Abstract
The β2-adrenergic receptor is an important member of the G-protein-coupled receptor (GPCR) superfamily, whose stability and function are modulated by membrane cholesterol. The recent high-resolution crystal structure of the β2-adrenergic receptor revealed the presence of possible cholesterol-binding sites in the receptor. However, the functional relevance of cholesterol binding to the receptor remains unexplored. We used MARTINI coarse-grained molecular-dynamics simulations to explore dimerization of the β2-adrenergic receptor in lipid bilayers containing cholesterol. A novel (to our knowledge) aspect of our results is that receptor dimerization is modulated by membrane cholesterol. We show that cholesterol binds to transmembrane helix IV, and cholesterol occupancy at this site restricts its involvement at the dimer interface. With increasing cholesterol concentration, an increased presence of transmembrane helices I and II, but a reduced presence of transmembrane helix IV, is observed at the dimer interface. To our knowledge, this study is one of the first to explore the correlation between cholesterol occupancy and GPCR organization. Our results indicate that dimer plasticity is relevant not just as an organizational principle but also as a subtle regulatory principle for GPCR function. We believe these results constitute an important step toward designing better drugs for GPCR dimer targets.
Introduction
The G-protein-coupled receptor (GPCR) superfamily comprises the largest group of integral membrane proteins in mammals and is involved in information transfer (signal transduction) across cellular membranes (1–3). GPCRs are typically seven-transmembrane-domain proteins and include >800 members that are encoded by ∼5% of human genes (4). GPCRs are important drug targets due to their central role in cellular signaling, and it is estimated that ∼50% of clinically prescribed drugs target GPCRs (5,6). An emerging and exciting area in GPCR research is oligomerization of GPCRs and the possible role of oligomerization in GPCR function and signaling (7–11). The potential implications of such oligomerization are far-reaching, particularly in the context of GPCRs as major drug targets (12–14). GPCR oligomerization implies an increased cross talk between receptors via homo- and/or heterodimers as well as higher-order oligomers (10,13,15). Interestingly, membrane lipids were recently implicated in the modulation of GPCR oligomerization (15,16).
Membrane cholesterol has been shown to influence the organization, stability, and function of several GPCRs, including rhodopsin (17), the β2-adrenergic receptor (18–20), and the serotonin1A receptor (21–26). The spatial organization of GPCRs such as the serotonin1A receptor, including both dimerization and higher-order oligomerization, has been reported to be influenced by membrane cholesterol (16,17). It has been speculated that cholesterol alters GPCR function and organization by either direct interactions with the receptor or indirect alterations of membrane biophysical properties (26,27). An interesting feature of a number of recently solved high-resolution crystal structures of GPCRs, such as the β1-adrenergic receptor (28), β2-adrenergic receptor (29,30), and A2A adenosine receptor (31), is the close association between cholesterol molecules and the receptor. Recent molecular-dynamics simulations revealed cholesterol-binding sites in GPCRs, although relatively weak interactions were observed (32–34). However, despite the increasing number of reports regarding a GPCR-cholesterol interaction, the exact role of cholesterol at the molecular level in GPCR structure and function continues to be elusive.
The β2-adrenergic receptor is an important member of the GPCR superfamily and serves as an excellent prototype for monitoring GPCR organization and function. This receptor type is expressed mainly in muscle tissues. The β2-adrenergic receptor is involved in muscle relaxation after activation (35) and dysfunction of this receptor is associated with cardiac diseases and asthma (36,37). It was reported that the β2-adrenergic receptor can exist as dimers in vivo (38), and dimerization is functionally important (39), although a monomeric receptor was shown to be the minimal functional unit necessary for signaling (40). A direct receptor-cholesterol interaction was revealed by the cocrystallization of cholesterol in the crystal structure of the β2-adrenergic receptor (30). In addition, receptor function was shown to be dependent on membrane cholesterol (19,20). Yet, the molecular relevance of cholesterol’s association with the receptor is still not clear.
In this work, we carried out coarse-grained molecular-dynamics simulations to analyze the molecular nature of the interaction between membrane cholesterol and the β2-adrenergic receptor, and explore the effect of cholesterol on the dimerization of the receptor. We simulated the β2-adrenergic receptor in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane bilayer in the presence of increasing concentrations of cholesterol, which were chosen to mimic the biological environment of the receptor. We used the MARTINI force field in our study since it has been shown to be suitable for applications such as membrane protein association (41–45) and partitioning of membrane proteins between membrane domains of varying compositions (46,47). Our results show that cholesterol binds to transmembrane helix IV and has an increased occupancy at that site, in both the monomeric and dimeric regimes. Interestingly, our results show that cholesterol occupancy modulates the dimerization process of the receptor, altering the dimer structure and the helices involved in the interface. These novel (to our knowledge) results constitute one of the first reports to explore the correlation between cholesterol’s association with the receptor and its oligomerization and organization at a molecular level.
Materials and Methods
System setup
Molecular-dynamics simulations of the β2-adrenergic receptor embedded in POPC membranes were carried out in the presence and absence of cholesterol. The systems were represented using the MARTINI coarse-grained force field (version 2.1) (48,49). A homology model of the β2-adrenergic receptor (amino acid residues 29–342) was generated from the crystal structure (29) (PDB: 2RH1) using SWISS-MODEL software (50). The PDB structure 2RH1 is a chimera of the β2-adrenergic receptor with the protein T4 lysozyme fused for crystallization. The homology model was built by removing the T4 lysozyme part and replacing it with the intracellular loop 3 of β2-adrenergic receptor. The atomistic structure obtained was minimized and then mapped to its coarse-grained representation. Bilayers containing POPC with increasing cholesterol concentration (0%, 9%, 30%, and 50%) were generated from an initial conformation of randomly placed POPC, cholesterol, and water beads. The bilayer formed in the simulation was equilibrated for 5 μs, leading to uniform distribution of cholesterol in the bilayer. Details regarding the number of lipids, cholesterol, and water used for each system are given in Table S1 of the Supporting Material. Two copies of the coarse-grained model of the β2-adrenergic receptor were inserted into each of the equilibrated bilayers such that the interreceptor distance (center of mass) was at least 6 nm (minimum distance of at least 3 nm). A top view of the β2-adrenergic receptor, in which the transmembrane helices are labeled, and the starting structure of a representative simulation are shown in Fig. 1, a and b.
Figure 1.

Schematic representation of the β2-adrenergic receptor. (a) Top view of the receptor with individual helices marked. (b) Starting structure of the two monomers of the β2-adrenergic receptor in the POPC bilayer. The two receptors are shown in shades of blue corresponding to panel a; lipid molecules are shown in gray, the phosphate bead of lipid is in orange, and the surrounding water molecules are in blue. (c) Time course of dimerization of the β2-adrenergic receptor in POPC bilayers with increasing cholesterol concentration. The minimum distance between two receptors (defined as the distance between the closest beads from two individual receptors, as shown in the figure) during the course of the simulation is plotted for receptor association in POPC bilayers alone (black); and in the presence of 9% (red), 30% (green) and 50% (blue) cholesterol concentration. A representative simulation from each of the four systems is plotted. See Materials and Methods for other details. To see this figure in color, go online.
Simulation parameters
All simulations were performed using the GROMACS simulation package, version 4.5.4 (51). The cutoff for nonbonded interactions was 1.2 nm, with electrostatic interactions shifted to zero in the range of 0–1.2 nm, and Lennard-Jones interactions shifted to zero in the range of 0.9–1.2 nm. A relative electrostatic screening of 15 was used. The temperature for each group was weakly coupled using the Berendsen thermostat algorithm (52) with a coupling constant of 0.1 ps to maintain a constant temperature of 300 K during simulation. Semi-isotropic pressure was maintained using the Berendsen barostat algorithm (52) with a pressure of 1 bar independently in the plane of the membrane and perpendicular to the membrane, a coupling constant of 0.5 ps, and a compressibility of 3 × 10−5 bar−1. The time step used in the simulations was 20 fs. Simulations were rendered using VMD software (53).
Analysis
A transmembrane helix of a receptor was considered to be at the dimer interface if it was within a cutoff of 0.5 nm of the other receptor. This value was chosen based on the minimum distance of approach between two beads in the MARTINI coarse-grained model (48,49). A lipid or cholesterol molecule was defined to be bound to a particular transmembrane helix or amino acid residue (site) if it was within 0.5 nm of that site. The maximum occupancy time was defined as the longest time a given cholesterol molecule was bound at a particular site. The values were normalized for all simulation lengths. A value of one implies that the same cholesterol molecule was present at the site during the entire simulation, and zero implies that cholesterol was never present at that site. The binding region for POPC/cholesterol on a given helix was determined by calculating the distance map. The distance cutoff was 0.5 nm as described above. The results showed no qualitative changes when a larger cutoff of 0.6 nm was used.
Results
The β2-adrenergic receptor dimerizes in the membrane bilayer
To analyze the effect of cholesterol on dimerization of the β2-adrenergic receptor, we carried out coarse-grained molecular-dynamics simulations of the receptor in POPC bilayers and POPC/cholesterol bilayers with increasing cholesterol concentrations (9%, 30%, and 50%). Multiple microsecond-timescale simulations were performed, for a total simulation time of ∼200 μs corresponding to ∼800 μs of effective time (atomistic simulation time) (48,49). Two copies of the receptor were initially placed in a POPC bilayer (with and without cholesterol) such that the interreceptor distance (center of mass) was at least 6 nm. Previous umbrella sampling calculations (46) showed that the free energy of association between two receptors is close to zero at a 1.5 nm distance separation (minimum distance between receptors). In our study, the receptors were initially placed at two times this distance, i.e., with a minimum distance of 3 nm. The initial setup of the system and the top view of the receptor are shown in Fig. 1. During the course of the simulation, the receptors diffused freely in the membrane and associated with each other on a microsecond timescale. The minimum distance between the two receptors during the course of representative simulations in each case is shown in Fig. 1 c. Several close associations were observed between the two receptors before the final dimerized structure was obtained. We calculated the phase space sampled by the receptors in the monomer regime in one of the representative systems (see Fig. S1). It is evident from the figure that all orientations around the central receptor were sampled, except for a narrow pathway leading to the central receptor. The sampling of the receptors in the monomer regime demonstrates that the dimer conformations were not biased according to the starting configurations, and adequate orientational and translational sampling was achieved. The time taken to form a stable dimer was variable and ranged from 2 to 15 μs. In most cases, the initial contact between two receptors that led to a stable contact was made by intracellular loops II and III. Several occurrences of the N-terminal region and the C-terminal helix VIII were also observed at the contact interface. Once the dimer was formed, it was stable during the course of the simulation, although small rearrangements of the two receptors relative to each other were observed. The final interreceptor distance (i.e., the distance between the center of mass of the two receptors in the final dimer) was calculated to be ∼3.2 nm.
Dimer interface of the β2-adrenergic receptor in the POPC bilayer
To further characterize the dimer structures, we analyzed the transmembrane helices involved at the dimer interface. A representation of the dimer interface (top and side views of the transmembrane helices) is shown in Fig. 2, a and e. Transmembrane helices IV and V from both receptors were present at the dimer interface in POPC bilayers. The simulations were repeated three times with different starting velocities, and each time transmembrane helices IV and V were observed to be present at the interface. A contact map of all helix-helix contacts, normalized over the time of occurrence, is shown in Fig. 3a. Since the most frequently observed dimer interface involved symmetric interhelical contacts, we termed such an interface a homo-interface (i.e., the same transmembrane helices from both receptors are involved at the dimer interface). In this conformation, an increased accessibility to membrane lipids was observed for transmembrane helices I and VII relative to transmembrane helices IV and V.
Figure 2.
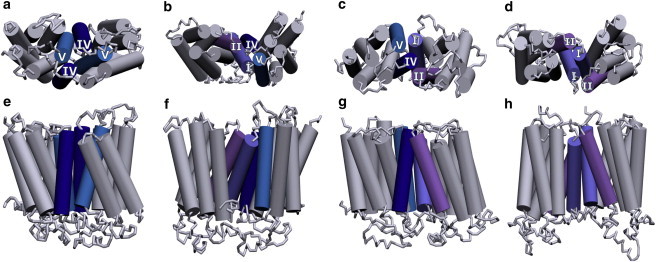
Schematic representation depicting the modulation of transmembrane helices of the β2-adrenergic receptor involved at the dimer interface with increasing cholesterol concentration. (a and e) POPC bilayer. (b–d and f–h) POPC bilayers containing 9% (b and f), 30% (c and g), and 50% (d and h) cholesterol. Representative top and side views of the transmembrane helices are shown for clarity. The transmembrane helices that comprise the dimer interface are colored and labeled. The remaining helices are colored gray. See Materials and Methods for other details. To see this figure in color, go online.
Figure 3.
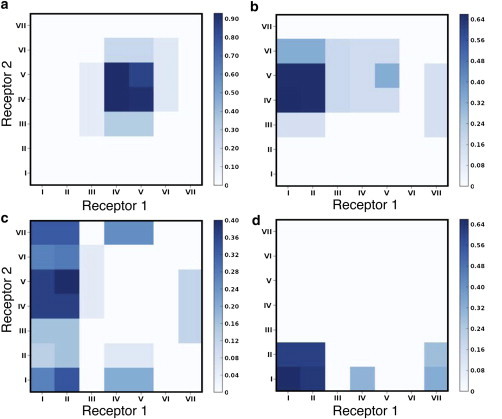
Contact maps depicting the helix-helix interactions between the two receptors. (a) POPC bilayer. (b–d) POPC bilayers containing 9% (b), 30% (c), and 50% (d) cholesterol. The values were calculated as an average over all simulations and normalized by the time of occurrence and simulation length. A cutoff distance of 0.5 nm was used to determine the contact residues. To see this figure in color, go online.
A visual inspection of the final dimer structures that formed in POPC bilayers revealed the presence of a phospholipid (POPC) molecule at the dimer interface (see Fig. S2). The POPC molecule was present at the interface in all three simulations. The binding of the POPC molecule to the groove region formed by transmembrane helices IV and V of the interacting monomers occurred simultaneously with the dimerization process. The POPC molecule remained at the contact interface during the remaining simulation time. Energetically favorable interactions with several aromatic amino acid residues on transmembrane helices IV and V were seen to stabilize the POPC molecule.
Cholesterol modulates helices involved at the dimer interface
Interestingly, the dimer interfaces observed in our simulations with POPC bilayers were altered in the presence of cholesterol. Fig. 2, b–d, show the progressive change in the dimer interface with increasing concentrations of cholesterol in the membrane. A contact map of the dimer interfaces with varying concentrations of cholesterol is shown in Fig. 3, b–d. In general, an increased presence of transmembrane helices I and II was observed with increasing cholesterol concentrations. We refer to an interface containing different transmembrane helices from the two receptors as a hetero-interface. The dimer interface in membranes containing 9% and 30% cholesterol was most often a hetero-interface, formed by transmembrane helices I and II of one receptor and transmembrane helices IV and V of the other. An example of a homo-interface was also observed in membranes containing 9% cholesterol involving transmembrane helices IV and V. At 30% membrane cholesterol, the rotation of the receptors around the contact point led to the occasional involvement of the adjacent transmembrane helices III and VII. At 50% membrane cholesterol, the dimer interface was formed mainly by transmembrane helices I and II from both receptors (see Fig. 2, d and h). A snapshot of the final dimer structure is shown in Fig. S3. The adjacent transmembrane helix VII was also observed to be occasionally involved at the dimer interface due to rotation of the receptors around the contact point (see Fig. 3 d). In this conformation, the C-terminal helix VIII was also occasionally found to be present at the dimer interface. The results obtained were consistent for the three replicate simulations performed. Taken together, these results show that the presence of cholesterol in the membrane increases the involvement of transmembrane helices I and II, and restricts the presence of transmembrane helix IV at the dimer interface of the β2-adrenergic receptor. This observation could have potential implications for designing drugs for GPCR targets (see below).
Cholesterol occupancy at transmembrane helix IV restricts its involvement in the dimer interface
To understand whether a direct receptor-cholesterol interaction or indirect effects (such as the alteration of membrane properties) are responsible for the modulation of the dimer interface with increasing membrane cholesterol content, we analyzed cholesterol density around the transmembrane helices. A density map of the cholesterol population around the receptor, analyzed over several z slices, is shown in Fig. S4. We identified three sites of high cholesterol density in the outer leaflet and four sites in the lower leaflet. Importantly, we identified a site on transmembrane helix IV with the highest density in the middle of the bilayer. Although a few hot spots of interactions could be identified, very few unbinding events at the highest density sites were observed since the cholesterol occupancy at these spots was of the order of microseconds. In the absence of sampling (i.e., in the absence of adequate binding/unbinding events at all interaction sites), measures based on population densities will necessarily be biased. To exclusively account for specific binding events, we calculated the maximum occupancy time of cholesterol around each of the transmembrane helices during the simulation. We defined the maximum occupancy time as the maximum time a given cholesterol molecule was continuously bound to a given site, normalized to the simulation length. A value of one implies that the cholesterol molecule was present at the given site throughout the entire simulation time, and zero implies it was never present at that site. The simulations were divided into two regimes: the first corresponding to monomeric receptors and the second corresponding to the receptor dimers. The maximum occupancy time of cholesterol around the transmembrane helices averaged over the three replicates performed for each cholesterol concentration is shown in Fig. 4. In general, we observed the longest occupancy of cholesterol at transmembrane helix IV. In the monomeric regime (Fig. 4, a, c, and e), the maximum occupancy at transmembrane helix IV increased with increasing cholesterol concentration. At the highest cholesterol concentration (50%), the maximum occupancy of cholesterol at transmembrane helix IV was considerably higher than in other transmembrane helices (Fig. 4, e and f). Taken together, these results point to a cholesterol-binding site on transmembrane helix IV whose occupancy is stochastic and dependent on the membrane cholesterol concentration. In the absence of several binding/unbinding events, the highest-occupancy sites observed in our simulations compare well with the highest-density sites, but need not correspond to binding sites with large favorable free energy.
Figure 4.

Cholesterol occupancy at the β2-adrenergic receptor (maximum occupancy time of cholesterol, i.e., the maximum time a given cholesterol molecule was continuously bound to each of the transmembrane helices). (a–f) The values shown are normalized and averaged for three simulations at increasing cholesterol concentrations: 9% (a and b), 30% (c and d), and 50% (e and f). The simulations were divided into two regimes: the monomer regime (a, c, and e) and the dimer regime (b, d, and f). A maximum occupancy time of one implies that a given cholesterol molecule was present at the given site throughout the entire simulation time, and a value of zero implies it was always absent from that site. The error bars represent the Standard Deviation (SD) between the simulations. See Materials and Methods for further details. To see this figure in color, go online.
Interestingly, the increased presence of a cholesterol molecule at transmembrane helix IV in the monomeric regime correlated well with the decreased presence of this helix at the final dimer interface. At 50% membrane cholesterol concentration, the maximum occupancy of cholesterol at transmembrane helix IV was much higher than at 9% cholesterol, and a lower involvement of transmembrane helix IV was observed at the dimer interface. At 9% and 30% membrane cholesterol concentrations, the occupancy of cholesterol at transmembrane helix IV was usually higher in one of the receptors compared with the other, and in general a hetero-interface involving transmembrane helices I and II from one receptor and transmembrane helices IV and V from the other was observed (see Fig. 3, b and c). At 9% cholesterol concentration, a single instance of homo-interface involving only transmembrane helices IV and V was observed. Analysis of that individual trajectory showed that the cholesterol occupancy around transmembrane helix IV in the monomeric regime was very low in both receptors under that condition. We therefore hypothesize that a high occupancy of cholesterol at transmembrane helix IV, i.e., a stable occupancy of cholesterol at that site, interferes with its subsequent participation in dimer interface formation.
Cholesterol occupancy site at transmembrane helix IV
To explore the molecular details of the cholesterol occupancy site at transmembrane helix IV, we calculated a residue-based distance map between the bound cholesterol and the amino acid residues on transmembrane helix IV. A high dynamics was observed at the site and a few representative snapshots are depicted in Fig. 5. One of these sites (Fig. 5, a and b) corresponds to the CCM site reported in the crystal structure (30). This site is at amino acid residues R151, I154, and W158 (the corresponding Ballesteros-Weinstein numbers are given in Table S2). The cholesterol molecule diffuses on a microsecond timescale (Fig. 5, c–f) to sample site m1 (as defined in Fig. S4) at amino acid residues W158 and I159. In the first site, the polar bead of the cholesterol molecule (modeling the terminal hydroxyl group), interacts with the charged residue R151. At site m1, the polar bead interacts mainly with the aromatic residue W158. The nonpolar beads of the cholesterol molecule interact mainly with nonpolar amino acid residues, such as I153, I154, V157, and I159. Although the most favorable interaction between the cholesterol molecule and the receptor occurs on transmembrane helix IV, several contacts are also observed with the adjacent transmembrane helices II and III.
Figure 5.

Cholesterol-binding sites on transmembrane helix IV of the β2-adrenergic receptor. (a) The CCM site observed in the crystal structure (30). (b) The cholesterol-binding site identified in the coarse-grained simulations that directly corresponds to the CCM site. (c–f) A high dynamics was observed for the cholesterol and additional conformations of the cholesterol near the site. The backbone of transmembrane helix IV is shown in blue, and the side chains of the amino acid residues R151, I154, and W158 are shown in gray. The bound cholesterol molecule is shown in magenta and the polar bead representing the -OH group is depicted in blue. For clarity, the surrounding receptors, lipid, cholesterol, and water molecules are not shown. See Materials and Methods for further details. To see this figure in color, go online.
POPC binding site at the dimer interface
As discussed above, the final dimer structures formed in POPC bilayers revealed the presence of a phospholipid molecule at the dimer interface (see Fig. S2). The binding of the POPC molecule at the dimer interface was also observed at 30% and 50% membrane cholesterol. Due to the variation in the transmembrane helices present at the interface in the absence and presence of cholesterol, we could not discern any consensus sequence. Two such POPC-binding sites are shown in Fig. S5 and correspond to the bilayers of POPC and POPC/30% cholesterol. The zwitterionic headgroup of the POPC interacts with either a charged residue such as E225 or polar aromatic residues such as Y199. The fatty acyl chain of the phospholipid molecule interacts mainly with nonpolar residues such as I205 or aromatic residues such as F217.
Discussion
GPCR organization represents a crucial determinant in cellular signaling (54). The heterogeneous distribution of GPCRs in membrane domains characterized by different lipid compositions has given rise to new challenges and complexities in receptor signaling. Therefore, cellular signaling has to be considered in the context of organization of various signaling components, including receptors, lipids, and G-proteins. In this context, several questions remain unexplored regarding the molecular details of receptor association and oligomerization. In this work, we analyzed the effect of membrane cholesterol on the dimerization of the β2-adrenergic receptor. The novel (to our knowledge) aspect of our work stems from the fact that we correlated the molecular-level receptor-cholesterol interaction with modulation of the receptor dimerization process. We performed multiple microsecond-timescale, coarse-grained simulations that allowed us to explore the interface for receptor dimers in lipid bilayers at several cholesterol concentrations. We showed that cholesterol modulates the dimer interface of the β2-adrenergic receptor by binding to a cholesterol occupancy site on transmembrane helix IV.
Exploring the energy landscape by unbiased simulations
It is difficult to explore the complex energetics of GPCR organization due to the limited methodologies available to probe these processes. Unbiased atomistic molecular-dynamics simulations have been used extensively to probe protein and membrane dynamics that occur on the nanosecond timescale (33,34,55–62). With increasing computational power, unbiased atomistic simulations have been used to study phenomena on the microsecond timescale (63–65). Unbiased coarse-grained simulations are increasingly being used to explore microsecond-timescale dynamics and organization (66–72). In particular, coarse-grained simulations using unbiased sampling have improved our understanding of GPCR association (41,73). Although unbiased molecular-dynamics simulations can better represent the equilibrium evolution of the system without any external bias (potential/force) or reaction coordinate, they are limited by the phase space they sample (64,74). Biased simulations such as umbrella sampling and force pulling are often performed to improve sampling, and with careful analysis can be used to estimate the underlying unbiased true energy landscape. In two previous studies, these methods were employed to calculate a potential of mean force (PMF) of GPCR association along a given reaction coordinate (interhelical distance) (43,75). In both studies, a 1D PMF was calculated for only limited dimer interfaces (e.g., a 1/7 interface) and the sampling of the other dimer interfaces was absent. Importantly, recent work suggested that even for the association of single transmembrane helices, 1D PMFs result in limited sampling and overestimate the energetics (76–78). The limited sampling arises from slow membrane dynamics, and biased simulations of membrane partitioning were recently shown to over- or underestimate the underlying (unbiased) energy landscape (79–81). Consequently, it still remains difficult to achieve a complete thermodynamic understanding of GPCR association in different membrane compositions. In this work, we carried out multiple unbiased coarse-grained simulations totaling ∼1 ms to analyze the association between two GPCRs and probe the role of the membrane lipid environment. The membrane’s effects in driving and modulating receptor association within membranes have been usually neglected and the focus of our work is to understand these effects.
The dimer interface and comparison with experimental data
Most studies of the dimer interface of the β2-adrenergic receptor have employed indirect methods, and several dimer interfaces have been proposed (39,82). Interfaces involving transmembrane helix VI (39) or helix VIII (82) have been proposed based on results obtained by different techniques. The most direct evidence for dimer interfaces comes from the related β1-adrenergic receptor (83). The crystal structure of the ligand-free basal structure of the β1-adrenergic receptor shows two distinct dimer structures: one involving transmembrane helices I and II and helix VIII, and the other involving transmembrane helices IV and V. The two dimer interfaces observed in the crystal structure correspond to the two homo-interfaces observed in our simulations (see Fig. 6), and no crystal contacts were observed that correspond to the hetero-interface. However, we propose that although both dimer interfaces are energetically favorable, the membrane environment (cholesterol) tunes the energetics to modulate the relative populations.
Figure 6.
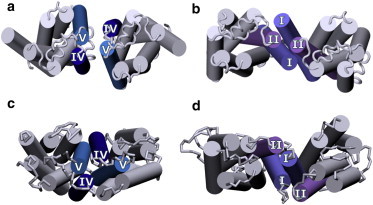
Comparison of the dimer interfaces of the β1-adrenergic receptor and the β2-adrenergic receptor. (a and b) Crystal structure of the β1-adrenergic receptor (83). (c and d) Dimer structures of the b2-adrenergic receptor obtained from coarse-grained simulations at 0% and 50% cholesterol concentration, respectively. The transmembrane helices that comprise the dimer interface are colored and labeled. The remaining helices are colored gray. To see this figure in color, go online.
Structural plasticity of the dimeric interface
The various dimer interfaces of the β2-adrenergic receptor revealed by various experimental methods (39,82,83) suggest that the energetics of association via the different dimer interfaces are comparable and perhaps dependent on the experimental conditions. In a recent biased molecular-dynamics simulation of the β2-adrenergic receptor, Johnston et al. (75) calculated the energetics of two interfaces and reported comparable stabilities for these two dimer interfaces. Although the dimer interfaces they considered were distinct from those observed in this report, their results confirmed a structural plasticity in the dimer structure. Similarly, relatively weak binding energetics was recently reported for different rhodopsin dimer interfaces and suggested to be relevant in the context of the supramolecular organization of GPCRs (43). Our data suggest a similar plasticity of the dimer interface for the β2-adrenergic receptor, further modulated by the membrane cholesterol. A total of 49 orientations are possible, considering a simple interaction matrix between the seven transmembrane helices. In our set of 12 simulations, several of these interfaces were sampled. To test the plasticity and relative stabilities of the dimer interfaces, we performed 10 additional shorter simulations at each cholesterol concentration. A comprehensive contact map with the increased sampling is shown in Fig. S6. The highest-population interfaces correspond to the most stable interfaces observed in the first set of simulations, i.e., a homo-interface involving transmembrane helices IV and V, a hetero-interface with transmembrane helices IV and V, and I and II, and another homo-interface involving transmembrane helices I and II. The plasticity in the dimer interfaces can be easily appreciated in terms of the relative populations of interfaces observed in the contact maps. Since the plasticity emerges from the comparable energetics of the interfaces, the modulation of the energetics by the environment results in changes in the relative populations of these interfaces.
Cholesterol occupancy sites
Lipid-receptor interactions are especially significant for GPCRs because they undergo conformational changes to carry out their function, giving rise to structural plasticity (84,85). These cooperative conformational changes involve the participation of surrounding lipid molecules, and various conformations are stabilized by the binding of different lipids. One of the first reports of a cholesterol-binding site in GPCRs was based on the crystal structure of the β2-adrenergic receptor (29,30), in which a stably bound cholesterol was reported. It should be noted that GPCRs are known to behave differently in cubic and lamellar lipidic mesophases (86), and the cholesterol-binding site, termed the cholesterol consensus motif (CCM), could be specific to the membrane lipid environment. Interestingly, one of the binding modes of cholesterol at helix IV we observed in our coarse-grained simulation correlates well with the CCM site reported in the crystal structure (30), indicating that cholesterol occupancy at this site is independent of the lipid packing arrangement (see Fig. 5). Cholesterol occupancy on helices V and VII, which contain another putative cholesterol-binding site (the CRAC motif (87)), is also relatively high, but much lower than on helix IV. These results are consistent with the dynamic nature of cholesterol binding to GPCRs predicted from previous molecular-dynamics studies (32–34). High occupancy of cholesterol at this site on helix IV was observed in both monomer and dimer states. More importantly, our results show that cholesterol occupancy at this site leads to modulation of the final dimer structures obtained. As stated above, this implies that the underlying energy landscape of receptor dimerization, i.e., the relative stabilities of these dimer interfaces, can be modulated by interaction with membrane cholesterol.
Functional significance of cholesterol-mediated dimer interfaces
The β2-adrenergic receptor was recently cocrystallized with the stimulatory G-protein (Gs), and the receptor-Gs protein interface was shown to be formed by transmembrane helices V and VI (88). Although monomers of the β2-adrenergic receptor were reported to be sufficient for signaling (40), the receptor was demonstrated to be present as a dimer in vivo (38), and receptor organization was shown to be important for its function (39). If the receptor-Gs protein interaction includes a receptor dimer, the dimer interface observed in our simulations in POPC bilayers could lead to potential steric hindrance with the Gs protein since it involves transmembrane helix V. In the presence of increasing membrane cholesterol, transmembrane helix V from either one receptor or both receptors becomes progressively available for association with the Gαs subunit. We speculate that in the altered dimer conformation in the presence of cholesterol, the β2-adrenergic receptor dimer can interact more favorably with the Gαs subunit. Due to the comparable energies of the different dimer interfaces of β2AR (43,75), the dimer interfaces observed in our simulations in the presence and absence of cholesterol only represent the most populated dimer interfaces and it is possible that other dimer conformations could be sampled under different conditions. To the best of our knowledge, our results constitute the first report of the modulation of the dimeric structure of a GPCR by the membrane cholesterol.
Knowledge about GPCR dimer models would allow the development of dimeric or bivalent drugs that could interact with both monomers in a dimer, and open up new possibilities in drug discovery (11). The modulation of GPCR dimer structure by membrane cholesterol could have interesting and far-reaching applications in cellular physiology and drug discovery. Cellular cholesterol is known to be developmentally regulated and its content increases with aging (89,90). This could imply that the organization of GPCR oligomers is age dependent. The efficacy of a specific drug designed to target a GPCR dimeric interface could therefore change with the process of aging. Interestingly, the membrane lipid environment of GPCRs has been implicated in disease progression during aging (91). In addition, there is a strong asymmetry in the manner in which cholesterol is distributed among various organs and tissues in the human body. In other words, the distribution of cholesterol among various tissues is not uniform. The central nervous system, which accounts for only ∼2% of the body mass, contains ∼25% of the free cholesterol present in the whole body (92). This means that GPCR oligomerization could be tissue specific. With the development of new technologies to detect GPCR oligomers in native tissues (10), this could represent an exciting possibility. Importantly, the fact that the same GPCR could be present in multiple tissues may pose a considerable challenge in designing drugs suitable for the dimer interface.
In conclusion, using multiple coarse-grained simulations of the β2-adrenergic receptor in membranes with varying lipid composition, we have shown that the dimerization process of the β2-adrenergic receptor is modulated by membrane cholesterol. More specifically, we showed that cholesterol occupancy at transmembrane helix IV restricts its involvement at the dimer interface and stabilizes a dimer interface with transmembrane helices I and II instead of transmembrane helices IV and V. Based on our results, it appears that dimer plasticity is relevant not just as an organizational principle but also as a regulatory principle for GPCR function. Understanding the cross talk between GPCRs and cholesterol represents an important step in our overall understanding of GPCR function in health and disease.
Acknowledgments
We thank members of our research groups and Sourav Haldar (NICHD, NIH) for critically reading the manuscript. We also thank the CSIR Fourth Paradigm Institute (Bangalore) for computational time.
This work was supported by the Council of Scientific and Industrial Research, Government of India. D.S. received a Ramalingaswami Fellowship from the Department of Biotechnology, Government of India. X.P. received a Junior Research Fellowship from the University Grants Commission (India). A.C. received a J.C. Bose Fellowship (Department of Science and Technology, Government of India). Computational time was supported by a DST Fast Track grant. A.C. is an Adjunct Professor at the Special Centre for Molecular Medicine of Jawaharlal Nehru University (New Delhi) and the Indian Institute of Science Education and Research (Mohali), and an Honorary Professor of the Jawaharlal Nehru Centre for Advanced Scientific Research (Bangalore).
Contributor Information
Amitabha Chattopadhyay, Email: amit@ccmb.res.in.
Durba Sengupta, Email: d.sengupta@ncl.res.in.
Supporting Material
References
- 1.Pierce K.L., Premont R.T., Lefkowitz R.J. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum D.M., Rasmussen S.G.F., Kobilka B.K. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkatakrishnan A.J., Deupi X., Babu M.M. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y., Devries M.E., Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLOS Comput. Biol. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlyer S., Horuk R. I want a new drug: G-protein-coupled receptors in drug development. Drug Discov. Today. 2006;11:481–493. doi: 10.1016/j.drudis.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Heilker R., Wolff M., Bieler M. G-protein-coupled receptor-focused drug discovery using a target class platform approach. Drug Discov. Today. 2009;14:231–240. doi: 10.1016/j.drudis.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Shanti K., Chattopadhyay A. A new paradigm in the functioning of G-protein-coupled receptors. Curr. Sci. 2000;79:402–403. [Google Scholar]
- 8.Lohse M.J. Dimerization in GPCR mobility and signaling. Curr. Opin. Pharmacol. 2010;10:53–58. doi: 10.1016/j.coph.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Palczewski K. Oligomeric forms of G protein-coupled receptors (GPCRs) Trends Biochem. Sci. 2010;35:595–600. doi: 10.1016/j.tibs.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albizu L., Cottet M., Durroux T. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat. Chem. Biol. 2010;6:587–594. doi: 10.1038/nchembio.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milligan G. The role of dimerisation in the cellular trafficking of G-protein-coupled receptors. Curr. Opin. Pharmacol. 2010;10:23–29. doi: 10.1016/j.coph.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Panetta R., Greenwood M.T. Physiological relevance of GPCR oligomerization and its impact on drug discovery. Drug Discov. Today. 2008;13:1059–1066. doi: 10.1016/j.drudis.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 13.González-Maeso J., Ang R.L., Sealfon S.C. Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature. 2008;452:93–97. doi: 10.1038/nature06612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casadó V., Cortés A., Canela E.I. GPCR homomers and heteromers: a better choice as targets for drug development than GPCR monomers? Pharmacol. Ther. 2009;124:248–257. doi: 10.1016/j.pharmthera.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganguly S., Clayton A.H.A., Chattopadhyay A. Organization of higher-order oligomers of the serotonin1A receptor explored utilizing homo-FRET in live cells. Biophys. J. 2011;100:361–368. doi: 10.1016/j.bpj.2010.12.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paila Y.D., Kombrabail M., Chattopadhyay A. Oligomerization of the serotonin1A receptor in live cells: a time-resolved fluorescence anisotropy approach. J. Phys. Chem. B. 2011;115:11439–11447. doi: 10.1021/jp201458h. [DOI] [PubMed] [Google Scholar]
- 17.Soubias O., Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta. 2012;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Z., Kobilka B. Using synthetic lipids to stabilize purified β2 adrenoceptor in detergent micelles. Anal. Biochem. 2005;343:344–346. doi: 10.1016/j.ab.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Paila Y.D., Jindal E., Chattopadhyay A. Cholesterol depletion enhances adrenergic signaling in cardiac myocytes. Biochim. Biophys. Acta. 2011;1808:461–465. doi: 10.1016/j.bbamem.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Pontier S.M., Percherancier Y., Bouvier M. Cholesterol-dependent separation of the β2-adrenergic receptor from its partners determines signaling efficacy: insight into nanoscale organization of signal transduction. J. Biol. Chem. 2008;283:24659–24672. doi: 10.1074/jbc.M800778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pucadyil T.J., Chattopadhyay A. Cholesterol modulates ligand binding and G-protein coupling to serotonin1A receptors from bovine hippocampus. Biochim. Biophys. Acta. 2004;1663:188–200. doi: 10.1016/j.bbamem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Pucadyil T.J., Chattopadhyay A. Cholesterol depletion induces dynamic confinement of the G-protein coupled serotonin1A receptor in the plasma membrane of living cells. Biochim. Biophys. Acta. 2007;1768:655–668. doi: 10.1016/j.bbamem.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Saxena R., Chattopadhyay A. Membrane cholesterol stabilizes the human serotonin1A receptor. Biochim. Biophys. Acta. 2012;1818:2936–2942. doi: 10.1016/j.bbamem.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 24.Jafurulla M., Chattopadhyay A. Membrane lipids in the function of serotonin and adrenergic receptors. Curr. Med. Chem. 2013;20:47–55. [PubMed] [Google Scholar]
- 25.Oates J., Watts A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr. Opin. Struct. Biol. 2011;21:802–807. doi: 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Paila Y.D., Chattopadhyay A. Membrane cholesterol in the function and organization of G-protein coupled receptors. Subcell. Biochem. 2010;51:439–466. doi: 10.1007/978-90-481-8622-8_16. [DOI] [PubMed] [Google Scholar]
- 27.Paila Y.D., Chattopadhyay A. The function of G-protein coupled receptors and membrane cholesterol: specific or general interaction? Glycoconj. J. 2009;26:711–720. doi: 10.1007/s10719-008-9218-5. [DOI] [PubMed] [Google Scholar]
- 28.Warne T., Moukhametzianov R., Tate C.G. The structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherezov V., Rosenbaum D.M., Stevens R.C. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson M.A., Cherezov V., Stevens R.C. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W., Chun E., Stevens R.C. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sengupta D., Chattopadhyay A. Identification of cholesterol binding sites in the serotonin1A receptor. J. Phys. Chem. B. 2012;116:12991–12996. doi: 10.1021/jp309888u. [DOI] [PubMed] [Google Scholar]
- 33.Lee J.Y., Lyman E. Predictions for cholesterol interaction sites on the A2A adenosine receptor. J. Am. Chem. Soc. 2012;134:16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cang X., Du Y., Jiang H. Mapping the functional binding sites of cholesterol in β2-adrenergic receptor by long-time molecular dynamics simulations. J. Phys. Chem. B. 2013;117:1085–1094. doi: 10.1021/jp3118192. [DOI] [PubMed] [Google Scholar]
- 35.Ferro A., Coash M., Queen L. Nitric oxide-dependent β2-adrenergic dilatation of rat aorta is mediated through activation of both protein kinase A and Akt. Br. J. Pharmacol. 2004;143:397–403. doi: 10.1038/sj.bjp.0705933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milano C.A., Allen L.F., Lefkowitz R.J. Enhanced myocardial function in transgenic mice overexpressing the β2-adrenergic receptor. Science. 1994;264:582–586. doi: 10.1126/science.8160017. [DOI] [PubMed] [Google Scholar]
- 37.Liggett S.B. The pharmacogenetics of β2-adrenergic receptors: relevance to asthma. J. Allergy Clin. Immunol. 2000;105:S487–S492. doi: 10.1016/s0091-6749(00)90048-4. [DOI] [PubMed] [Google Scholar]
- 38.Angers S., Salahpour A., Bouvier M. Detection of β2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc. Natl. Acad. Sci. USA. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hebert T.E., Moffett S., Bouvier M. A peptide derived from a β2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J. Biol. Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 40.Whorton M.R., Bokoch M.P., Sunahara R.K. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. USA. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Periole X., Huber T., Sakmar T.P. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J. Am. Chem. Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- 42.Sengupta D., Marrink S.J. Lipid-mediated interactions tune the association of glycophorin A helix and its disruptive mutants in membranes. Phys. Chem. Chem. Phys. 2010;12:12987–12996. doi: 10.1039/c0cp00101e. [DOI] [PubMed] [Google Scholar]
- 43.Periole X., Knepp A.M., Huber T. Structural determinants of the supramolecular organization of G protein-coupled receptors in bilayers. J. Am. Chem. Soc. 2012;134:10959–10965. doi: 10.1021/ja303286e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasanna X., Praveen P.J., Sengupta D. Sequence dependent lipid-mediated effects modulate the dimerization of ErbB2 and its associative mutants. Phys. Chem. Chem. Phys. 2013;15:19031–19041. doi: 10.1039/c3cp52447g. [DOI] [PubMed] [Google Scholar]
- 45.Sengupta D., Rampioni A., Marrink S.J. Simulations of the c-subunit of ATP-synthase reveal helix rearrangements. Mol. Membr. Biol. 2009;26:422–434. doi: 10.3109/09687680903321073. [DOI] [PubMed] [Google Scholar]
- 46.Schäfer L.V., de Jong D.H., Marrink S.J. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc. Natl. Acad. Sci. USA. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sengupta D. Cholesterol modulates the structure, binding modes, and energetics of caveolin-membrane interactions. J. Phys. Chem. B. 2012;116:14556–14564. doi: 10.1021/jp3077886. [DOI] [PubMed] [Google Scholar]
- 48.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 49.Monticelli L., Kandasamy S.K., Marrink S.-J. The MARTINI coarse grained forcefield: extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 50.Arnold K., Bordoli L., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 51.Van Der Spoel D., Lindahl E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 52.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 53.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 54.Saxena R., Chattopadhyay A. Membrane organization and dynamics of the serotonin1A receptor in live cells. J. Neurochem. 2011;116:726–733. doi: 10.1111/j.1471-4159.2010.07037.x. [DOI] [PubMed] [Google Scholar]
- 55.Lindahl E., Sansom M.S.P. Membrane proteins: molecular dynamics simulations. Curr. Opin. Struct. Biol. 2008;18:425–431. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 56.Klepeis J.L., Lindorff-Larsen K., Shaw D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Khalili-Araghi F., Gumbart J., Schulten K. Molecular dynamics simulations of membrane channels and transporters. Curr. Opin. Struct. Biol. 2009;19:128–137. doi: 10.1016/j.sbi.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Risselada H.J., Grubmüller H. How SNARE molecules mediate membrane fusion: recent insights from molecular simulations. Curr. Opin. Struct. Biol. 2012;22:187–196. doi: 10.1016/j.sbi.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 59.Lemmin T., Soto C.S., Dal Peraro M. Assembly of the transmembrane domain of E. coli PhoQ histidine kinase: implications for signal transduction from molecular simulations. PLoS Comput. Biol. 2013;9:e1002878. doi: 10.1371/journal.pcbi.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skjaerven L., Grant B., Martinez A. Conformational sampling and nucleotide-dependent transitions of the GroEL subunit probed by unbiased molecular dynamics simulations. PLoS Comput. Biol. 2011;7:e1002004. doi: 10.1371/journal.pcbi.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larsson D.S.D., Liljas L., van der Spoel D. Virus capsid dissolution studied by microsecond molecular dynamics simulations. PloS Comput. Biol. 2012;7:e1002502. doi: 10.1371/journal.pcbi.1002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olausson B.E., Grossfield A., Vogel A. Molecular dynamics simulations reveal specific interactions of post-translational palmitoyl modifications with rhodopsin in membranes. J. Am. Chem. Soc. 2012;134:4324–4331. doi: 10.1021/ja2108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arkhipov A., Shan Y., Shaw D.E. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dror R.O., Dirks R.M., Shaw D.E. Biomolecular simulation: a computational microscope for molecular biology. Annu. Rev. Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 65.Shan Y., Eastwood M.P., Shaw D.E. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 66.Baaden M., Marrink S.J. Coarse-grain modelling of protein-protein interactions. Curr. Opin. Struct. Biol. 2013;23:878–886. doi: 10.1016/j.sbi.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 67.Mim C., Cui H., Unger V.M. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Jong D.H., Lopez C.A., Marrink S.J. Molecular view on protein sorting into liquid-ordered membrane domains mediated by gangliosides and lipid anchors. Faraday Discuss. 2013;161:347–363. doi: 10.1039/c2fd20086d. discussion 419–459. [DOI] [PubMed] [Google Scholar]
- 69.Hall B.A., Armitage J.P., Sansom M.S.P. Mechanism of bacterial signal transduction revealed by molecular dynamics of Tsr dimers and trimers of dimers in lipid vesicles. PLoS Comput. Biol. 2012;8:e1002685. doi: 10.1371/journal.pcbi.1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei P., Zheng B.K., Luo S.Z. The association of polar residues in the DAP12 homodimer: TOXCAT and molecular dynamics simulation studies. Biophys. J. 2013;104:1435–1444. doi: 10.1016/j.bpj.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chng C.P., Tan S.M. Leukocyte integrin αLβ2 transmembrane association dynamics revealed by coarse-grained molecular dynamics simulations. Proteins. 2011;79:2203–2213. doi: 10.1002/prot.23044. [DOI] [PubMed] [Google Scholar]
- 72.Janosi L., Li Z., Gorfe A.A. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc. Natl. Acad. Sci. USA. 2012;109:8097–8102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mondal S., Johnston J.M., Weinstein H. Membrane driven spatial organization of GPCRs. Sci. Rep. 2013;3:2909. doi: 10.1038/srep02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Gunsteren W.F., Daura X., Mark A.E. Computation of free energy. Helv. Chim. Acta. 2002;85:3113–3129. [Google Scholar]
- 75.Johnston J.M., Wang H., Provasi D., Filizola M. Assessing the relative stability of dimer interfaces in G protein-coupled receptors. PLoS Comput. Biol. 2012;8:e1002649. doi: 10.1371/journal.pcbi.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park S., Im W. Two dimensional window exchange umbrella sampling for transmembrane helix assembly. J. Chem. Theory Comput. 2013;9:13–17. doi: 10.1021/ct3008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park S., Kim T., Im W. Transmembrane helix assembly by window exchange umbrella sampling. Phys. Rev. Lett. 2012;108:108102. doi: 10.1103/PhysRevLett.108.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li P.C., Miyashita N., Sugita Y. Multidimensional umbrella sampling and replica-exchange molecular dynamics simulations for structure prediction of transmembrane helix dimers. J. Comput. Chem. 2014;35:300–308. doi: 10.1002/jcc.23494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neale C., Madill C., Pomès R. Accelerating convergence in molecular dynamics simulations of solutes in lipid membranes by conducting a random walk along the bilayer normal. J. Chem. Theory Comput. 2013;9:3686–3703. doi: 10.1021/ct301005b. [DOI] [PubMed] [Google Scholar]
- 80.Jämbeck J.P.M., Lyubartsev A.P. Exploring the free energy landscape of solutes embedded in lipid bilayers. J. Phys. Chem. Lett. 2013;4:1781–1787. doi: 10.1021/jz4007993. [DOI] [PubMed] [Google Scholar]
- 81.Kopelevich D.I. One-dimensional potential of mean force underestimates activation barrier for transport across flexible lipid membranes. J. Chem. Phys. 2013;139:134906. doi: 10.1063/1.4823500. [DOI] [PubMed] [Google Scholar]
- 82.Fung J.J., Deupi X., Kobilka B.K. Ligand-regulated oligomerization of β2-adrenoceptors in a model lipid bilayer. EMBO J. 2009;28:3315–3328. doi: 10.1038/emboj.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang J., Chen S., Huang X.-Y. Crystal structure of oligomeric β1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat. Struct. Mol. Biol. 2013;20:419–425. doi: 10.1038/nsmb.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deupi X., Kobilka B.K. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (Bethesda) 2010;25:293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Unal H., Karnik S.S. Domain coupling in GPCRs: the engine for induced conformational changes. Trends Pharmacol. Sci. 2012;33:79–88. doi: 10.1016/j.tips.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khelashvili G., Albornoz P.B.C., Weinstein H. Why GPCRs behave differently in cubic and lamellar lipidic mesophases. J. Am. Chem. Soc. 2012;134:15858–15868. doi: 10.1021/ja3056485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jafurulla M., Tiwari S., Chattopadhyay A. Identification of cholesterol recognition amino acid consensus (CRAC) motif in G-protein coupled receptors. Biochem. Biophys. Res. Commun. 2011;404:569–573. doi: 10.1016/j.bbrc.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 88.Rasmussen S.G.F., DeVree B.T., Kobilka B.K. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martin M., Dotti C.G., Ledesma M.D. Brain cholesterol in normal and pathological aging. Biochim. Biophys. Acta. 2010;1801:934–944. doi: 10.1016/j.bbalip.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 90.Stranahan A.M., Cutler R.G., Mattson M.P.J. Diet-induced elevations in serum cholesterol are associated with alterations in hippocampal lipid metabolism and increased oxidative stress. J. Neurochem. 2011;118:611–615. doi: 10.1111/j.1471-4159.2011.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alemany R., Perona J.S., Ruiz-Gutierrez V. G protein-coupled receptor systems and their lipid environment in health disorders during aging. Biochim. Biophys. Acta. 2007;1768:964–975. doi: 10.1016/j.bbamem.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 92.Chattopadhyay A., Paila Y.D. Lipid-protein interactions, regulation and dysfunction of brain cholesterol. Biochem. Biophys. Res. Commun. 2007;354:627–633. doi: 10.1016/j.bbrc.2007.01.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


