Abstract
Huntington's disease (HD) is a late-onset and progressive neurodegenerative disorder that is caused by aggregation of mutant huntingtin protein which contains expanded-polyglutamine. The molecular chaperones modulate the aggregation in early stage and known for the most potent protector of neurodegeneration in animal models of HD. Over the past decades, a number of studies have demonstrated molecular chaperones alleviate the pathogenic symptoms by polyQ-mediated toxicity. Moreover, chaperone-inducible drugs and anti-aggregation drugs have beneficial effects on symptoms of disease. Here, we focus on the function of molecular chaperone in animal models of HD, and review the recent therapeutic approaches to modulate expression and turn-over of molecular chaperone and to develop anti-aggregation drugs.
Keywords: protein aggregation, Molecular chaperone, Huntington, anti-aggregation drug
INTRODUCTION
Huntington' disease (HD) is a fatal and late-onset neurodegenerative disorder with an autosomal dominant manner of inheritance [1]. The most obvious symptoms of HD are chorea, psychiatric impairment and cognitive deficits due to region-specific neuronal degeneration of medium-size spiny GABAergic neurons in striatum and pyramidal neurons in cerebral cortex [2]. It is reported that 5 to 10 per 100,000 people suffers from HD in worldwide [3]. However, no efficacious treatment has been developed with HD yet, for now, there are some medications which can only retard the progression or alleviate symptoms of the disease [4].
HD is a monogenic disorder caused by the expansion of CAG triplet in the first exon of gene which encodes a protein called huntingtin (Htt) [2]. The Htt with abnormally expanded polyglutamine (polyQ) stretch (>36) is prone to be cleaved by caspases or other proteases and it releases N-terminal fragments. These fragments aggregate with each other easily and become toxic [5, 6, 7, 8, 9]. The mutant Htt (mHtt) oligomer or aggregate exerts a toxic gain-of-function in transcriptional regulation and axonal transport by sequestrating other proteins aberrantly [10]. In HD, aggregation of misfolded mHtt is considered a main cause of pathogenesis, thus it would be rational if we approach to inhibit the misfolding of aggregation-prone proteins to cure the disease. Here, we review recent therapeutic approaches to modulate expression and turnover of molecular chaperone, and anti-aggregation agents to prevent formation of mHtt aggregation or its toxic oligomer.
THE NORMAL HTT AND MHTT AGGREGATION
Normal Htt is ubiquitously expressed, but its level of expression is higher in brain. This soluble protein is mainly localized in cytoplasm, and is also found in nucleus and vesicular membranes [11]. Because Htt is a 348-kDa protein which has polyQ and proline-rich region at the N-terminus and numerous protein interaction motifs including HEAT domain, it may act as a scaffold in various cellular mechanisms. In vitro molecular studies showed that normal Htt interacts with huntingtin-associated protein 1 (HAP1), which is an important factor for vesicle trafficking in the neuronal cells [12]. Htt also binds to repressor element 1 transcription factor/neuron restrictive silencer factor (REST/NRSF), which is involved in transcriptional repression [13]. In the huntingtin knockout mice study, wild-type Htt is suggested to have an anti-apoptotic function in embryonic development [14].
In contrast to normal Htt, mHtt has longer polyQ (>36) stretch in N-terminus. The length of polyQ stretch shows inverse correlation with the onset of symptom [1]. Recent study suggested that the number of polyQ tract in mHtt is implicated in flexibility, which is required for close proximity between N17 (the first 17 amino acids of Htt) and proline-rich domain [15]. N17 and proline-rich domain have opposite effects on mHtt aggregation [16, 17]. The deletion of proline-rich domain leads to a rapid transition into aggregates, in contrast, the deletion of N17 domain decreases the mHtt aggregation [18]. It seems that the proline-rich domain prevents polyQ aggregation through inhibition of N17 at proximal position. When the flexibility of polyQ tract is reduced because of long polyQ length, N17 is critical to induce mHtt aggregation. In the formation of protein aggregation, different threshold of polyQ length may be determined by different intradomain composition of disease protein in several types of polyQ disorders.
The expanded polyQ-tract containing mHtt is easily misfolded, and tends to self-aggregation [19]. It leads to formation of a toxic soluble oligomer, and gradually accumulates into intracellular aggregates, consisting of insoluble β-sheet rich amyloid deposits [20]. Previous studies have shown that mHtt aggregates are detected earlier than pathogenic symptoms in human patients and mouse model of HD [10]. It is doubtful that mHtt aggregates could induce the degeneration of neuron. A number of studies suggest that insoluble aggregates seem to play a protective role, leading to autophagy-mediated clearance [21, 22]. Moreover, R6/2 chimera and shortstop stain of YAC128 HD mouse models show no correlation of mHtt inclusion and pathogenic phenotype [23, 24]. Although there are still controversies which one is more toxic between soluble oligomer and insoluble aggregates, the contributions of both putative toxic insults on HD are reported. Thus, it would be better therapeutic direction to cope with misfolded monomer, which inhibits aggregation with itself.
THE MOLECULAR PATHOGENESIS OF MHTT IN HD
Although mHtt has similar expression and distribution with normal Htt protein in HD patient, toxicity of mHtt is region-specific, especially striatum and cerebral cortex are vulnerable regions. Only a single report explains the striatum-specific neuronal degeneration resulting from mHtt sumoylation by Rhes (Ras homologue enriched in striatum) [25].
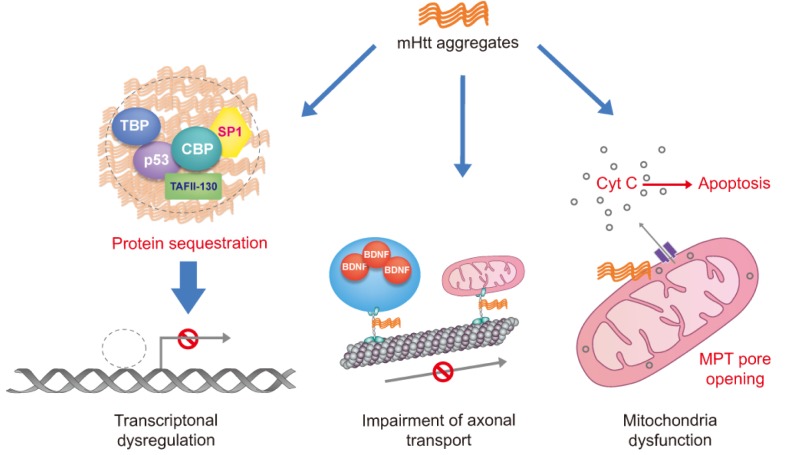
A prominent pathological feature of HD is accumulation of mHtt aggregates in neurons. mHtt is toxic to cells by affecting transcription, mitochondrial function, synaptic transmission and axonal transport (Fig. 1) [10]. mHtt interacts aberrantly and sequesters with many other cellular proteins into nuclear aggregates and cytoplasmic inclusion.
Fig. 1.
Potential molecular pathogenesis of toxicity of mHtt aggregates. Mutant huntingtin may affect the aberrant interaction with or sequester transcription factors leading to transcriptional dysregulation of many genes. Moreover, mutant huntingtin causes defects in trafficking of vesicle and cellular organelle such as mitochondria through long dendritic and axonal projections by affecting both molecular motors and microtubules. Finally, mutant huntingtin directly influence to decrease the Ca2+ threshold for MPT pore opening by interaction with the outer mitochondrial membrane, leading to Cyt c release and apoptosis. Mutant huntingtin (mHtt), cAMP response element binding protein (CREB) binding protein (CBP), TATA-binding protein (TBP), specificity protein 1 (SP1) and TBP-associated factor, 135 kDa (TAFII-130), brain-derived neurotrophic factor (BDNF), mitochondrial permeability transition (MPT), Cytochrome c (Cyt c).
In HD patients and mouse models of HD, transcriptional dysregulation is an early feature in pathogenic mechanism. The transcriptions of key neuronal genes are progressively repressed by sequestering selective transcription factors and co-activator, including cAMP response element binding protein (CREB) binding protein (CBP), TATA-binding protein (TBP), p53, SP1 and TAFII-130 into aggregates of mHtt [26, 27, 28]. The mHtt also loses its ability to retain of cytoplasmic REST/NRSF, leading to transcriptional repression of neuron restrictive silencing element (NRSE) containing gene such as brain-derived neurotrophic factor (BDNF) [13, 29]. Nuclear localized REST/NRSF controls transcriptional repression of NRSE containing target genes through recruitment of co-repressor mSin3a, HDAC1 and HDAC2. Consistent with microarray data, ChIP-seq analysis reveals that the histone acetylation at promoter of key neuronal gene is reduced [30]. H3K4 trimethylation at transcriptionally repressed promoters is also decreased in human HD and in brain of HD model mice [31]. Thus therapeutic approach targeting histone methylation and acetylation is thought to rescue transcriptional dysregulation by mHtt. Many of HDAC inhibitors such as suberoylanilide hydroxamic acid (SAHA), sodium butyrate (SB) and trichostatin A (TSA) have been addressed to mouse model of HD, and have been demonstrated alleviation of mHtt toxicity [32, 33, 34].
The aberrant interaction of mHtt also affects axonal-transport of vesicle and cellular organelles. Cortical BDNF which is transported to striatum is critical to striatal neuronal activity implicated in cortico-striatal connection, but mHtt aberrantly interacts with HAP1 and p150Glued subunit of dynactin leads to impairment of retrograde transport of BDNF [35]. The mHtt not only damages retrograde transport, but also impairs anterograde transport due to reduction of α-tubulin acetylation, which is important for kinesin 1 binding to microtubules [36, 37].
In HD pathogenesis, mitochondrial dysfunction is one of risk factors for neuronal survival. Mitochondria produce ATP, which is the cellular energy source, using oxidative phosphorylation, and mitochondria handle the calcium homeostasis in the brain. The mHtt not only inhibits the axonal-transport of mitochondria by affecting both molecular motors and microtubules [36], but also represses the expression of PPARγ co-activator-1α (PGC-1α) via interfering with CREB function [38]. The transcriptions of nuclear-encoded mitochondrial genes related to mitochondrial biogenesis and respiration are downregulated by impaired PGC-1α transcriptional activity in mouse model of HD. The mHtt also affects the outer mitochondrial membrane, and induces mitochondrial permeability transition (MPT) pore opening via reduction of Ca2+ threshold to trigger opening, which is implicated in excitotoxicity mediated neuronal cell death [39].
MOLECULAR CHAPERONE IN HD
Within cells, misfolded proteins are refolded into their correct conformation by molecular chaperones or are degraded by proteasomal and lysosomal pathway. Molecular chaperones recognize misfolded protein through exposed hydrophobic surfaces and capture it to refolding. Molecular chaperones are also the first line of defense against misfolded protein aggregates. The molecular chaperones prevent inappropriate interactions between misfolded proteins or aberrant interactions with nearby proteins [40, 41, 42].
The common features of several types of neurodegeneration are the aggregation of misfolded causative proteins. Because most of neurodegenerative diseases occur in late-onset manners, many researchers consider that the accumulation of aggregates in neurons is attributed to reduction of functional capacity of molecular chaperones and proteasomal activity during the normal aging process. The level of molecular chaperones shows a biphasic response in neurodegenerative disease. In early stage, the expression of molecular chaperones is increased by aggregates-induced cellular stress, whereas amount of chaperones are decreased due to sequestration into aggregates in the late pathogenesis, leading to progressive accumulation of protein aggregation [43, 44, 45]. Because molecular chaperone functions in prevention of the earliest aberrant protein interactions, which trigger pathogenic cascades, therapeutic approaches modulating chaperone expression and function can be promising for treatment of neurodegenerative diseases.
Several types of molecular chaperones are implicated in suppression of protein aggregation in HD model systems. In Drosophila HD disease models, HSP70 and HSP40 have been identified as genetic suppression factors against the neurotoxicity caused by mHtt [46, 47]. These studies have demonstrated that Hsp70 and Hsp40 directly interact with polyQ-containing exon1 of mHtt in vivo and in vitro. However, the formation of polyQ containing aggregates is not reduced by overexpression of Hsp70 in fly eyes. In the yeast system, polyQ-induced neurodegeneration is alleviated by co-expression of Hsp70 or Hsp40 with mHtt, and the results clearly showed that the inhibition of large-insoluble polyQ aggregates [47]. In the mouse model of HD, R6/2 HD transgenic mice was crossed with Hsp70-overexpressing transgenic mice, and the resulting R6/2-Hsp70 transgenic mouse exhibited the modest effects on disease progression [48]. Conversely, the deletion of Hsp70 in R6/2 transgenic HD mice exacerbates the behavioral and neuropathological defects, including decreased lifespan, weight loss, tremor, limb clasping and motor dysfunction. Although the lack of Hsp70 have no correlation with levels of fibrillar aggregates, the size of inclusion bodies formed by mHtt is increased in the neocortex of R6/2tg/- -Hsp70-/- mice [49]. Similarly, Hsp104 overexpressed transgenic mice with expressing the first 171 residues of mHtt not only ameliorate neurotoxicity via reduction of aggregate formation, but also prolong lifespan of HD mice by 20% [50]. In the lentivirus-based rat model of HD, Hsp104 and Hsp27 rescue the down-regulated dopamine and cAMP-regulated phosphoprotein 32 (DARPP-32) levels that lead to prevent striatal neuronal degeneration [51]. Other molecular chaperones have been reported to suppress detrimental effects induced mHtt aggregation. HSP84 co-localizes with mHtt aggregate in vitro and in vivo, and reduces the polyQ-mediated cellular toxicity [52]. ER chaperone glucose-regulated protein 78 (GRP78) inhibits the formation of mHtt aggregates and blocks cell death via inhibition of caspase-12 activation [53]. Prefoldin also reduces aggregates and cell death by mHtt through suppression of aggregation at the small oligomer stage [54].
Since the genome-wide RNA interference analysis identified the genes that suppress the polyQ aggregation in C. elegans, the eukaryotic chaperonin TCP-1 Ring Complex [TRiC, also called to chaperonin containing TCP-1 (CCT)], a member of HSP60 family, have been suggested to have a significant role in protecting against polyQ aggregation [55]. Unlike HSP70, TRiC/CCT consists of eight subunits and forms ring-shaped complex that sequesters non-native state of protein in the cavity and properly folds in an ATP-dependent manner [56]. Several lines of evidence suggested that TRiC/CCT inhibits polyQ aggregation and alleviates the cytotoxicity of mHtt during the early stage of the aggregation process. In mammalian cell system, disruption of TRiC/CCT by RNA interference results in cellular toxicity caused by the appearance of soluble mHtt aggregates [57]. In yeast model system, knockdown of TRiC/CCT also increases the mHtt aggregation and toxicity. Notably, TRiC/CCT cooperates with the Hsp70 system to promote the assembly of mHtt into soluble oligomers about 500 kDa [58]. Interestingly, specific TRiC/CCT subunit, including CCT1 and CCT4, modulates the polyQ aggregation to non-pathogenic conformations. Moreover, overexpression of a single TRiC/CCT subunit CCT1 is sufficient to rescue mHtt-aggregate formation [59]. Apical domain of CCT1 directly interacts with N17 domain of mHtt and prevents the aggregation with inter- and intramolecular interactions within mHtt [18]. Recently, it is demonstrated that exogenous apical domain of CCT1 (ApiCCT1) is delivered to striatal neuronal cells prepared from full-length knock-in HD mice, and ApiCCT1 is sufficient to alleviate mHtt-mediated toxicity and delays the onset of inclusion body formation [60]. In addition to subunit specific effect, TRiC/CCT complex also affects the aggregation of mHtt via capture of the smaller mHtt oligomers within its cavity [61].
THERAPEUTIC APPROACH TO INDUCE THE MOLECULAR CHAPERONE LEVELS
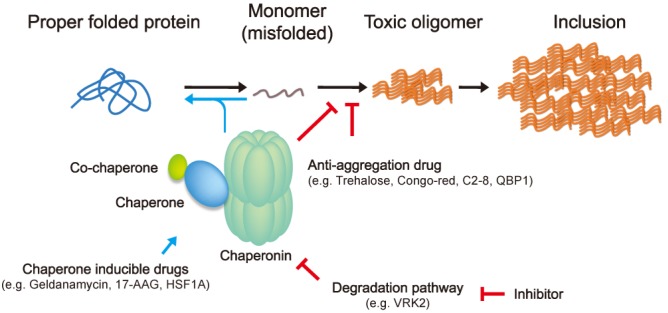
Over the past decade, a number of studies have revealed that molecular chaperones alleviate neurodegeneration through modulation of the aberrant protein interactions by mHtt in the early stage of aggregation (Fig. 2). Heat-shock factor protein 1 (HSF1) is known to induce a set of HSP proteins under the stress condition such as heat-shock [62]. Overexpression of active form of HSF1 elevates the expression of HSP proteins and suppresses the mHtt aggregates formation in cultured cells, and HSF1-overexpression transgenic mouse showed prolonged lifespan and restoration of weight loss [63]. Pharmacological agents have been reported to potentiate chaperone expression by HSF1 activation. It is well-characterized that these drugs inhibit the HSP90 action against negative regulation of HSF1 activation. Geldanamycin, known as a HSP90 inhibitor, binds to ATP-binding site of HSP90 and blocks the interaction between HSP90 and HSF1, leading to HSF1 trimerization and activation of HSPs synthesis [64, 65]. Additionally, 17-allylamino-17-demethoxygeldanamycin (17-AAG), a geldanamycin derivative, suppresses neurodegeneration in a fly HD model [66]. Recently, HSF1A, small benzyl pyrazole-based molecule, has been developed as an activator of HSF1 without inhibition of HSP90 to avoid undesirable proteotoxic activity [67]. Since TRiC/CCT expression is not affected by HSF1, it is necessary to develop another approach elevating the TRiC/CCT protein levels such as inhibition of degradation pathway. Although the turnover mechanism of TRiC/CCT still remains unclear, TRiC/CCT is degraded by ubiquitin-proteasome system [68]. Interestingly, a recent study demonstrated that vaccinia-related kinase 2 (VRK2) has a role in degradation of TRiC/CCT, which was dependent on its kinase activity and enhanced the accumulation of mHtt aggregates in cultured cell lines [69]. It would be possible approach to develop therapeutic inhibitors targeting VRK2 for HD.
Fig. 2.
Therapeutic approach to inhibit mHtt aggregation. An initiating event in aggregation may conversion of mutant huntingtin to an abnormal conformation. It leads to progress through oligomeric intermediates to the formation of large aggregates. Although there are still controversies which one is more toxic between soluble oligomer and insoluble aggregates, inhibition early in the aggregation pathway would be beneficial to the cells because it may prevent the formation of putative toxic insults. Accordingly, molecular chaperones and anti-aggregation drugs are shed the light in this therapeutic intervention. Particularly, molecular chaperones not only induce proper folding of misfolded proteins by interacting with exposed hydrophobic surfaces, but also inhibit aggregation with mutant huntingtin itself. A number of HSF1 activating drugs have been developed to induce the Heat-shock proteins, but transcription of TRiC/CCT was not affected by HSF1. Thus, another pathway is necessary to modulate the TRiC/CCT levels such as inhibition of degradation pathway. Recently, one report reveals that VRK2 facilitates the TRiC/CCT protein degradation through increase of its ubiquitination. 17-allylamino-17-demethoxygeldanamycin (17-AAG), polyglutamine binding peptide 1 (QBP1), vaccinia-related kinase 2 (VRK2).
ANTI-AGGREGATION AGENTS
A number of studies to search small molecules inhibiting oligomerization of β-sheet containing peptide have demonstrated that these molecules can successfully alleviate the symptoms of disease (Fig. 2). When the azo-dye congo-red was intraperitoneally injected to R6/2 HD transgenic mice after onset of symptoms, it promoted the clearance of mHtt aggregates in vivo and exerted protective effects on survival, weight loss and motor function [70]. The disaccharide trehalose also prevented the polyQ aggregation in vitro and in vivo, and had beneficial effects on striatal atrophy, weight loss, survival and motor function in R6/2 transgenic mouse model [71]. Because both congo-red and trehalose cannot penetrate blood-brain barrier (BBB), some reports said difficulties to confirm the beneficial effects of congo-red [72] and trehalose when administrated high concentration (2% in water) to HD mouse. To solve this problem, cell-penetrating peptide like guanidine residues have been introduced to trehalose. Accordingly, this trehalose derivative crosses BBB even at lower concentration (0.4% in water) and shows the similar beneficial effects on disease symptoms [73]. Moreover, BBB-permeable C2-8 small inhibitor has also been screened using yeast-based aggregation assay, and it improves the motor functions and reduces harmful effects on neuronal atrophy in HD model mouse at non-toxic dose [74]. In addition to small molecules, polyglutamine binding peptide 1 (QBP1) also significantly suppresses polyQ aggregation and neurodegeneration in fly HD model [75].
CONCLUSIONS
There are promising evidences leading us to understand the effects of mHtt, and the critical step of HD pathogenesis would be aggregation of mHtt, which results in a number of neuronal protein sequestrations into intranuclear and cytoplasmic aggregates by aberrant interaction. Accordingly, depletion of transcriptional factor and molecular motor cause the transcriptional dysregulation of important neuronal genes such as BDNF, neurotransmitters and its cognate receptors, and axonal transport of vesicles and mitochondria, leading to neurodegeneration. Post-mitotic neurons are vulnerable against toxic aggregates because neuron cannot dilute the toxic insults during cell division, consequently, it will be a heavy load to protein quality control machinery.
According to our understating of the pathogenesis, it would be rational therapeutic approach to treat HD through inhibition of mHtt aggregation, followed by subsequent alleviation of its downstream harmful effects. Molecular chaperones may have a central role in this process, and anti-aggregation drugs are also shed the light in this approach. Since molecular chaperone inducing drugs have been demonstrated beneficial effects on neurodegeneration in animal models, it is necessary to understand the upstream pathway of modulating the molecular chaperone for the first effective treatment for HD.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Research Foundation of Korea (NRF) (Nos. 20110027957 and 20120005830), the Next-Generation BioGreen 21 Program (No. PJ00950301), Rural Development Administration. This research also supported by BK21 plus funded by the Ministry of Education, Korea (10Z20130012243).
References
- 1.Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 2.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series. EMBO Rep. 2004;5:958–963. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer PO, Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- 5.Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, Warby SC, Doty CN, Roy S, Wellington CL, Leavitt BR, Raymond LA, Nicholson DW, Hayden MR. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 6.Wellington CL, Ellerby LM, Gutekunst CA, Rogers D, Warby S, Graham RK, Loubser O, van Raamsdonk J, Singaraja R, Yang YZ, Gafni J, Bredesen D, Hersch SM, Leavitt BR, Roy S, Nicholson DW, Hayden MR. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease. J Neurosci. 2002;22:7862–7872. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellington CL, Singaraja R, Ellerby L, Savill J, Roy S, Leavitt B, Cattaneo E, Hackam A, Sharp A, Thornberry N, Nicholson DW, Bredesen DE, Hayden MR. Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J Biol Chem. 2000;275:19831–19838. doi: 10.1074/jbc.M001475200. [DOI] [PubMed] [Google Scholar]
- 8.Sánchez I, Xu CJ, Juo P, Kakizaka A, Blenis J, Yuan J. Caspase-8 is required for cell death induced by expanded polyglutamine repeats. Neuron. 1999;22:623–633. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 9.Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, Qin ZH, Aronin N, DiFiglia M. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc Natl Acad Sci U S A. 2001;98:12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 11.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 12.Li XJ, Li SH, Sharp AH, Nucifora FC, Jr, Schilling G, Lanahan A, Worley P, Snyder SH, Ross CA. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 13.Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 14.Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 15.Caron NS, Desmond CR, Xia J, Truant R. Polyglutamine domain flexibility mediates the proximity between flanking sequences in huntingtin. Proc Natl Acad Sci U S A. 2013;110:14610–14615. doi: 10.1073/pnas.1301342110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharyya A, Thakur AK, Chellgren VM, Thiagarajan G, Williams AD, Chellgren BW, Creamer TP, Wetzel R. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 17.Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJ, Anjum DH, Kodali R, Creamer TP, Conway JF, Gronenborn AM, Wetzel R. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, Frydman J. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16:1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci. 2003;60:1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- 21.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A, Yamamoto A. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiner A, Del Mar N, Deng YP, Meade CA, Sun Z, Goldowitz D. R6/2 neurons with intranuclear inclusions survive for prolonged periods in the brains of chimeric mice. J Comp Neurol. 2007;505:603–629. doi: 10.1002/cne.21515. [DOI] [PubMed] [Google Scholar]
- 24.Slow EJ, Graham RK, Osmand AP, Devon RS, Lu G, Deng Y, Pearson J, Vaid K, Bissada N, Wetzel R, Leavitt BR, Hayden MR. Absence of behavioral abnormalities and neurodegeneration in vivo despite widespread neuronal huntingtin inclusions. Proc Natl Acad Sci U S A. 2005;102:11402–11407. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 28.van Roon-Mom WM, Reid SJ, Jones AL, MacDonald ME, Faull RL, Snell RG. Insoluble TATA-binding protein accumulation in Huntington's disease cortex. Brain Res Mol Brain Res. 2002;109:1–10. doi: 10.1016/s0169-328x(02)00450-3. [DOI] [PubMed] [Google Scholar]
- 29.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 30.McFarland KN, Das S, Sun TT, Leyfer D, Xia E, Sangrey GR, Kuhn A, Luthi-Carter R, Clark TW, Sadri-Vakili G, Cha JH. Genome-wide histone acetylation is altered in a transgenic mouse model of Huntington's disease. PLoS One. 2012;7:e41423. doi: 10.1371/journal.pone.0041423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vashishtha M, Ng CW, Yildirim F, Gipson TA, Kratter IH, Bodai L, Song W, Lau A, Labadorf A, Vogel-Ciernia A, Troncosco J, Ross CA, Bates GP, Krainc D, Sadri-Vakili G, Finkbeiner S, Marsh JL, Housman DE, Fraenkel E, Thompson LM. Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci U S A. 2013;110:E3027–E3036. doi: 10.1073/pnas.1311323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, Sathasivam K, Ghazi-Noori S, Mahal A, Lowden PA, Steffan JS, Marsh JL, Thompson LM, Lewis CM, Marks PA, Bates GP. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y, Chopra V, Chopra R, Locascio JJ, Liao Z, Ding H, Zheng B, Matson WR, Ferrante RJ, Rosas HD, Hersch SM, Scherzer CR. Transcriptional modulator H2A histone family, member Y (H2AFY) marks Huntington disease activity in man and mouse. Proc Natl Acad Sci U S A. 2011;108:17141–17146. doi: 10.1073/pnas.1104409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 36.Dompierre JP, Godin JD, Charrin BC, Cordelières FP, King SJ, Humbert S, Saudou F. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 40.Muchowski PJ. Protein misfolding, amyloid formation, and neurodegeneration: a critical role for molecular chaperones? Neuron. 2002;35:9–12. doi: 10.1016/s0896-6273(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 41.Söti C, Csermely P. Chaperones and aging: role in neurodegeneration and in other civilizational diseases. Neurochem Int. 2002;41:383–389. doi: 10.1016/s0197-0186(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 42.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 43.Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, Mahal A, Smith DL, Woodman B, Bates GP. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 44.Huen NY, Chan HY. Dynamic regulation of molecular chaperone gene expression in polyglutamine disease. Biochem Biophys Res Commun. 2005;334:1074–1084. doi: 10.1016/j.bbrc.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 45.Yamanaka T, Miyazaki H, Oyama F, Kurosawa M, Washizu C, Doi H, Nukina N. Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J. 2008;27:827–839. doi: 10.1038/emboj.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazemi-Esfarjani P, Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 47.Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci U S A. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansson O, Nylandsted J, Castilho RF, Leist M, Jäättelä M, Brundin P. Overexpression of heat shock protein 70 in R6/2 Huntington's disease mice has only modest effects on disease progression. Brain Res. 2003;970:47–57. doi: 10.1016/s0006-8993(02)04275-0. [DOI] [PubMed] [Google Scholar]
- 49.Wacker JL, Huang SY, Steele AD, Aron R, Lotz GP, Nguyen Q, Giorgini F, Roberson ED, Lindquist S, Masliah E, Muchowski PJ. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington's disease. J Neurosci. 2009;29:9104–9114. doi: 10.1523/JNEUROSCI.2250-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum Mol Genet. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 51.Perrin V, Régulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, Luthi-Carter R, Déglon N. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington's disease. Mol Ther. 2007;15:903–911. doi: 10.1038/mt.sj.6300141. [DOI] [PubMed] [Google Scholar]
- 52.Mitsui K, Nakayama H, Akagi T, Nekooki M, Ohtawa K, Takio K, Hashikawa T, Nukina N. Purification of polyglutamine aggregates and identification of elongation factor-1alpha and heat shock protein 84 as aggregate-interacting proteins. J Neurosci. 2002;22:9267–9277. doi: 10.1523/JNEUROSCI.22-21-09267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang Y, Lv H, Liao M, Xu X, Huang S, Tan H, Peng T, Zhang Y, Li H. GRP78 counteracts cell death and protein aggregation caused by mutant huntingtin proteins. Neurosci Lett. 2012;516:182–187. doi: 10.1016/j.neulet.2012.03.074. [DOI] [PubMed] [Google Scholar]
- 54.Tashiro E, Zako T, Muto H, Itoo Y, Sörgjerd K, Terada N, Abe A, Miyazawa M, Kitamura A, Kitaura H, Kubota H, Maeda M, Momoi T, Iguchi-Ariga SM, Kinjo M, Ariga H. Prefoldin protects neuronal cells from polyglutamine toxicity by preventing aggregation formation. J Biol Chem. 2013;288:19958–19972. doi: 10.1074/jbc.M113.477984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J. Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell. 2011;144:240–252. doi: 10.1016/j.cell.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 58.Behrends C, Langer CA, Boteva R, Böttcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, Hartl FU. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 59.Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sontag EM, Joachimiak LA, Tan Z, Tomlinson A, Housman DE, Glabe CG, Potkin SG, Frydman J, Thompson LM. Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes. Proc Natl Acad Sci U S A. 2013;110:3077–3082. doi: 10.1073/pnas.1222663110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shahmoradian SH, Galaz-Montoya JG, Schmid MF, Cong Y, Ma B, Spiess C, Frydman J, Ludtke SJ, Chiu W. TRiC's tricks inhibit huntingtin aggregation. Elife. 2013;2:e00710. doi: 10.7554/eLife.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 63.Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S, Nakai A. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem. 2005;280:34908–34916. doi: 10.1074/jbc.M506288200. [DOI] [PubMed] [Google Scholar]
- 64.Roe SM, Prodromou C, O'Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 65.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 66.Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T. Heat shock transcription factor 1-activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J Biol Chem. 2008;283:26188–26197. doi: 10.1074/jbc.M710521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yokota S, Kayano T, Ohta T, Kurimoto M, Yanagi H, Yura T, Kubota H. Proteasome-dependent degradation of cytosolic chaperonin CCT. Biochem Biophys Res Commun. 2000;279:712–717. doi: 10.1006/bbrc.2000.4011. [DOI] [PubMed] [Google Scholar]
- 69.Kim S, Park DY, Lee D, Kim W, Jeong YH, Lee J, Chung SK, Ha H, Choi BH, Kim KT. Vaccinia-related kinase 2 mediates accumulation of polyglutamine aggregates via negative regulation of the chaperonin TRiC. Mol Cell Biol. 2014;34:643–652. doi: 10.1128/MCB.00756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sánchez I, Mahlke C, Yuan J. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M, Nukina N. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 72.Wood NI, Pallier PN, Wanderer J, Morton AJ. Systemic administration of Congo red does not improve motor or cognitive function in R6/2 mice. Neurobiol Dis. 2007;25:342–353. doi: 10.1016/j.nbd.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 73.Im J, Kim S, Jeong YH, Kim W, Lee D, Lee WS, Chang YT, Kim KT, Chung SK. Preparation and evaluation of BBB-permeable trehalose derivatives as potential therapeutic agents for Huntington's disease. Medchemcomm. 2013;4:310–316. [Google Scholar]
- 74.Chopra V, Fox JH, Lieberman G, Dorsey K, Matson W, Waldmeier P, Housman DE, Kazantsev A, Young AB, Hersch S. A small-molecule therapeutic lead for Huntington's disease: preclinical pharmacology and efficacy of C2-8 in the R6/2 transgenic mouse. Proc Natl Acad Sci U S A. 2007;104:16685–16689. doi: 10.1073/pnas.0707842104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nagai Y, Fujikake N, Ohno K, Higashiyama H, Popiel HA, Rahadian J, Yamaguchi M, Strittmatter WJ, Burke JR, Toda T. Prevention of polyglutamine oligomerization and neurodegeneration by the peptide inhibitor QBP1 in Drosophila. Hum Mol Genet. 2003;12:1253–1259. doi: 10.1093/hmg/ddg144. [DOI] [PubMed] [Google Scholar]