Abstract
Presenilin 1 (PSEN1) gene mutations are found in 30 to 70% of familial early onset Alzheimer disease (EOAD) cases (onset <60 years). Prevalence of these mutations is highly variable including ethnic differences worldwide. No Peruvian kindred with familial AD (FAD) have been described. Standardized clinical evaluation and cognitive assessment was completed in a Peruvian family with severe EOAD. Clinical course was characterized by very early onset (before age 35 years), progressive cognitive impairment with early memory loss, spatial disorientation and executive dysfunction. We sequenced all exons of PSEN1 in the proband and identified a c.475C>G DNA change resulting in a p.L153V missense mutation in the transmembrane domain 2 of the gene. This mutation is also present in the three additional affected siblings but not in a non-affected family member consistent with segregation of this mutation with the disease.
This is the first report of a Peruvian family affected with EOAD associated with a PSEN1 mutation. This same mutation has been reported previously in English and French families, but a novel variants very close to the mutation and ancestry informative markers analysis suggests the mutation might be of Amerindian or African origin in this Peruvian family.
INTRODUCTION
Alzheimer's disease is considered the leading cause of dementia in the elderly, characterized by gradual decline of memory and other cognitive domains until symptoms become incapacitating. Early onset forms (onset before 65 years old) accounts for up to 6% of all cases, wherein the genetic component plays an important role [1]. Autosomal dominant familial forms of Alzheimer's disease (AD) are mostly associated with three causative genes APP, PSEN1 and PSEN2. Presenilin 1 mutations are found in 30 to 70% of familial cases with onset before age of 55 years, becoming the most common cause of early onset familial Alzheimer's disease (EOFAD)[2, 3].
PSEN1 gene is located on chromosome 14q24.2 and encodes a membrane protein that constitutes the catalytic subunit of the γ-secretase complex [4]. Mutant γ-secretase containing PSEN1 mutations produce more Aβ42 increasing abnormal amyloid aggregation; post-mortem studies of EOFAD-PSEN1 cases showed more neocortical senile plaques, higher Aβ42/Aβ40 ratio and similar amount of neurofibrillary degeneration compared to sporadic cases[5]. There are 197 different mutations published to date, mostly single-nucleotide substitutions and some insertions/deletions have been described. (http://www.molgen.ua.ac.be/ADmutations/). PSEN1 cases have been reported mostly in several Caucasian populations around the world [6, 7]. Some founder effects have been suggested for the most prevalent mutations. A possible founder effect for non-Caucasian Mexicans affected with the PSEN1 Ala431Glu mutation also have been proposed [8]. PSEN1 missense mutations also have been reported in Latin-American families coming from Mexico, Colombia and Argentina [9]. Our goal was to describe a Latino EOFAD family carrying a quite rare missense PSEN1 mutation and propose a possible ancestral origin for this mutation.
MATERIAL AND METHODS
Individuals
Four self-described “mestizo” (Spanish European and Amerindian ancestry) family members with progressive cognitive impairment originating in the northern coast of Peru consulted at Instituto Nacional de Ciencias Neurologicas (Figure 1). Standardized workup for cognitive impairment including neurological examination, Minimental State Examination Test, general neuropsychological assessment based on Barcelona test [10], neuroimaging and laboratory blood tests were performed on the proband and three affected siblings. Basic neurologic and cognitive assessment was also performed in one asymptomatic sibling. All participants were followed up on for regular consultations for the past two years.
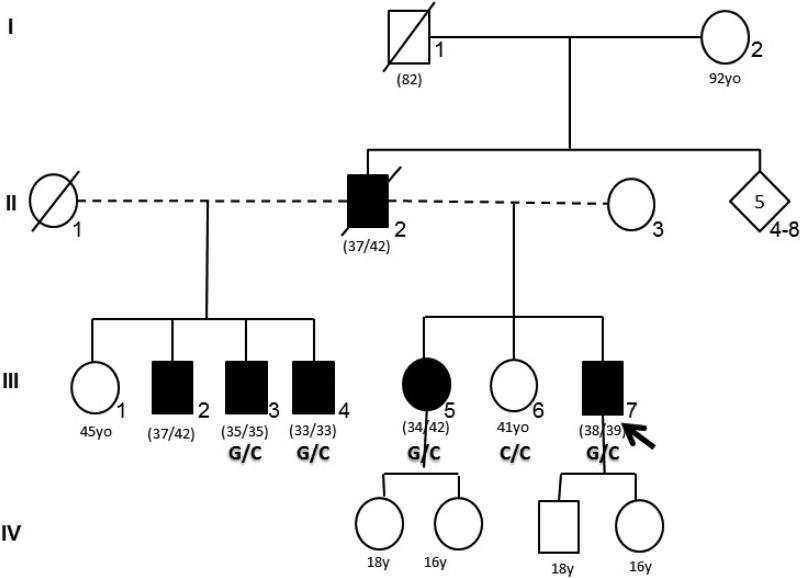
Figure 1.
Pedigree shows family members with Alzheimer's phenotype (in black) with segregation of the Leu153Val missense mutation in PSEN1 gen (G/C). (nn/nn): Age at onset /age at examination of age at death.
DNA Sequencing Analysis
Genetic analysis of the proband, three affected siblings and one non affected family member was undertaken after institutional review board approval and proper written informed consent.
The exons of PSEN1 and at least 60-bp of their flanking introns were fully sequenced in both directions. These fragments were PCR amplified from genomic DNA using primers that were selected by Primer3 software[11]. PCR was carried out by mixing 20 pmol of primers and 40 ng of genomic DNA with 10μl HotStarTaq DNA Polymerase master mixture (Qiagen) in a final volume of 20 μl. All exon amplifications were carried out with PCR profile as following: 1 cycle at 95°C for 15 min, 32 cycles at 98°C/20sec, 60°C/30sec, 72°C/2min, and a final extension at 72°C for 5 min. After PCR, 1 μl of ExoSAP-IT (USB) were added to the PCR products and digested for overnight at 37°C to remove the residual primers and dNTPs. Four microliter of the ExoSAP-treated PCR fragments were then sequenced using BigDye terminator cycle sequencing kit (Applied Biosystems) in a final volume of 10 μl and 30 cycles. The sequence information was collected and analyzed on a 3130 Genetic Analyzer (Applied Biosystems), and nucleotide variants were inspected and identified by Sequencer software (Gene Codes Corp.) alignment.
RESULTS
Clinical characteristics
Proband (III-7), a 39 year-old right-handed man with twelve years of education came to consultation with one year history characterized by memory loss (losing objects, forgetting meetings and telephone numbers), word-finding difficulties, diminished language fluency and mild behavioral abnormalities with anxiety and depression. There were no pyramidal or extrapyramidal signs or gait disturbances. Mini Mental State Examination (MMSE) showed a score of 15 and the Montreal Cognitive Assessment (MoCA) a score of 7. A neuropsychological assessment showed severe alteration of working memory, anomia, verbal and visual episodic and semantic memory, severe executive dysfunction and spatial disorientation. Bilateral global atrophy with parieto-temporal predominance was found on brain Magnetic Resonance Imaging. At a 12 month follow-up, we noticed occasional myoclonic jerks, he was unable to draw a clock, and the MMSE score declined to 10/30.
Family history shows a dominantly inherited pattern of the disease (Fig. 1). Proband's older sister (III-5, 42 years old) is fully dependent and was confined to a wheelchair after five years of rapidly progressive cognitive deterioration starting at age 34. She has diffuse myoclonic jerks, lower limbs spasticity and epilepsy. Father developed a progressive cognitive impairment at 37 years old and died at age 42 due to respiratory complications. Three other paternal siblings were also affected, with ages at onset of 37, 35 and 33 years. Clinical assessment in III-3 and III-4 showed MMSE of 15 and 22 respectively. All affected subjects fulfilled the NINCDS-ADRDA criteria for probable AD.
Genetics
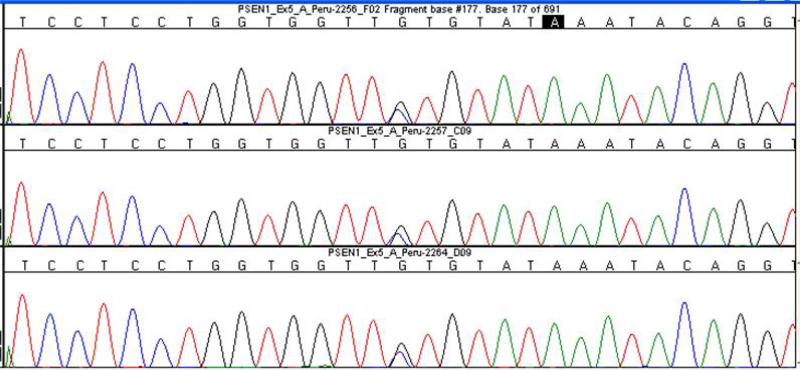
Sequence analysis identified a heterozygous C to G transversion in nucleotide 457 of exon 5 (c.457C>G) leading to the Leu153Val missense mutation in the transmembrane domain 2 of PSEN1 in all four affected siblings (Fig. 2).
Figure 2.
Color chromatogram showing the Leu153Val missense mutation in PSEN1 gen (G/C) in three affected individuals. 2256 (Proband, III7), 2257(II3) and 2264(III4).
We also identified a novel SNP (c.480+501 C>T, chr14:73640916, hg19) cosegregating with the mutation and located 524-bp downstream in intron 5. The T allele of this novel SNP is on the same chromosome haplotype with the mutation G allele. This novel SNP is not found in the public genetic variant database dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/). This includes information in more than 1,000 samples from all major populations (including all 426 European, 400 Asian, 200 African and 271 from the Americas, part of the 1000 genomes project). The variant was also absent when we genotyped additional 752 individuals (including 349 Spanish subjects, 298 Peruvian ”mestizos”, 50 “pure “Amerindians and 55 African Americans, see Table 1).
Table 1.
Subjects screened for novel PSEN1-L153V linked SNP
| Ancestry | n | PD | Controls |
|---|---|---|---|
| African American | 55 | 0 | 55 |
| Peruvian mestizos | 298 | 248 | 50 |
| Amerindians Puno | 50 | 0 | 50 |
| Spain | 349 | 0 | 349 |
| Total | 752 | 248 | 504 |
PD: Parkinson Disease
To determine the possible origin of the mutation in members of this family we genotyped 29 Ancestry Informative Markers (AIMs) that are used by our group routinely to separate the four major continental groups (African, European, Amerindian and Asian).Continental ancestry proportions were then estimated using STRUCTURE (http://pritch.bsd.uchicago.edu/structure.html) with reference to the HDGP+HapMap phase III groups similar to previous ancestry analysis[12]. Results showed that this family has a complex admixture of all 4 continental groups with a major influence of African and Amerindian ancestry (29.4% and 41.9% on average respectively). Overall these results are compatible with the hypothesis that the PSEN1 mutation in this family is unique and may have an Amerindian or African origin.
DISCUSSION
This is the first report of a Peruvian family with a very early onset form of AD associated with a PSEN1 mutation. Mean age at onset for all 6 affected family members is 35 years. The L153Vco-segregated with the disease in four individuals where DNA was available. This missense mutation alters a conserved residue in the second transmembrane domain of the PSEN1 protein, lying in one of the most important clusters of mutations of the gene. [13]. Although the underlying mechanisms for how PSEN1 mutations cause familial AD remain unresolved, a recent study suggested that these mutations confer a gain of negative function that impairs the activity of both mutant and coexpressed wildtype proteins, in a phenomenon called dominant-negative effect [14]
One English [15, 16] and two French [13, 17] families have been previously reported carrying the same mutation. The English family had one family member with myoclonus and none with seizures or spasticity. This is in contrast to our family in which myoclonus, seizures and spasticity have occurred (See Table 2). The reports of the French families give no clinical details other than ages and dementia with apraxia and language deterioration. In all four reported families (including ours) the age of onset is uniformly early, usually in the 30's (range 33-44). To our knowledge there are no previous reports of this mutation in other Latin-American populations. The large Colombian family has an E280A mutation in PSEN1 [9, 18]. To determine the possible origin of this mutation, and due to the unavailability of samples from previously reported families with the same mutation, we decided to screen the novel variant found very close to the mutation and that also segregates with the disease.
Table 2.
Reported Families with the PSEN1 L153V Mutation
| Family Name | 177 | ALZ 148 | ALZ 180 | ALZ PE |
|---|---|---|---|---|
| Alzheimer's criteria | Definitive | Probable | Probable | Probable |
| Inheritance | AD | AD | AD | AD |
| # Affected individuals | 3 | 5 | 4 | 6 |
| Age at onset | 35.3 [35-36] | [34,40] | [39,44] | 35.7 [33,38] |
| Age of death | 44 [41,49] | ... | ... | 42 |
| Disease duration | 8.6 [6,13] | ... | ... | 3.5 [1,8] |
| Myoclonus | Yes | ... | ... | Yes |
| Seizures | No | ... | ... | Yes |
| Spasticity | No | ... | ... | Yes |
| Ethnic origin | Causasian (UK) | French | French | Mestizo (Peru) |
| Segregation proven | Yes | Yes | Yes | Yes |
| References | Janssen. 2002 | Raux, G.2005 | Raux,G.2005 | This study,2013 |
We did not identify this variant in any public databases (including the NHLBI Exome sequencing project) nor in 752 available individuals from different ethnic populations. Interestingly, where in Peru most individuals are an admixed of Spanish and Amerindian ancestry, this family presents influence mostly from Amerindian and African ancestry although markers from all 4 major continental groups can be found based on analysis of 29 AIMs, . Due to this fact, and the absence of this variant in more than 3,000 chromosomes from individuals worldwide we cannot make a definitive statement regarding the origin of the mutation. Although our data is consistent with an Amerindian or African origin, we cannot discard the possibility of this being a recent variant that occurred in a European background.
In conclusion, we report the first autosomal dominant early-onset Alzheimer's disease family in the Peruvian population associated with rapid clinical progression. The novel SNP linked to the mutation is very rare, especially in white populations, increasing the probability that it is of Amerindian or African origin. A comprehensive ancestry analysis all cases reported with this mutation might clarify this issue.
Highlights.
We report the first Peruvian family with EOAD carrying a PSEN1 mutation (L153V).
This is the fourth family carrying this PSEN1 mutation worldwide.
Our data suggests a different origin in this family, possibly on an African or Amerindian haplotype.
ACKNOWLEDGEMENTS
We want to thank the Latin American Research Consortium on the Genetics of Parkinson's disease (LARGE PD) for providing healthy control samples from Peru. This project was supported by NIH Research Training Grant # R25 TW009345 funded by the Fogarty International Center, the National Institute of Mental Health, and the NIH Office of the Director Office of Research on Women's Health and the Office of AIDS Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bettens K, Sleegers K, Van Broeckhoven C. Genetic insights in Alzheimer's disease. Lancet Neurol. 2013;12:92–104. doi: 10.1016/S1474-4422(12)70259-4. [DOI] [PubMed] [Google Scholar]
- 2.Bekris LM, Yu CE, Bird TD, Tsuang DW. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol. 2010;23:213–227. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filley CM, Rollins YD, Anderson CA, Arciniegas DB, Howard KL, Murrell JR, Boyer PJ, Kleinschmidt-DeMasters BK, Ghetti B. The genetics of very early onset Alzheimer disease. Cogn Behav Neurol. 2007;20:149–156. doi: 10.1097/WNN.0b013e318145a8c8. [DOI] [PubMed] [Google Scholar]
- 4.Lessard CB, Wagner SL, Koo EH. And four equals one: presenilin takes the gamma-secretase role by itself. Proc Natl Acad Sci U S A. 2010;107:21236–21237. doi: 10.1073/pnas.1016284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schellenberg GD, Montine TJ. The genetics and neuropathology of Alzheimer's disease. Acta neuropathologica. 2012;124:305–323. doi: 10.1007/s00401-012-0996-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruni AC, Bernardi L, Colao R, Rubino E, Smirne N, Frangipane F, Terni B, Curcio SA, Mirabelli M, Clodomiro A, Di Lorenzo R, Maletta R, Anfossi M, Gallo M, Geracitano S, Tomaino C, Muraca MG, Leotta A, Lio SG, Pinessi L, Rainero I, Sorbi S, Nee L, Milan G, Pappata S, Postiglione A, Abbamondi N, Forloni G, St George Hyslop P, Rogaeva E, Bugiani O, Giaccone G, Foncin JF, Spillantini MG, Puccio G. Worldwide distribution of PSEN1 Met146Leu mutation: a large variability for a founder mutation. Neurology. 2010;74:798–806. doi: 10.1212/WNL.0b013e3181d52785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lleo A, Blesa R, Queralt R, Ezquerra M, Molinuevo JL, Pena-Casanova J, Rojo A, Oliva R. Frequency of mutations in the presenilin and amyloid precursor protein genes in early-onset Alzheimer disease in Spain. Arch Neurol. 2002;59:1759–1763. doi: 10.1001/archneur.59.11.1759. [DOI] [PubMed] [Google Scholar]
- 8.Yescas P, Huertas-Vazquez A, Villarreal-Molina MT, Rasmussen A, Tusie-Luna MT, Lopez M, Canizales-Quinteros S, Alonso ME. Founder effect for the Ala431Glu mutation of the presenilin 1 gene causing early-onset Alzheimer's disease in Mexican families. Neurogenetics. 2006;7:195–200. doi: 10.1007/s10048-006-0043-3. [DOI] [PubMed] [Google Scholar]
- 9.Arango D, Cruts M, Torres O, Backhovens H, Serrano ML, Villareal E, Montanes P, Matallana D, Cano C, Van Broeckhoven C, Jacquier M. Systematic genetic study of Alzheimer disease in Latin America: mutation frequencies of the amyloid beta precursor protein and presenilin genes in Colombia. Am J Med Genet. 2001;103:138–143. doi: 10.1002/1096-8628(20011001)103:2<138::aid-ajmg1529>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 10.Quintana M, Pena-Casanova J, Sanchez-Benavides G, Langohr K, Manero RM, Aguilar M, Badenes D, Molinuevo JL, Robles A, Barquero MS, Antunez C, Martinez-Parra C, Frank-Garcia A, Fernandez M, Blesa R. Spanish multicenter normative studies (Neuronorma project): norms for the abbreviated Barcelona Test. Arch Clin Neuropsychol. 2011;26:144–157. doi: 10.1093/arclin/acq098. [DOI] [PubMed] [Google Scholar]
- 11.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 12.Heinz T, Alvarez-Iglesias V, Pardo-Seco J, Taboada-Echalar P, Gomez-Carballa A, Torres-Balanza A, Rocabado O, Carracedo A, Vullo C, Salas A. Ancestry analysis reveals a predominant Native American component with moderate European admixture in Bolivians, Forensic science international. Genetics. 2013;7:537–542. doi: 10.1016/j.fsigen.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Raux G, Gantier R, Martin C, Pothin Y, Brice A, Frebourg T, Campion D. A novel presenilin 1 missense mutation (L153V) segregating with early-onset autosomal dominant Alzheimer's disease. Hum Mutat. 2000;16:95. doi: 10.1002/1098-1004(200007)16:1<95::AID-HUMU28>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 14.Heilig EA, Gutti U, Tai T, Shen J, Kelleher RJ., 3rd Trans-dominant negative effects of pathogenic PSEN1 mutations on gamma-secretase activity and Abeta production. J Neurosci. 2013;33:11606–11617. doi: 10.1523/JNEUROSCI.0954-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen JC, Beck JA, Campbell TA, Dickinson A, Fox NC, Harvey RJ, Houlden H, Rossor MN, Collinge J. Early onset familial Alzheimer's disease: Mutation frequency in 31 families. Neurology. 2003;60:235–239. doi: 10.1212/01.wnl.0000042088.22694.e3. [DOI] [PubMed] [Google Scholar]
- 16.Janssen JC, Lantos PL, Fox NC, Harvey RJ, Beck J, Dickinson A, Campbell TA, Collinge J, Hanger DP, Cipolotti L, Stevens JM, Rossor MN. Autopsy-confirmed familial early-onset Alzheimer disease caused by the l153V presenilin 1 mutation. Arch Neurol. 2001;58:953–958. doi: 10.1001/archneur.58.6.953. [DOI] [PubMed] [Google Scholar]
- 17.Raux G, Guyant-Marechal L, Martin C, Bou J, Penet C, Brice A, Hannequin D, Frebourg T, Campion D. Molecular diagnosis of autosomal dominant early onset Alzheimer's disease: an update. J Med Genet. 2005;42:793–795. doi: 10.1136/jmg.2005.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lendon CL, Martinez A, Behrens IM, Kosik KS, Madrigal L, Norton J, Neuman R, Myers A, Busfield F, Wragg M, Arcos M, Arango Viana JC, Ossa J, Ruiz A, Goate AM, Lopera F. E280A PS-1 mutation causes Alzheimer's disease but age of onset is not modified by ApoE alleles. Hum Mutat. 1997;10:186–195. doi: 10.1002/(SICI)1098-1004(1997)10:3<186::AID-HUMU2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]