid, 28 mM sodium phosphate, and 4 mM ammonium molybdate). Reaction mixture was incubated at 95°C for 90 min in a water bath. Absorbance of all the sample mixtures was measured at 695 nm. Ascorbic acid (100 μg/mL) was used as standard control.
2.6.3. Qualitative Test for Free Radical Scavenging Activity
10–15 μL of each fraction was spotted on the baseline of the silica gel plates (Himedia) as a spot for chromatographic separation and identification of the fractions using methanol : chloroform (95 : 5, v/v) as mobile phase. It was allowed to develop the chromatogram for 30 minutes. After completion of the chromatogram the whole plate was sprayed with 0.15% (w/v) DPPH solution using an atomizer [22].
2.6.4. Quantitative Test for Free Radical Scavenging Activity by DPPH
Free radical scavenging activity of each fraction was assayed by DPPH (1,1-diphenyl-2-picrylhydrazyl) [23]. 2 mL of DPPH solution (0.002% in methanol) was mixed with 2 mL of different concentrations (5–200 μg/mL) of each fraction and standard (ascorbic acid) in separate tubes. The tubes were incubated in dark at room temperature for 30 minutes and the optical density was measured at 517 nm using UV-Vis spectrophotometer. The absorbance of the DPPH control (without extract/standard) was noted. The scavenging activity was calculated using the formula
Scavenging activity (%) = [(A − B)/A] × 100, where A is absorbance of DPPH control and B is absorbance of DPPH in the presence of extract/standard.
2.6.5. Total Reducing Power
Total reducing capacity of the extracts was determined according to the method of Subramaniam [24]. The bioactive compound containing fractions (100 μg/mL) were mixed with 1% potassium ferricyanide and the mixture was incubated at 50°C for 20 min; 2.5 mL of 10% trichloroacetic acid (TCA) was added to the mixture and centrifuged at 5000 rpm for 10 min. The upper layer of solution (2.5 mL) was mixed with 2.5 mL of distilled water and 0.5 mL of 0.1% ferric chloride and the color developed was measured at 700 nm. Ascorbic acid (100 μg/mL) was used as standard control.
2.7. Structure Elucidation
Among the four fractions F2 was more potent based on preliminary screening, that is, antioxidant capacity, so the purity of the F2 was confirmed by HPLC by using hexane and ethyl acetate as mobile phase at a flow rate of 1 mL min−1; detection was carried out by UV detector with 209 nm. Then the pure compound F2 structure was predicted by spectral analysis. All solvents used for spectroscopic and other physical studies were reagent grade and were further purified by standard methods [25]. Melting points (mp) were determined using a calibrated thermometer by Guna Digital MeltingPoint apparatus and expressed in degrees centigrade (°C). Infrared spectra (IR) were obtained on a Perkin-Elmer Model 281-B spectrophotometer. Samples were analyzed as potassium bromide (KBr) disks. Absorption was reported in wavenumbers (cm−1). 1H and 13C NMR spectra were recorded as solutions in DMSO-d 6 on a Bruker AMX 400 MHz spectrometer operating at 400 MHz for 1H and 100 MHz for 13C. The 1H and 13C chemical shifts were expressed in parts per million (ppm) with reference to tetramethylsilane (TMS). LCMS mass spectra were recorded on a Jeol SX 102 DA/600 mass spectrometer.
2.8. Statistical Analysis
Experimental results are the mean ± standard deviation (SD). Statistical comparisons using one way analysis of variance (ANOVA) followed by Duncan's test for comparison of results between lichens samples and standard antioxidant with P < 0.05 was regarded as significance and P < 0.01 as very significant.
3. Results and Discussion
3.1. Isolation and Screening of Actinomycetes
Two morphologically different strains along with clear zones around the colony were observed in the starch casein medium after 5 days of incubation (Figure 1). These two strains, GN1 and GN2, were screened for the production of bioactive compounds; both the strains exhibited good antimicrobial activity against different pathogenic bacteria as shown in Table 1. But the second strain which showed maximum activity was selected for morphological, cultural, molecular characterization and bioactive compound production. These results were correlated with the bioactive compounds from N. pseudobrasiliensis and N. mediterranei [26, 27].
Figure 1.
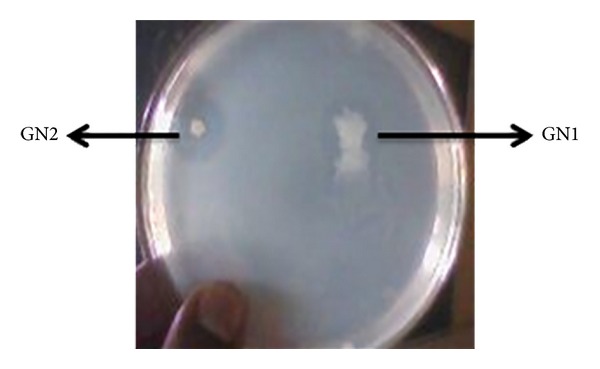
Isolated colonies of actinomycetes showing clear zone on SCA medium.
Table 1.
Screening of actinomycetes by antimicrobial activity.
| S. number | Test organisms | Zone of inhibition in cm | |
|---|---|---|---|
| GN1 | GN2 | ||
| 1 | E. coli | 0.9 | 1.5 |
| 2 | Staphylococcus aureus | 0.7 | 1.2 |
| 3 | Bacillus subtilis | 0.7 | 1.0 |
| 4 | Pseudomonas aeruginosa | 0.6 | 1.0 |
3.2. Morphological and Biochemical Test
Micro- and macroscopic characteristics: the aerial mycelium formed monopodially branched spore-bearing hyphae with the shape of loops, open or compact spirals with 3–6 curves (Figure 2). The strain was clearly polymorph and the colonies were completely covered by aerial mycelium and it formed a clear zone around the colony (Figure 1). These morphological characters are closely agreed with the findings of Goodfellow [28] and Hoshino et al. [29]. In carbon assimilation test the isolate GN2 grew poorly in the presence of sucrose and fructose as a sole carbon source and formed abundant mycelium on the media with glucose and maltose, whereas in nitrogen utilization test, it has grown abundantly on glutamic acid and poorly on histidine, methionine, and leucine. It also showed other biochemical characters like starch, casein, and gelatin hydrolysis as shown in Table 2.
Figure 2.
Microscopic picture of isolated strain.
Table 2.
Macroscopic and microscopic tests of GN2 strain.
| Characters | GN2 |
|---|---|
| Colony appearance | Mycelial (cottony) |
| Sporulation of aerial mycelia | Long chains |
| Motility | Nonmotile |
| Colony colour | White |
| Gram's staining | + |
| Starch hydrolysis | + |
| Gelatin hydrolysis | + |
| Casein hydrolysis | + |
|
| |
| Carbon utilization | |
| Glucose | ++ |
| Sucrose | + |
| Fructose | + |
| Maltose | ++ |
|
| |
| Nitrogen utilization | |
| Glutamic acid | ++ |
| Histidine | + |
| Methionine | + |
| Leucine | + |
3.3. Identification of Actinomycetes
The taxonomic identification of the GN2 was based on 16s rDNA analysis. The 16s rDNA sequence of the strain was compared with the sequences in GenBank using BLAST and aligned with the sequences retrieved from NCBI GenBank database using the Clustal W method. The phylogenetic tree was constructed based on neighbor joining tree method and illustrated in Figure 3. The database was deposited in NCBI GenBank with an accession number KC710971. Based on the cultural, morphological, physiological, and molecular analysis, the GN2 was identified as Nocardiopsis alba.
Figure 3.
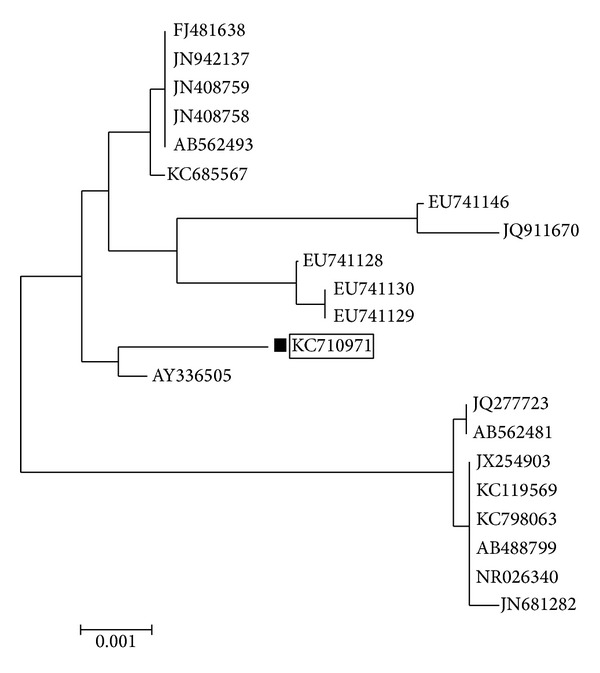
Phylogenetic tree of isolated actinomycetes GN2.
3.4. Determination of Biological Activities of the Bioactive Compounds
3.4.1. Determination of Total Phenolics and Total Antioxidants
The antioxidant activities of bioactive compounds are mainly due to their redox properties, which can play an important role in absorbing and neutralizing free radicals [30]. The phenolic content of each of the fractions was estimated as 13.62 ± 1.12; 13.94 ± 0.98; 12.52 ± 1.39; and 14.37 ± 1.47 mgGA/g (F1, F2, F3, and F4), respectively, which is very similar to that of phenolic activity of the standard (ascorbic acid) (19.48 ± 1.37 mgGA/g) shown in Figure 4. The highest antioxidant capacity of bioactive compound could be attributed to the presence of high total polyphenol contents, since a positive correlation between phenolic composition and antioxidant activity was proved by Katalinic et al. [31]. The presence of the phenolic groups in the secondary metabolites is considered to be a key element for the antioxidative efficiency [32].
Figure 4.
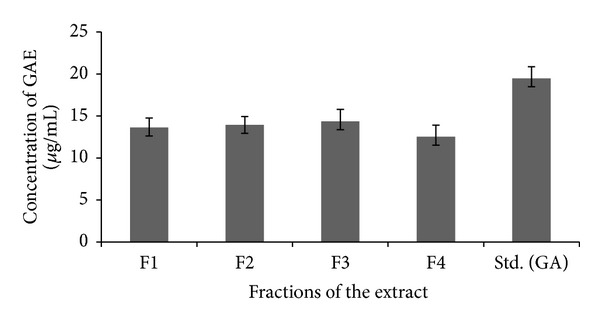
Total phenolics in the fractions from GN2 strain compared with standard gallic acid.
The bioactive compounds extracted from fractions showed very potent total antioxidant capacity. The results of experimental samples and the standard antioxidant (ascorbic acid) equivalents are presented in Figure 5. The results showed that the total antioxidant capacity of extracted samples was 2.72 ± 0.4, 2.95 ± 1.18, 3.05 ± 0.98, and 1.62 ± 0.4 AA/g, respectively, and the standard antioxidant ascorbic acid shows 10.63 ± 0.85 AA/g antioxidant activity. In the present experiment, total antioxidant activity was increased if total phenolics in terms of gallic acid equivalents were increased (Figure 6)
Figure 5.
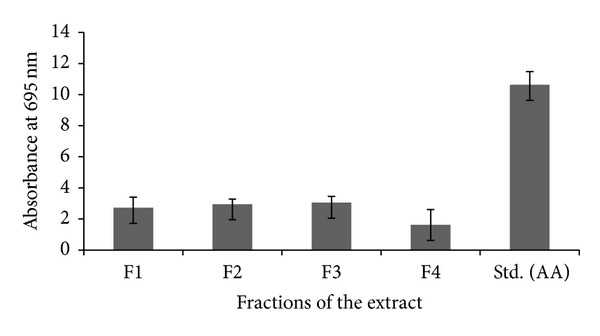
Total antioxidant activities of the fractions and standard antioxidant ascorbic acid.
Figure 6.
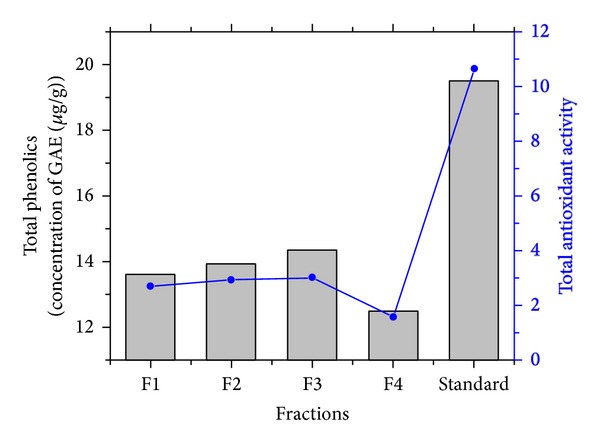
Variation of antioxidant capacity as a function of total phenolics content.
3.4.2. Qualitative and Quantitative Test for Free Radical Scavenging Activity by DPPH
The DPPH assay is one of the most common and relatively quick methods used for testing radical scavenging activity of biological active particles [33]. The chromatogram the plate was sprayed with DPPH (0.15% W/V) solution. The yellow color spots (Figure 7) indicated the presence of antioxidant nature of collected fractions with R f values of 15, 12, and 10 cm. The results of radical scavenging effect of four fractions and ascorbic acid have exhibited dependent scavenging activity of DPPH radicals. Though the fractions were able to scavenge DPPH* (free radical) and convert it into DPPHH, the scavenging effect of the fractions was lesser than that of ascorbic acid. The radical scavenging effect of F2, F3, and F4 was greater than 50% at concentration of 50 μg/mL and that fraction 1 was greater than 50% at concentration of 100 μg/mL.
Figure 7.
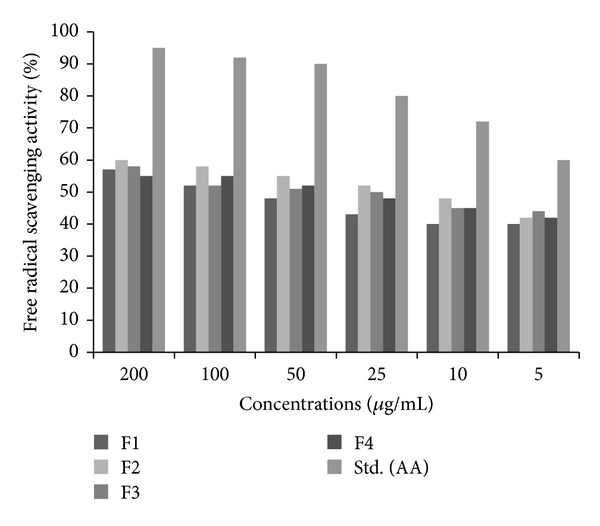
Free radical scavenging activities of four fractions and ascorbic acid.
3.4.3. Reducing Power
The reducing power of the four fractions was determined by the reduction of Fe3+ to Fe2+ in the presence of different concentrations of each fraction and ascorbic acid. The absorbance of reaction mixture at 700 nm increased with the increase in concentration of extract indicating reducing potential of extract. Measured values of absorbance varied from 0.68 to 1.56. Among the tested fractions, fraction 2 gave the highest reducing power, although the reducing activity was lower than the standard ascorbic acid as shown in Figure 8. The reducing capacity of the tested fractions decreased in the following order: fractions 2, 3, and 4 and fraction 1, respectively.
Figure 8.
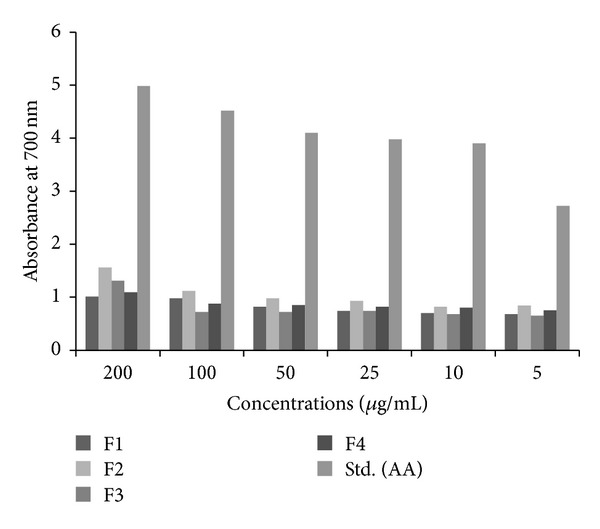
Ferric reducing activities of four fractions and ascorbic acid.
3.5. Structure Elucidation
The isolated pure compound showed a well developed peak having a retention time at 5 min (Figure 9). The molecular weight of this compound (F2) was determined by a mass spectrum and was found to be m/z 306.09 as a molecular ion (Figure 10). By the elemental analysis, the molecular formula of fraction 2 was identified as C19H14O4. Based on the molecular formula (C19H14O4) the double bond equivalents (DBE) value of fraction 2 was calculated as thirteen. After assuming a (DBE) value the two IR signal at signals observed at 1682 and 1667 cm−1 indicates the presence of two C=O groups indicates two double bonds equivalency. IR signal at 1612 cm−1 represents the conjugated C=C bond represents two double bonds equivalency. The IR signal at 3410 cm−1 represents the presence of aliphatic OH group. IR signal at 3013 cm−1 represents the presence of aromatic protons. Two signals at 2928 cm−1 and 2857 cm−1 and one signal at 1612 cm−1 represent the presence of conjugated =C–H signal (Figure 11). It was further supported by the presence of twelve aromatic carbons, two carbonyl carbons, and four conjugated double bonded carbons in its 13C NMR spectrum of sample. 13C signals at δ 176.7 and 176.0 are supporting to the two carbonyl carbons (Figure 12).
Figure 9.
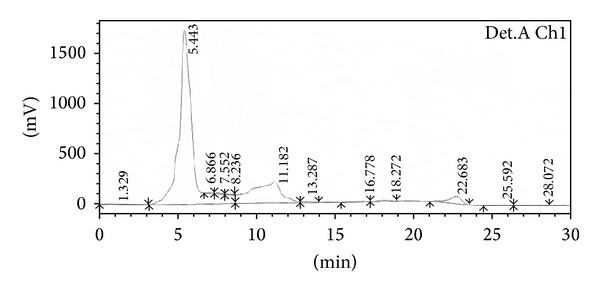
HPLC of isolated bioactive compound (fraction 2).
Figure 10.
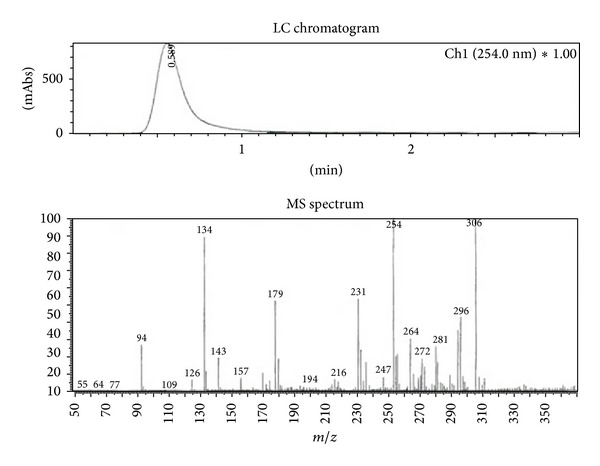
Mass spectrum of the F2 fraction.
Figure 11.
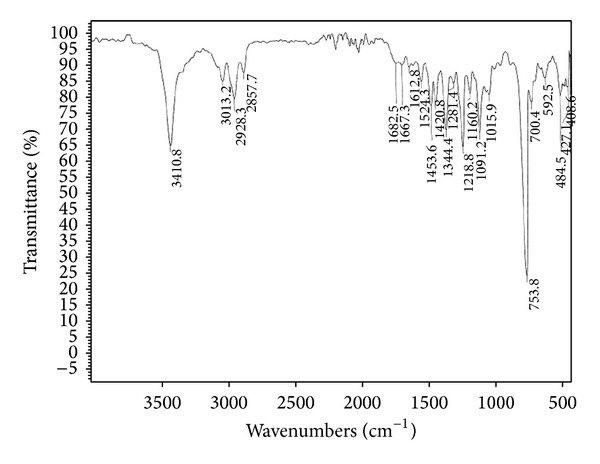
IR spectrum of the F2 fraction.
Figure 12.
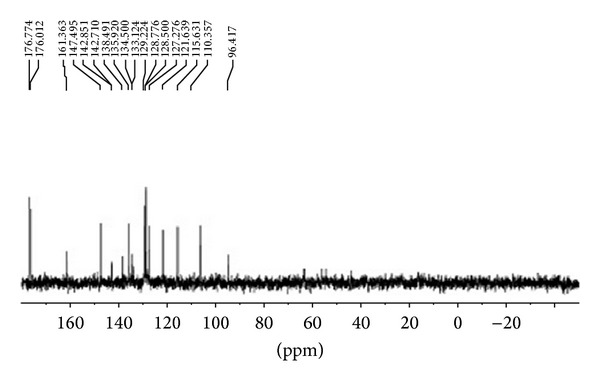
13C NMR spectrum of the F2 fraction.
In 1H NMR the proton signals between δ 7.54 and 8.22 indicate the presence of seven aromatic protons. 1H NMR singlet signal at δ 5.23 represents the presence of aliphatic OH which is attached to ethereal carbon. The remaining 1H NMR signals between δ 3.11 and δ 6.2 characterize the aliphatic protons on the resonated carbons (Figure 13). By all the above observations the structure of the F2 is predicted and the name of compound is noted as “(Z)-1-((1-hydroxypenta-2,4-dien-1-yl)oxy)anthracene-9,10-dione” (Figure 14).
Figure 13.
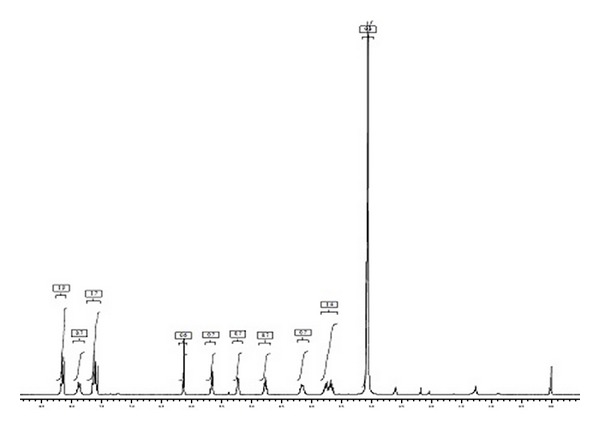
1H NMR of the F2 fraction.
Figure 14.
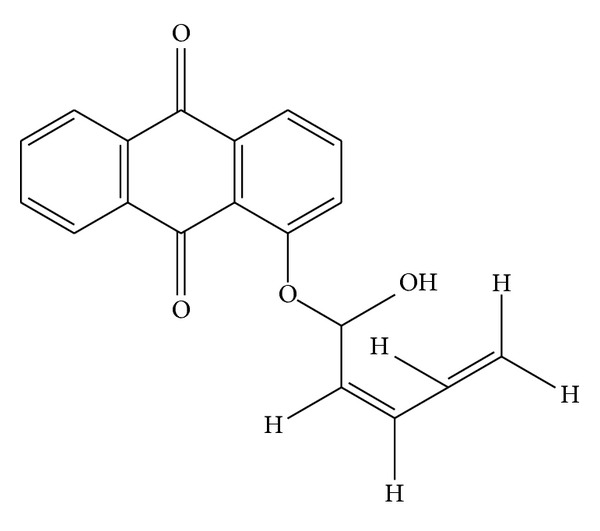
Structure of F2 fraction.
3.6. Spectral Data
Melting point (MP): 174–176°C; infrared (IR) (KBr): 1682 and 1667 (C=O), 2928 cm−1 and 2857 (=C–H), 1612 (C=C), 3410 (O–H), 3013 (ArC–H) cm−1; Proton NMR (1H NMR (400 MHz, DMSO-d 6): d 8.12–8.22 (3H, m), 7.84–7.92 (1H, d), 7.54–7.67 (3H, m), 6.12–6.17 (1H, d), 5.61–5.7 (1H, d), 5.23 (1H, s, OH), 4.72–4.81 (1H, m), 4.1–4.21 (1H, m), 3.32 3.11 (2H, m). Carbon NMR (13C NMR) (100 MHz, DMSO-d 6): d 176.7, 176.1, 161.3, 147.4, 142.8, 138.4, 135.9, 134.5, 133.7, 129.2, 128.7, 127.2, 121.6, 115.6, 110.3, 96.4. LC MS (%): m/z 306 (100%) [MH+•]; elemental analysis. C19H14O4.
4. Conclusion
Marine environments are particularly complex and have varied group of life forms, which occur in environments with extreme variations in pressure, salinity, and temperature. Owing to this nature, marine microorganisms have developed exceptional metabolic and physiological capabilities to be able to survive in such intense habitats that led them to produce different kind of metabolites, which could not be produced by the terrestrial microorganisms. Extensive research on marine natural products over the past three decades has revealed that marine actinomycetes are most prolific sources of novel and diverse metabolites. In the present study, an actinomycete, N. alba, was isolated from mangrove soil and screened for its ability to produce bioactive compound. The bioactive compounds produced by this strain were purified by column chromatography. The potential fraction obtained by column chromatography was subjected to FTIR, NMR, and mass spectroscopy to elucidate its chemical structure and the structure of the compound was predicted as “(Z)-1-((1-hydroxypenta-2,4-dien-1-yl)oxy)anthracene-9,10-dione”. The extracted bioactive compound has shown good antioxidant properties and further research is in progress to produce bioactive compound in large quantities and to make the compound an industrially important one.
Acknowledgments
The authors are thankful to Dr. C. Nagaraju and Mr. K. Chandra Sekhar, Department of Chemistry, Sri Venkateswara University, for their assistance while predicting the structure.
Conflict of Interests
The authors declare that they have no conflict of interests regarding the publication of this paper.
References
- 1.Castillo UF, Strobel GA, Ford EJ, et al. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology. 2002;148(9):2675–2685. doi: 10.1099/00221287-148-9-2675. [DOI] [PubMed] [Google Scholar]
- 2.Singh MP, Petersen PJ, Weiss WJ, et al. Mannopeptimycins, new cyclic glycopeptide antibiotics produced by Streptomyces hygroscopicus LL-AC98: antibacterial and mechanistic activities. Antimicrobial Agents and Chemotherapy. 2003;47(1):62–69. doi: 10.1128/AAC.47.1.62-69.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Shatoury SA, El-Shenawy NS, Abd El-Salam IM. Antimicrobial, antitumor and in vivo cytotoxicity of actinomycetes inhabiting marine shellfish. World Journal of Microbiology and Biotechnology. 2009;25(9):1547–1555. [Google Scholar]
- 4.Bentley SD, Chater KF, Cerdeño-Tárraga AM, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417(6885):141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 5.Sosio M, Bossi E, Bianchi A, Donadio S. Multiple peptide synthetase gene clusters in actinomycetes. Molecular and General Genetics. 2000;264(3):213–221. doi: 10.1007/s004380000336. [DOI] [PubMed] [Google Scholar]
- 6.Nolan R, Cross T. Isolation and screening of actinomycete. In: Goodfellow M, Williams ST, Mordarski M, editors. Actinomycetes in Biotechnology. London, UK: Academic Press; 1998. pp. 33–67. [Google Scholar]
- 7.Thakur D, Bora TC, Bordoloi GN, Mazumdar S. Influence of nutrition and culturing conditions for optimum growth and antimicrobial metabolite production by Streptomyces sp. 201. Journal de Mycologie Medicale. 2009;19(3):161–167. [Google Scholar]
- 8.Hopwood DA. Therapeutic treasures from the deep. Nature Chemical Biology. 2007;3(8):457–458. doi: 10.1038/nchembio0807-457. [DOI] [PubMed] [Google Scholar]
- 9.Zengler K, Parakar A, Keller M. New methods to access microbial diversity for small molecule discover. In: Zhang L, Demain AL, editors. Natural Producherapeutic Medicine. Totowa, NJ, USA: Humana Press; 2005. pp. 275–294. [Google Scholar]
- 10.Bibb MJ. Regulation of secondary metabolism in Streptomyces. Current Opinion in Microbiology. 2005;8(2):208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Forar LR, Amany K, Ali E, Arab BC. Taxonomy, Identification and biological activities of a novel isolate of Streptomyces tendae . Journal of Biotechnology. 2006;9:427–436. [Google Scholar]
- 12.Hacène H, Daoudi-Hamdad F, Bhatnagar T, Baratti JC, Lefebvre G. H107, a new aminoglycoside anti-Pseudomonas antibiotic produced by a new strain of Spirillospora . Microbios. 2000;102(402):69–77. [PubMed] [Google Scholar]
- 13.Schweder T, Lindequist U, Lalk M. Screening for new metabolites from marine microorganisms. Advances in Biochemical Engineering/Biotechnology. 2005;96:1–48. doi: 10.1007/b135781. [DOI] [PubMed] [Google Scholar]
- 14.Sharma SK, Gupta VK. In vitro antioxidant studies of Ficus racemosa linn root. Pharmacognosy Magazine. 2008;13:70–74. [Google Scholar]
- 15.Suriyavathana M, Nandhini K. In vitro antioxidant profile of Liv-PRO-08 oral ayurvedic formulation. Journal of Pharmacy Research. 2010;3:873–876. [Google Scholar]
- 16.El-Nakeeb MA. Selective isolation of aerobic actinomycetes [M.S. thesis] NewBrunswick, NJ, USA: Rutgers, The State University; 1961. [Google Scholar]
- 17.Küster E, Williams ST. Selection of media for isolation of Streptomycetes. Nature. 1964;202(4935):928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- 18.Nonomura H. Key for classification and identification of 458 species of the Streptomycetes included in ISP. Journal of Fermentation Technology. 1974;52:78–92. [Google Scholar]
- 19.Buchanan RE, Gibbons NE. Bergey’s Manual of Determinative Bacteriology. 8th edition. Baltimore, Md, USA: Williams & Wilkins; 1974. [Google Scholar]
- 20.Junaid S, Rakesh KN, Dileep N, Poornima G, Kekuda TRP, Mukunda S. Total phenolic content and antioxidant activity of seed extract of Lagerstroemia speciosa L. Chemical Science Transactions. 2013;2:75–80. [Google Scholar]
- 21.Prieto P, Pineda M, Aguilar M. Spectrophotometric quantitation of antioxidant capacity through the formation of a phosphomolybdenum complex: specific application to the determination of vitamin E. Analytical Biochemistry. 1999;269(2):337–341. doi: 10.1006/abio.1999.4019. [DOI] [PubMed] [Google Scholar]
- 22.Hwang BK, Ahn SJ, Moon SS. Production, purification, and antifungal activity of the antibiotic nucleoside, tubercidin, produced by Streptomyces violaceoniger . Canadian Journal of Botany. 1994;72(4):480–485. [Google Scholar]
- 23.Kekuda PTR, Shobha KS, Onkarappa R. Studies on antioxidant and anthelmintic activity of two Streptomyces species isolated from Western Ghat soil of Agumbe, Karnataka. Journal of Pharmacy Research. 2010;3:26–29. [Google Scholar]
- 24.Subramaniam P, Valliappan K, Sivakumar K, Lakshmanan K. Marine actinobacteria of the coral reef environment of the gulf of mannar biosphere reserve, India: a search for antioxidant property. International Journal of Pharmacy and Pharmaceutical Sciences. 2012;4:316–321. [Google Scholar]
- 25.Armarego WLF, Perrin DD. Purification of Laboratory Chemicals. 4th edition. Oxford, UK: Butterworth Heinemann; 1997. [Google Scholar]
- 26.Tanaka Y, Gräefe U, Yazawa K, Mikami Y. Production of nocardicyclins by clinical isolates of Nocardia pseudobrasiliensis and in vivo antitumor activity of the antibiotic. Journal of Antibiotics. 1998;51(6):589–591. doi: 10.7164/antibiotics.51.589. [DOI] [PubMed] [Google Scholar]
- 27.Sun CH, Wang Y, Wang Z, et al. Chemomicin A, a new angucyclinone antibiotic produced by Nocardia mediterranei subsp. kanglensis 1747–64. Journal of Antibiotics. 2007;60(3):211–215. doi: 10.1038/ja.2007.25. [DOI] [PubMed] [Google Scholar]
- 28.Goodfellow M. Nocardia and related genera. In: Balows A, Duerden BI, editors. Topley and Wilson’s Microbiology and Microbial Infections. London, UK: Arnold; 1998. pp. 463–489. [Google Scholar]
- 29.Hoshino Y, Mukai A, Yazawa K, et al. Transvalencin A, a thiazolidine zinc complex antibiotic produced by a clinical isolate of nocardia transvalensis: I. Taxonomy, fermentation, isolation and biological activities. Journal of Antibiotics. 2004;57(12):797–802. doi: 10.7164/antibiotics.57.797. [DOI] [PubMed] [Google Scholar]
- 30.Saha MR, Alam MA, Akter R, Jahangir R. In-vitro free radical scavenging activity of Ixora coccinea L. Bangladesh Journal of Pharmacology. 2008;3:90–96. [Google Scholar]
- 31.Katalinic V, Milos M, Kulisic T, Jukic M. Screening of 70 medicinal plant extracts for antioxidant capacity and total phenols. Food Chemistry. 2006;94(4):550–557. [Google Scholar]
- 32.Marković ZS, Manojlović NT. Analytical characterization of lichexanthone in lichen: HPLC, UV spectroscopic, and DFT analysis of lichexanthone extracted from Laurera benguelensis (Mull. Arg.) Zahlbr. Monatshefte für Chemie—Chemical Monthly. 2010;141(9):945–952. [Google Scholar]
- 33.Elmastas M, Isildak O, Turkekul I, Temur N. Determination of antioxidant activity and antioxidant compounds in wild edible mushrooms. Journal of Food Composition and Analysis. 2007;20(3-4):337–345. [Google Scholar]


