Abstract
Transfer messenger RNA (tmRNA; also known as 10Sa RNA or SsrA RNA) is a small RNA molecule that is conserved among bacteria. It has structural and functional similarities to tRNA: it has an upper half of the tRNA-like structure, its 5’ end is processed by RNase P, it has typical tRNA-specific base modifications, it is aminoacylated with alanine, it binds to EF-Tu after aminoacylation and it enters the ribosome with EF-Tu and GTP. However, tmRNA lacks an anticodon, and instead it has a coding sequence for a short peptide called tag-peptide. An elaborate interplay of actions of tmRNA as both tRNA and mRNA with the help of a tmRNA-binding protein, SmpB, facilitates trans-translation, which produces a single polypeptide from two mRNA molecules. Initially alanyl-tmRNA in complex with EF-Tu and SmpB enters the vacant A-site of the stalled ribosome like aminoacyl-tRNA but without a codon–anticodon interaction, and subsequently truncated mRNA is replaced with the tag-encoding region of tmRNA. During these processes, not only tmRNA but also SmpB structurally and functionally mimics both tRNA and mRNA. Thus trans-translation rescues the stalled ribosome, thereby allowing recycling of the ribosome. Since the tag-peptide serves as a target of AAA+ proteases, the trans-translation products are preferentially degraded so that they do not accumulate in the cell. Although alternative rescue systems have recently been revealed, trans-translation is the only system that universally exists in bacteria. Furthermore, it is unique in that it employs a small RNA and that it prevents accumulation of non-functional proteins from truncated mRNA in the cell. It might play the major role in rescuing the stalled translation in the bacterial cell.
Keywords: tmRNA, SmpB, ribosome, trans-translation, molecular mimicry
INTRODUCTION
Translation often stalls in various situations in a cell, sometimes in a programmed fashion and other times unexpectedly. For example, translation of mRNA lacking a stop codon (non-stop mRNA) does not terminate efficiently because peptide release factor does not function. Thus, the cell should have a system to cope with such emergencies. However, little attention was given to this issue until the mid-1990s, and therefore the discovery of tmRNA was a big surprise. Initially the tRNA-like structure and function of tmRNA were elucidated. Both terminal regions of tmRNA can form a secondary structure resembling the upper half of the cloverleaf-like structure of tRNA (Komine et al., 1994; Ushida et al., 1994), which includes several tRNA-specific consensus sequences and base modifications (Figure 1A; Felden et al., 1997, 1998). Like that of tRNA, the 3′ end of tmRNA can be aminoacylated with an amino acid (alanine) by an aminoacyl-tRNA synthestase (alanyl-tRNA synthetase; Komine et al., 1994; Ushida et al., 1994). Other tRNA-like functions, such as 5′ processing by RNase P (Komine et al., 1994), binding to EF-Tu (Rudinger-Thirion et al., 1999; Barends et al., 2000, 2001; Hanawa-Suetsugu et al., 2001) and interaction with 70S ribosome (Ushida et al., 1994; Komine et al., 1996; Tadaki et al., 1996), have also been revealed. Although it is about fivefold larger than tRNA, tmRNA has no apparent anticodon, making it difficult to clarify whether and how tmRNA is involved in translation. A few years later, it was found that tmRNA has functions not only as tRNA but also as mRNA: a short peptide is encoded by the middle part of tmRNA (Tu et al., 1995), which is surrounded by four pseudoknot structures (Figure 1A; Nameki et al., 1999b,c). Intriguingly, these two functions cooperate, rather than being independent, to produce a chimeric polypeptide from two mRNAs, a C-terminally truncated polypeptide encoded by mRNA fusing the tmRNA-encoded short peptide with an alanine residue of unknown origin in between them (Keiler et al., 1996; Muto et al., 1996; Himeno et al., 1997). This acrobatic translation involving co-translational mRNA swapping produces a single polypeptide from two mRNAs, and thus it has been called trans-translation (Figure 2; Muto et al., 1998). This system provides a stop codon to allow completion of translation of a non-stop mRNA and consequently recycling of the ribosome. In addition, trans-translation has been regarded as a quality control system that prevents non-functional polypeptides derived from truncated mRNAs from accumulating in the cell, as the tag-peptide consisting of the first alanine residue and the tmRNA-encoded short peptide, especially the sequence of the last four hydrophobic amino acids (ALAA), serves as the target for cellular ATP-dependent proteases (Gottesman et al., 1998; Herman et al., 1998; Flynn et al., 2001; Choy et al., 2007).
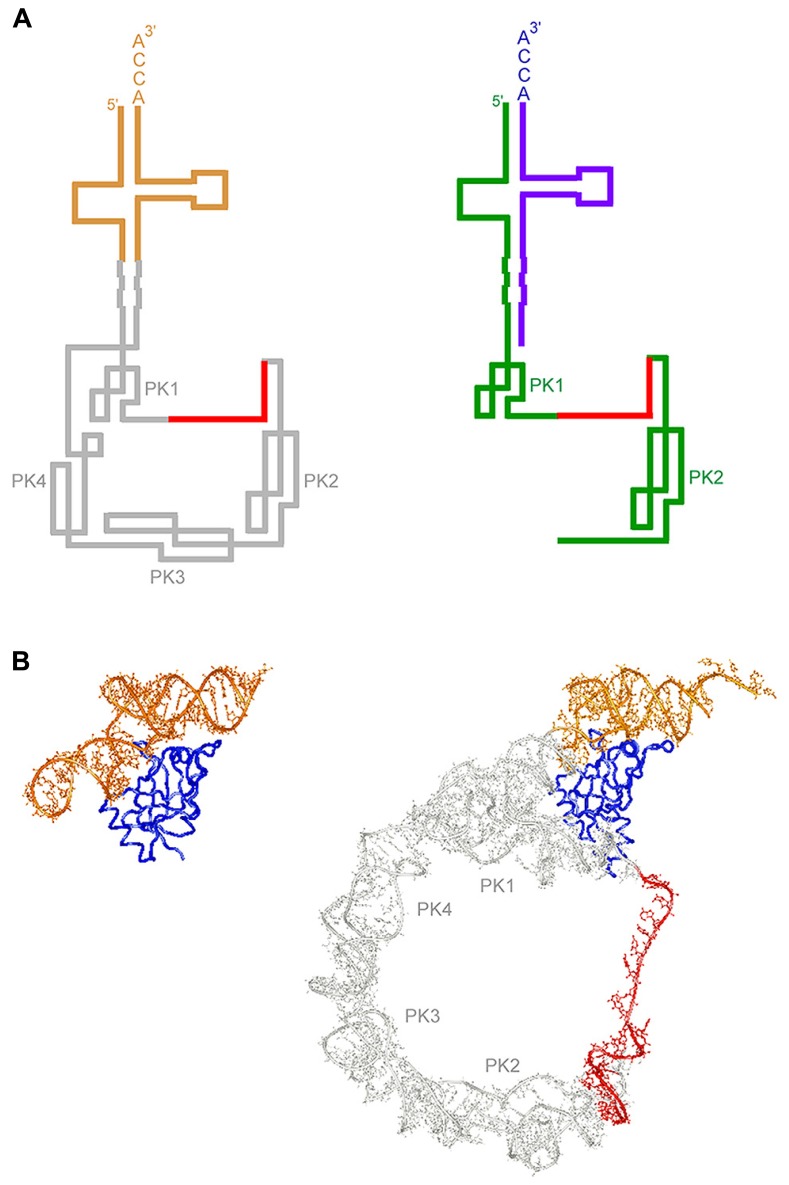
FIGURE 1.
Secondary and tertiary structures of tmRNA. (A) Secondary structure of tmRNA. (Left) The most typical secondary structure of tmRNA is shown, with TLD and the tag-encoding region designated in brown and red, respectively. (Right) A secondary structure of two-piece tmRNA. The 5’-coding piece and 3’-amino acid acceptor piece are designated in green and violet, respectively. (B) Tetiary structure of tmRNA or its fragment in complex with SmpB. (Left) A crystal structure of a complex of a TLD fragment lacking the 3’CCA end of T. thermophilus tmRNA (the 5’ 25 residues and 3’ 34 residues connected by a UUCG loop) and the globular domain (N-terminal 123 of a total of 144 residues) of SmpB (PDB ID: 2CZJ; Bessho et al., 2007). TLD (brown) and SmpB (dark blue) mimic the upper and lower halves of the L-shaped structure of tRNA, respectively. (Right) A cryo-EM structure of tmRNA·SmpB in a T. thermophilus post-translocated state complex of trans-translation (PDB ID: 3YIR; Weis et al., 2010b) is shown, with TLD, the tag-encoding region and SmpB designated in brown, red, and dark blue, respectively. The N-terminal globular domain of SmpB mimicking the lower half of the L-shaped structure of tRNA is in close contact with the upstream region of the tag-encoding sequence.
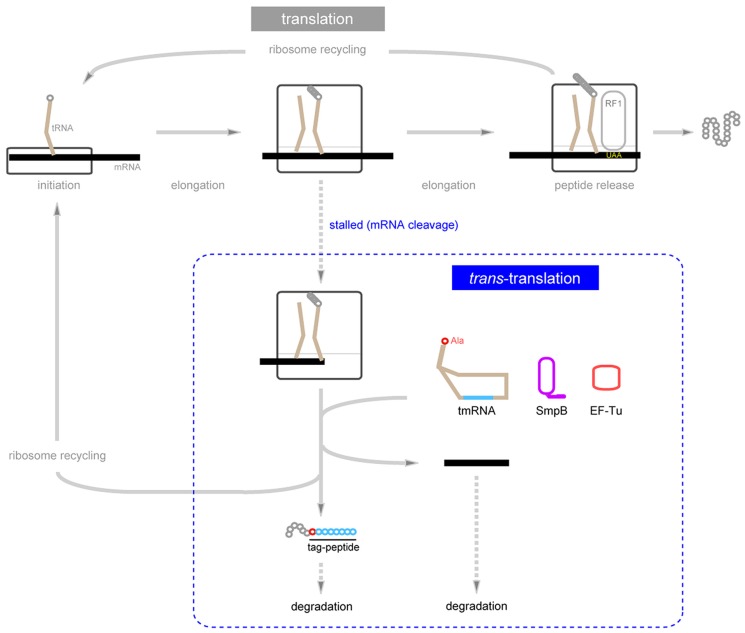
FIGURE 2.
Schematic representation of trans-translation. When the translating ribosome is stalled due to mRNA truncation, it is rescued by trans-translation, which produces a chimeric protein consisting of the C-terminally truncated polypeptide (gray circles) encoded by 3’-truncated mRNA fusing the tmRNA-encoded short peptide (blue circles) with an alanine residue (red circle) originating from alanine aminoacylated to the 3’-end of tmRNA in between them. Trans-translation promotes degradation of both truncated mRNAs and aberrant polypeptides from truncated mRNAs.
Trans-translation requires a tmRNA binding protein called SmpB in addition to canonical elongation factors (Karzai et al., 1999). SmpB consists of a globular domain and an unstructured C-terminal tail (Dong et al., 2002; Someya et al., 2003). It binds to the tRNA-like domain (TLD) of tmRNA to prevent tmRNA from degradation and enhance aminoacylation of tmRNA (Barends et al., 2001; Hanawa-Suetsugu et al., 2002; Shimizu and Ueda, 2002; Nameki et al., 2005). SmpB also plays a crucial role in the ribosomal process of trans-translation.
The trans-translation system is universally present in bacterial cells and is present in organelles of some eukaryotes but not in the cytoplasm of eukaryotes or archaebacteria. There is accumulating evidence indicating that the bacterial cell is equipped with additional systems to cope with stalled translation. Here, we review the current understanding of the molecular mechanism and the cellular functions of tmRNA-mediated trans-translation as well as other ribosome rescue systems.
Molecular Mechanism of Trans-Translation
An outline of the process of trans-translation is as follows (Figure 3): initially, tmRNA in complex with SmpB is aminoacylated with alanine by alanyl-tRNA synthetase. Ala-tmRNA enters the A-site of the stalled ribosome on a truncated mRNA to receive the nascent polypeptide from peptidyl-tRNA in the P-site. Then peptidyl-Ala-tmRNA translocates to the P-site, which exchanges the template from truncated mRNA to the tag-encoding region on tmRNA. It can reasonably explain the missing origin of the alanine residue connecting the truncated polypeptide encoded by mRNA with the tmRNA-encoded tag-peptide: it is derived from the alanine moiety aminoacylated to tmRNA. This model was proposed on the basis of the results of an in vivo study showing that truncated polypeptides fusing the tmRNA-encoding tag-peptide in its C-termini accumulate in the cell when a truncated mRNA is expressed (Keiler et al., 1996), and the model was supported by the results of an in vitro study showing that the tag-peptide is synthesized using Escherichia coli cell extract depending on the addition of poly(U) and on the aminoacylation capacity of tmRNA (Muto et al., 1996; Himeno et al., 1997). However, several questions have been raised. How does tmRNA find the stalled ribosome? How does tmRNA enter the A-site of the ribosome without an anticodon? How does the tag-encoding region of tmRNA substitute for truncated mRNA? How is the resuming point on tmRNA determined? SmpB has emerged as the key molecule to solve these questions.
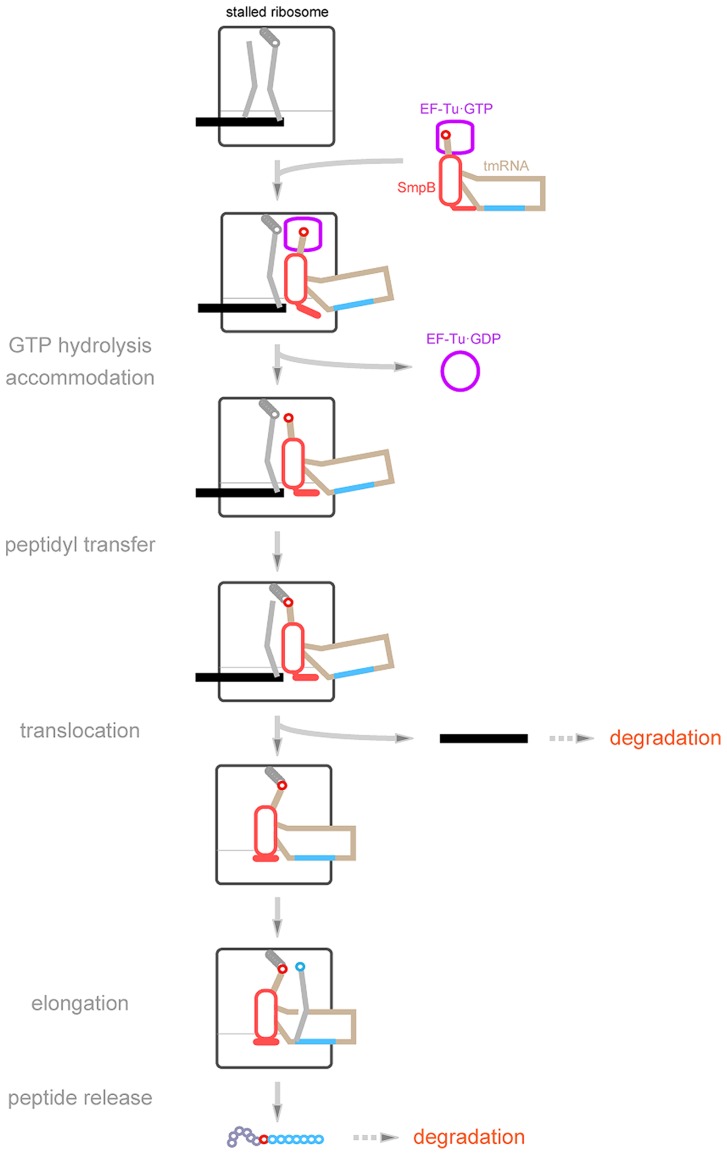
FIGURE 3.
A current model of trans-translation. Ala-tmRNA·SmpB·EF-Tu·GTP enters the vacant A-site of the stalled ribosome to trigger trans-translation. GTP hydrolysis by EF-Tu may induce a conformation change of the stalled complex, allowing the C-terminal tail of SmpB to interact with the mRNA path with an extended structure. After peptidyl-transfer, peptidyl-Ala-tmRNA·SmpB translocates from the A-site to the P-site with the help of EF-G to drive out truncated mRNA from the ribosome. During this process, the extended C-terminal tail somehow folds. Then the resume codon of tmRNA is set on the decoding region. SmpB and the tag-encoding region are shown in red and blue, respectively.
Besides canonical translation factors and tmRNA, SmpB is the minimal requirement for in vitro trans-translation (Shimizu and Ueda, 2002; Takada et al., 2007; Kurita et al., 2012). SmpB has been thought to continue binding to tmRNA throughout the process of trans-translation (Ivanova et al., 2007). In a crystal structure of a complex of SmpB and a model RNA fragment corresponding to TLD of tmRNA, the globular domain of SmpB binds to TLD so that it compensates for the lack of the lower half of the L-form structure in tmRNA (Figure 1B; Gutmann et al., 2003; Bessho et al., 2007). Thus, TLD in complex with the globular domain of SmpB would structurally mimic a whole tRNA molecule. A directed hydroxyl radical probing study has revealed two SmpB binding sites in an E. coli ribosome, one in the A-site and the other in the P-site, both of which can be superimposed on the lower half of the tRNA molecules in the translating ribosome (Kurita et al., 2007). An additional mimicry of the upper half of tRNA by TLD could complete two whole translating tRNA mimics at the A-site and P-site so that their aminoacyl ends are oriented to the peptidyl-transferase center. The pattern of cleavage of 16S rRNA by hydroxyl radicals from the C-terminal tail residues has suggested two binding sites of the C-terminal tail of SmpB with two different modes of conformation in the ribosome in addition to the unstructured conformation in solution: an extended conformation from the A-site to the downstream tunnel along the mRNA path as an α-helical structure and a folded conformation around the mRNA path in the P-site (Kurita et al., 2007, 2010).
On the basis of these SmpB properties, the trans-translation process can be described in more detail (Figure 3). Ala-tmRNA·SmpB·EF-Tu·GTP enters the vacant A-site of the stalled ribosome. GTP hydrolysis induces a conformational change of the stalled complex to release EF-Tu.GDP, allowing accommodation of Ala-TLD·SmpB in the A-site. During this process, the C-terminal tail of SmpB interacts with the mRNA path extending towards the mRNA entry channel. Subsequently, Ala-TLD in the A-site receives the nascent polypeptide chain from peptidyl-tRNA in the P-site, and the resulting peptidyl-Ala-TLD·SmpB translocates from the A-site to the P-site. During this process, the C-terminal tail of SmpB dissociates from the mRNA entry channel and binds around the site of codon–anticodon interaction in the P-site with change in its conformation from the extended structure to the folded structure, which in turn promotes release of mRNA from the ribosome. The conformational change of the C-terminal tail concomitant with translocation makes the A-site free, thereby allowing introduction of the resume codon of tmRNA into the decoding region. This model has been supported by results of structural studies of several trans-translation intermediates. Cryo-EM studies have revealed three kinds of intermediates in the pre-accommodated, accommodated, and translocated states (Kaur et al., 2006; Cheng et al., 2010; Fu et al., 2010; Weis et al., 2010a,b). In all of them, SmpB and TLD occupy the lower and upper halves, respectively, of the tRNA-binding sites. Although the C-terminal tail of SmpB has not been identified in these maps due to low resolution, its interaction with the mRNA path has clearly been shown in a crystal structure of a pre-accommodation state complex of trans-translation containing kirromycin (Neubauer et al., 2012).
This model can explain why trans-translation preferentially occurs at the ribosome stalled on mRNA with a shorter 3′-extension, which has been exemplified in vitro (Ivanova et al., 2004; Asano et al., 2005): the C-terminal tail of SmpB competes with the 3′-extension of mRNA for the mRNA entry channel. A chemical footprinting study has suggested that SmpB interacts with A1492, A1493, and G530 in 16S rRNA, which form the decoding region (Nonin-Lecomte et al., 2009). However, these nucleotides can be changeable without loss of both peptidyl-transferase and GTP hydrolytic activities in trans-translation, indicating much lower significance of these decoding nucleotides for trans-translation (Miller et al., 2011). In a crystal structure of a Thermus thermophilus pre-accommodation state complex of trans-translation, G530 stacks with a residue (Y127) around the start of the C-terminal tail of SmpB (Neubauer et al., 2012). Recently, Miller and Buskirk (2014) have found that the corresponding residue in E. coli SmpB (H136) has a crucial role in GTP hydrolysis, leading to the proposal that stacking of this residue with G530 triggers GTP hydrolysis.
EF-G promotes release of truncated mRNA from the stalled ribosome after peptidyl transfer to Ala-tmRNA, suggesting the presence of a canonical translocation-like step in the trans-translation process (Ivanova et al., 2005). During this event, SmpB must pass through the barrier between the A-site and P-site, and tmRNA must enter the inside of the mRNA entry channel to set the resume codon in the decoding region. Consistently, in a cryo-EM map of a translocational intermediate complex containing EF-G and fusidic acid, both bridge B1a, which serves as a barrier between the A-site and P-site, and latch, which is usually closed by the interaction between the head (helix 34) and body (G530 region) to form the mRNA tunnel, are open (Ramrath et al., 2012). Precise positioning of the resume codon at the decoding region requires the sequence just upstream of the resume codon at positions –6 to +1 (Williams et al., 1999; Lee et al., 2001), and this sequence is recognized by the globular domain of SmpB (Konno et al., 2007), suggesting that SmpB bridges two separate domains of tmRNA in the P-site to determine the resume codon for tag-translation presumably just after translocation. This is in agreement with cryo-EM maps of translocated state complexes of trans-translation (Figure 1B; Fu et al., 2010; Weis et al., 2010b). Another study has suggested the importance of the C-terminal tail of SmpB and its interaction with the start GCA codon on tmRNA for determination of the start point of tag-peptide translation (Camenares et al., 2013). Taken together, the results suggest that the interaction between tmRNA and SmpB is more important for resume point determination than the interaction between tmRNA and the ribosome. It should be noted that some kinds of aminoglycosides that bind the decoding region shift the resume point of tag-translation (Takahashi et al., 2003; Konno et al., 2004).
Although several examples of the potential molecular mimicry of tRNA by a translation factor have been reported, SmpB is the sole molecule that has been assumed to mimic the dynamic behavior of tRNA throughout all of the classical and hybrid states, A/T, A/A, A/P, P/P, and P/E, in the translating ribosome. The ribosomal protein S1, which has been identified as a tmRNA-binding protein (Wower et al., 2000), is not thought to participate in the early stage of trans-translation, at least until the first translocation (Qi et al., 2007; Takada et al., 2007).
Requirement of mRNA Cleavage for Trans-Translation
Because the tag-sequence serves as a degradation signal, trans-translation products are hardly detected in the cell or its extract, although they become accumulated and thus detectable when the tag-encoding sequence of tmRNA is engineered. It has long been believed that trans-translation occurs around the 3′-end of truncated mRNA lacking a stop codon (non-stop mRNA) in the stalled ribosome since publication of the results of an earlier in vivo study using an artificial mRNA (Keiler et al., 1996). Non-stop mRNA can be produced either unexpectedly or in a programmed fashion, and a similar situation can also arise when the normal termination codon is read through in the presence of a non-sense suppressor tRNA (Ueda et al., 2002) or a miscoding drug (Abo et al., 2002). Proteomic analyses have identified endogenous trans-translation products from various bacterial sources, indicating that trans-translation preferentially occurs at specific sites of specific mRNAs (Roche and Sauer, 2001; Collier et al., 2002; Fujihara et al., 2002; Hong et al., 2007; Barends et al., 2010). Consequently, several situations that promote trans-translation in the middle of mRNA have been focused on: translational pausing due to a rare codon (Roche and Sauer, 1999), an inefficient termination codon (Roche and Sauer, 2001; Hayes et al., 2002a; Sunohara et al., 2002) and a programmed stalling sequence (Collier et al., 2004) induces trans-translation. However, whether trans-translation actually occurs in the middle of mRNA without cleavage has been controversial. It has been found that a bacterial toxin, RelE, cleaves an mRNA specifically at the A-site in the stalled ribosome (Pedersen et al., 2003). RelE is usually inactivated by an antitoxin, RelB, and it is activated by degradation of RelB by Lon protease upon amino acid starvation. Yet, the finding of an A-site-specific endoribonuclease has supported the idea that mRNA cleavage is the prerequisite for trans-translation. Ribosome stalling induces cleavage of mRNA at the A-site even in a cell lacking RelE or several other endoribonucleases (Hayes and Sauer, 2003; Sunohara et al., 2004a,b; Li et al., 2008), indicating the involvement of an as-yet-unidentified ribonuclease or the ribosome itself in mRNA cleavage. It has also been reported that the 3′–5′ exoribonulease activity of RNase II is an important prerequisite for A-site-specific mRNA cleavage (Garza-Sánchez et al., 2009). Besides RelE, several kinds of ribosome-dependent endoribonucleases, each having a specific antitoxin, have been identified in E. coli (Feng et al., 2013).
In vitro studies have clearly shown that trans-translation can occur in the middle of mRNA, although the efficiency of trans-translation is drastically decreased with increasing length of the 3′-extension from the stalled position (Ivanova et al., 2004; Asano et al., 2005). This is in agreement with results of structural studies showing that the C-terminal tail of SmpB occupies the mRNA path in the early stage of trans-translation so that it competes with the 3′-extension of mRNA (Kurita et al., 2010; Neubauer et al., 2012).
Proteomic studies have shown that trans-translation preferentially occurs at the proline codon just preceding the stop codon (Hayes et al., 2002a,b). Asp–Pro, Glu–Pro, Pro–Pro, Ile–Pro, and Val–Pro are favorable C-terminal dipeptides for trans-translation, suggesting an additional importance of the penultimate residue. In fact, Asp–Pro and Pro–Pro are unusually underrepresented at the C-terminus in most bacterial proteins. Due to the structural irregularity of proline having a secondary amine instead of the primary amine, peptidyl-Pro-tRNAPro in the A-site would interfere with the access of peptide release factor (Janssen and Hayes, 2009), rather than that of the Ala-tmRNA·SmpB·EF-Tu·GTP complex. Consistently, limited amounts of aminoacyl-tRNA or release factor induce trans-translation, indicating competition of trans-translation with aminoacyl-tRNA and release factor for sense and non-sense codons, respectively, in the stalled ribosome (Ivanova et al., 2004; Asano et al., 2005; Li et al., 2007). Consecutive proline residues also affect peptidyl-transfer during the elongation process to cause translational arrest, which can be rescued by EF-P (Doerfel et al., 2013; Ude et al., 2013).
These in vivo and in vitro studies together have settled the controversy shown above: translation can stall even in the middle of mRNA in some situations, and this kind of stalled ribosome can be a potential target for trans-translation, although it would substantially occur only after cleavage of mRNA around the A-site by RelE or another as-yet-unidentified ribonuclease with the help of a 3′–5′ exoribonuclease RNase II.
Trans-Translation as Quality Control Systems of Protein and mRNA
As described above, the most significant role of trans-translation is to promote recycling of stalled ribosomes in the cell. Trans-translation is thought to have additional roles as quality control systems of protein and mRNA.
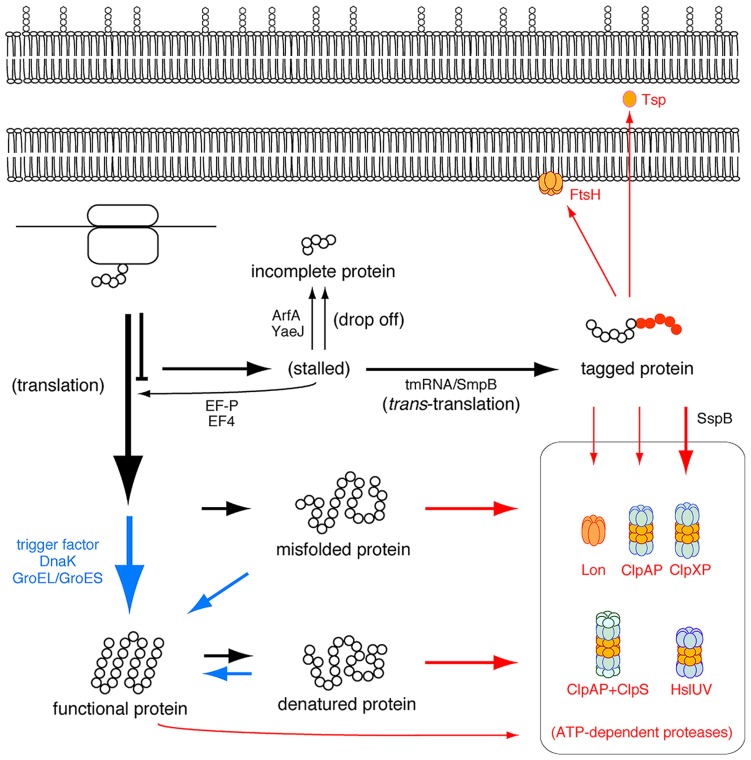
Most trans-translation products would be non-functional, and thus their accumulation might be deleterious for the cell. To avoid this situation, the tag-peptide and in turn the trans-translation products are promptly degraded in the cell by cytoplasmic ATP-dependent proteases (AAA+ proteases), including ClpXP, ClpAP, Lon and FtsH, and the periplasmic protease Tsp (Prc; Figure 4). ClpX or ClpA recognizes the C-terminal ALAA sequence of the tag-peptide to unfold the trans-translation products in an ATP-dependent fashion for degradation by its partner ClpP peptidase (Gottesman et al., 1998). The tag-peptide specifically binds to a protein, SspB, to increase its affinity to ClpX, and consequently ClpXP is thought to play the dominant role in degradation of trans-translation products at least in β- and γ-proteobacteria (Flynn et al., 2001) and perhaps in α-proteobacteria (Lessner et al., 2007). Lon participates in degradation of trans-translation products under stressful conditions (Choy et al., 2007). FtsH is anchored to the cytoplasmic side of the inner membrane to degrade the membrane-associated trans-translation products (Herman et al., 1998). The C-terminal ALAA sequence of the tag-peptide required for ClpXP and ClpAP is highly conserved among bacteria except Mycoplasma, in which the tag-peptide terminates with AFA instead of ALAA. This can be addressed by the absence of ClpXP, ClpAP, and Tsp in Mycoplasma (Gur and Sauer, 2008; Ge and Karzai, 2009).
FIGURE 4.
Ribosome rescue pathways together with folding and degradation pathways in an E. coli cell. Folding and degradation pathways are shown in blue and red, respectively.
While trans-translation is induced by cleavage of mRNA in the stalled ribosome as described above, trans-translation also promotes further degradation of non-stop mRNA (Yamamoto et al., 2003). Trans-translation would expose the 3′ end of non-stop mRNA sequestered by the stalled ribosome, facilitating the access of 3′–5′ exoribonuclease for degradation of non-stop mRNA. It should be a great advantage for the cell, considering that ribosome stalling would be repeated until non-stop mRNA is degraded, even if the stalled ribosome at the 3′ end of the polysome is rescued by trans-translation. RNase R is a likely candidate for such an exoribonuclease (Oussenko et al., 2005; Mehta et al., 2006; Richards et al., 2006; Ge et al., 2011). E. coli RNase R makes a complex with tmRNA and SmpB (Karzai and Sauer, 2001) via direct interaction with SmpB (Liang and Deutscher, 2010). It is induced under stress conditions in E. coli (Chen and Deutscher, 2005) and is involved in cell cycle-regulated degradation of tmRNA in Caulobacter crescentus (Hong et al., 2005). E. coli RNase R is acetylated in exponential phase, resulting in the exponential phase-specific degradation via tighter binding to tmRNA·SmpB (Liang et al., 2011).
Physiological Significance of Trans-Translation
The apparent universality of the trans-translation system among bacteria suggests some biological significance of this system. Indeed, it is essential for some bacteria including Neisseria gonorrhoeae (Huang et al., 2000), Mycoplasma genitalium (Hutchison et al., 1999), Haemophilus influenzae (Akerley et al., 2002), Helicobacter pylori (Thibonnier et al., 2008), and Shigella flexneri (Ramadoss et al., 2013a), and its depletion causes a wide variety of disorders. Since its lack causes avirulence of some infectious bacteria, the trans-translation system has been focused on as an effective target for antibiotics (Shi et al., 2011; Ramadoss et al., 2013b).
Many of these defective phenotypes are caused by a defect in the trans-translation reaction rather than degradation of the trans-translation products (Keiler, 2008). This suggests that ribosome recycling is more important for the cell than preventing accumulation of non-functional proteins. Upon starvation of amino acids, supply of amino acids from trans-translation products should become important for new protein synthesis (Pedersen et al., 2003; Li et al., 2008).
Trans-translation is often employed for regulation of gene expression. In E. coli, tmRNA-mediated trans-translation targets mRNA for LacI, a repressor of the lactose operon, to accelerate its degradation upon glucose depletion, leading to derepression of the lac operon (Abo et al., 2000). In Bacillus subtilis, trans-translation occurs around the catabolite responsive element (cre) sequence, a binding site of the repressor protein catabolite control protein A (CcpA), within the coding region of several mRNAs including TreP mRNA for trehalose phosphorylase (Fujihara et al., 2002). Binding of CcpA to the cre sequence would induce a transcriptional roadblock to produce truncated mRNA (Ujiie et al., 2009). In C. crescentus, the cell cycle (Keiler and Shapiro, 2003a) and the initiation of DNA replication (Keiler and Shapiro, 2003b; Hong et al., 2007) are controlled by trans-translation.
There is accumulating evidence for increased importance of tmRNA under stressful conditions, such as high or low temperature (Oh and Apirion, 1991; Muto et al., 2000; Shin and Price, 2007), nutrient starvation (Oh and Apirion, 1991; Okan et al., 2006; Abe et al., 2008), ethanol treatment (Muto et al., 2000), cadmium treatment (Muto et al., 2000), and acid exposure (Thibonnier et al., 2008). Stresses might increase the frequency of aberrant translation in cells, which can be rescued by trans-translation. Consistently, the total amount of trans-translation products increases under stressful conditions (Fujihara et al., 2002). Perhaps in response to the increased requirement of the trans-translation system, the intracellular level of tmRNA or SmpB increases with an increase in stress in some bacteria (Muto et al., 2000; Palecková et al., 2007; Rezzonico et al., 2007).
The trans-translation system sometimes regulates other stress response systems possibly via expression of a global regulator. For example, depletion of tmRNA induces heat shock response in E. coli (Munavar et al., 2005). The expression level of the sigma factor RpoS (sigma S), which controls the expression of a series of genes involved in general stress response, is positively controlled by trans-translation in E. coli (Ranquet and Gottesman, 2007). The involvement of trans-translation in the extracellular stress-response pathway via another sigma factor, RpoE (sigma E), has also been suggested (Ono et al., 2009). Other stress-related proteins including the molecular chaperone DnaK are regulated by trans-translation in streptomycetes (Barends et al., 2010). Interestingly, the expression of ArfA, an alternative ribosome rescue system (described in a later subsection), is regulated by trans-translation (Chadani et al., 2011a; Garza-Sánchez et al., 2011).
Evolutionary Aspects of Trans-Translation
Although it is not essential for viability in most bacteria, tmRNA is present universally in the bacterial kingdom and in some plastids or mitochondria of some protists. The tRNA-like secondary structure of TLD as well as several tRNA-specific consensus sequences is highly conserved except for the deformed D-arm structure, which has an extensive interaction with SmpB. In addition, the third base-pair position of the amino acid acceptor stem is completely conserved as G–U, which serves as a potent identity determinant for recognition by alanyl-tRNA synthetase (AlaRS). Alanine might not be an absolute prerequisite for trans-translation as the amino acid aminoacylated to tmRNA as exemplified by Nameki et al. (1999a). However, AlaRS might be the most preferable aminoacyl-tRNA synthetase for tmRNA, considering the unique recognition mode of AlaRS depending largely on the acceptor stem instead of the anticodon. The tRNA-like structure would also be significant for recognition by EF-Tu and RNase P as well as for ribosome binding. In contrast to the high degree of conservation of TLD, there is variation in the pseudoknot-rich region (Nameki et al., 1999b; Williams, 2002). Plastid tmRNA has fewer or no pseudoknot structures (Gueneau de Novoa and Williams, 2004). Indeed, at least the last three of four pseudoknots of E. coli tmRNA are dispensable for trans-translation in vitro (Nameki et al., 2000), although they participate in proper folding and processing of tmRNA (Wower et al., 2004). TLD and the pseudoknot-rich region are linked by a long degenerated helix, which protrudes from TLD in a direction corresponding to that of the long variable arm of class II tRNA (Figure 1B; Bessho et al., 2007). This direction should be conserved within the constraints of the tRNA-like dynamic behavior of tmRNA in the limited space of the ribosome, although the sequence of the connector helix is less conserved.
In some lineages of α-proteobacteria, β-proteobacteria, cyanobacteria, and mitochondria of lower eukaryotes, tmRNA is separated into two pieces, a 5′-coding piece typically including only two pseudoknots (PK1 and PK2) with the tag-encoding region in between and a 3′-amino acid acceptor piece, and the two pieces join together by base-pairing to form into a tRNA-like structure similar to that in one-piece tmRNA (Figure 1A; Keiler et al., 2000; Williams, 2002; Sharkady and Williams, 2004). The gene for two-piece tmRNA is circularly permuted (Keiler et al., 2000; Williams, 2002; Mao et al., 2009), and the permuted precursor might be processed into a mature two-piece tmRNA probably with the help of RNase P and tRNase Z. A similar processing strategy has been found in the circularly permuted tRNA gene in some primitive eukaryotes or archaebacteria (Soma et al., 2007). The two-piece tmRNA in C. crescentus belonging to α-proteobacteria has been shown to actually function in trans-translation (Keiler et al., 2000). Either one-piece or two-piece tmRNA is present in mitochondrial genome of some groups of protists (jakobids; Jacob et al., 2004). They lack a tag-encoding sequence as well as pseudoknots, arguing against their capacity for bacterial type of trans-translation.
SmpB together with tmRNA is universally present in bacteria. Plastid tmRNA is encoded by the plastid genome, while plastid SmpB is encoded by the nuclear genome and it is imported from the cytoplasm (Jacob et al., 2005). Up to now, there has been no report about mitochondrial SmpB. Both tmRNA and SmpB should have been required at the birth of trans-translation. The gene for tmRNA might have been formed by insertion of something into a tRNAAla gene. In contrast, no one can envisage the origin of SmpB because of the absence of its homologue.
Diversity of Rescue Systems of Stalled Translation
As described above, trans-translation targets various kinds of translational pausing due to a rare codon, an inefficient termination codon or a programmed stalling sequence, but after cleavage of mRNA. The bacterial cell has alternative mechanisms to rescue the stalled ribosome (Figure 4; Himeno, 2010).
Peptidyl-tRNA hydrolase (Pth) has an activity to hydrolyze the linkage between tRNA and the nascent polypeptide of peptidyl-tRNA after it drops off from the ribosome. Drop-off is enhanced by RRF alone, RRF together with RF3 (Heurgué-Hamard et al., 1998; Gong et al., 2007) or RRF, IF3, and EF-G (Singh et al., 2008). Drop-off was initially assumed to occur in the earlier cycles of elongation. This seems reasonable considering that a longer nascent polypeptide chain would prevent release of peptidyl-tRNA from the peptide channel of the translating ribosome. However, drop-off has been shown to efficiently occur at the 3′ end of non-stop mRNA in the absence of tmRNA (Kuroha et al., 2009). Overexpression of tmRNA suppresses the temperature-sensitive phenotype of Pth (Singh and Varshney, 2004), suggesting that Pth contributes not only to hydrolyzing the dropped-off peptidyl-tRNA but also to rescuing the stalled ribosome or suggesting that trans-translation can substitute for spontaneous or factor-promoting drop-off and the following peptidyl-tRNA hydrolysis by Pth.
It has recently been found that two proteins, ArfA (YhdL) and YaeJ (ArfB), facilitate rescue of the stalled ribosome. A single knockout of either E. coli ArfA or tmRNA is viable, whereas a double knockout is lethal, explaining why tmRNA is not essential in many bacteria (Chadani et al., 2010). The ribosome rescue activity of ArfA was initially shown using E. coli crude extract (Chadani et al., 2010). However, ArfA alone does not have an activity to hydrolyze peptidyl-tRNA in the P-site possibly due to the absence of a typical GGQ catalytic motif, and it requires the help of RF-2 (Chadani et al., 2012; Shimizu, 2012). RF-2 usually acts as a UAA or UGA codon-dependent release factor, while it serves as a stop codon-independent release factor in the presence of ArfA. Intriguingly, translation for ArfA protein is stalled near the 3′-terminus of its mRNA due to cleavage by RNase III, and consequently ArfA is usually degraded via the trans-translation system, and only when the cellular trans-translation activity becomes diminished, is C-terminally truncated but active ArfA synthesized via spontaneous drop-off or ArfA-mediated release of the nascent polypeptide (Garza-Sánchez et al., 2011; Chadani et al., 2011a). Thus, the ArfA-mediated ribosome rescue system is considered to be a backup system for trans-translation. YaeJ has also been shown to rescue the ribosomes stalled at the 3′ end of a non-stop mRNA in vitro (Handa et al., 2011) and in vivo (Chadani et al., 2011b). A double knockout of E. coli ArfA and tmRNA is lethal as described above, whereas overexpression of YaeJ makes it viable (Chadani et al., 2011a). Unlike ArfA, YaeJ alone has an activity to hydrolyze peptidyl-tRNA in the P-site of the stalled ribosome, as expected from the similar sequence and structure to those of the catalytic domain of bacterial class I release factor having a GGQ motif. YaeJ is likely to act as a stop codon-independent peptide chain release factor since it lacks a stop codon-recognition domain and instead it is replaced by a C-terminal basic-residue-rich extension that might be unstructured in solution (Handa et al., 2010). In a crystal structure of E. coli YaeJ in complex with the stalled ribosome from T. thermophilus, the C-terminal extension of YaeJ, like that of SmpB, binds along the mRNA path of the stalled ribosome extending to the downstream mRNA tunnel with an α-helical structure (Gagnon et al., 2012). ArfA as well as Ala-tmRNA·SmpB·EF-Tu(GTP) does not favor the long 3′ extension of mRNA from the decoding region upon entrance to the stalled ribosome, while YaeJ is less sensitive (Shimizu, 2012). Thus bacterial cells are equipped with multiple systems to cope with stalled translation, and they are therefore often still viable even when they lose the trans-translation system. Judging from phenotypes of factor-depleted cells, the trans-translation system must be the primary ribosome rescue system.
There are some reports about stress-specific ribosome rescue systems. The heat shock protein HSP15 has been shown to bind to the dissociated 50S subunit with a nascent protein (Korber et al., 2000). Upon exposure to a high temperature, a fraction of translating ribosomes might prematurely be dissociated into subunits, although peptidyl-tRNA remains bound to the dissociated 50S subunit unless the nascent peptide is short. In this 50S subunit, HSP15 fixes peptidyl-tRNA at the P-site to make the A-site free presumably for entrance of a peptide release factor (Jiang et al., 2009). High intracellular magnesium ion concentration or low temperature causes translational arrest after defective translocation. It also promotes release of a GTPase, EF4 (LepA), which is usually stored in the cell membrane, into the cytoplasm to rescue the translational arrest by back translocation (Pech et al., 2011).
Translation often stalls at a specific site on an mRNA in a programmed fashion. As in the case of ArfA expression described above (Chadani et al., 2011a; Garza-Sánchez et al., 2011), translational arrest is sometimes used for repression of gene expression via trans-translation. On the other hand, a stalled ribosome often prevents access of rescue machineries to keep translational arrest for regulation of gene expression. E. coli tryptophanase (tna) operon is induced by tryptophan via the translational arrest of the leader peptide (TnaC) by inhibiting hydrolysis of peptidyl-tRNAPro by RF2 (Yanofsky, 2007). This stalled ribosome is not rescued by trans-translation in the presence of tryptophan, although it is rescued slowly by RRF and RF3, leading to drop-off (Gong et al., 2007). The ribosome is also stalled at an internal proline codon of E. coli secM mRNA, which up-regulates the translation of the downstream SecA-encoding sequence presumably by disrupting the secondary structure that sequesters the ribosome binding site (Muto et al., 2006). This translational arrest is caused by inefficient peptidyl-transfer of Pro-tRNAPro in the A-site to the nascent peptidyl-tRNA in the P-site, which inhibits entrance of Ala-tmRNA to the A-site and the A-site specific cleavage of mRNA (Garza-Sánchez et al., 2006). The translation of B. subtilis yidC mRNA is regulated by translational arrest at multiple sites on the upstream mifM mRNA (Chiba et al., 2009; Chiba and Ito, 2012). Puromycin is less reactive to this translational arrest, suggesting that ribosome rescue machineries including Ala-tmRNA·SmpB·EF-Tu(GTP) are also less accessible to the A-site of this stalled ribosome. Consecutive proline codons cause a translational arrest due to inefficient peptidyl-transfer between peptidyl-(Pro)n-tRNA in the P-site and Pro-tRNA in the A-site (Doerfel et al., 2013; Ude et al., 2013). In this case, the A-site is occupied by Pro-tRNAPro, and in turn Ala-tmRNA·SmpB·EF-Tu(GTP), ArfA or YaeJ would fail to access this stalled ribosome. Instead, the peptidyltransferase center is modulated by EF-P binding to the region between the P-site and E-site to resume peptidyl-transfer (Blaha et al., 2009). Pro–Pro–Pro and Gly–Pro–Pro arrest sequences, which can be rescued by EF-P, are often found in bacterial genes, and they might be programmed for regulation of gene expression.
Pth is essential for bacteria and is widely distributed among the other domains. While the trans-translation system universally exists in bacteria, YaeJ is distributed among Gram-negative bacteria and ArfA shows more limited distribution within enterobacteria. EF-P is conserved among bacteria and its homologue (a/eIF5A) is universally present in archaea and eukaryotes. EF4 is universally conserved among bacteria. Neither tmRNA nor SmpB is present in the cytoplasm of eukaryotes, where a complex of Dom34p (Pelota) and the GTP-binding protein Hbs1 promotes subunit dissociation of the stalled ribosome and drop-off of peptidyl-tRNA (Shoemaker et al., 2010) in concert with an ATPase, ABCE1 (Pisareva et al., 2011). The Dom34p·Hbs1 complex is structurally similar to the eRF1·eRF3 complex or the aminoacyl-tRNA·EF-Tu complex (Chen et al., 2010), although the GGQ motif is absent in Dom34, and peptidyl-tRNA hydrolysis is assumed to be catalyzed by Pth after drop-off. In mitochondria, two protein factors partially similar to the bacterial class I release factor, ICT1 (a homologue of YaeJ) and C12orf65, both lacking a stop codon-recognition domain while retaining the catalytic GGQ motif, participate in ribosome rescue (Handa et al., 2010; Richter et al., 2010; Kogure et al., 2012, 2014). ICT1 (YaeJ), but not C12orf65, has an insertion of an α-helix in the GGQ domain, and thus ICT1 is less similar to class I release factor. C12orf65 has been found in very limited bacteria. Collectively, various kinds of release factor homologues, YaeJ, ICT1, C12orf65, ArfA/RF2, Dom34p (Pelota), and Hbs1, have been found to participate in ribosome rescue. The trans-translation system is unique in that it employs a small RNA and that it prevents accumulation of non-functional proteins from truncated mRNA in the cell.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to all those who have been involved in this work. This work was supported by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan to Hyouta Himeno, grants-in-aid for Scientific Research (B) and (C) from the Japan Society for the Promotion of Science to Akira Muto and grants-in-aid for Young Scientists from the Japan Society for the Promotion of Science to Daisuke Kurita.
REFERENCES
- Abe T., Sakaki K., Fujihara A., Ujiie H., Ushida C., Himeno H., et al. (2008). tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Mol. Microbiol. 69 1491–1498 10.1111/j.1365-2958.2008.06381.x [DOI] [PubMed] [Google Scholar]
- Abo T., Inada T., Ogawa K., Aiba H. (2000). SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J. 19 3762–3769 10.1093/emboj/19.14.3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo T., Ueda K., Sunohara T., Ogawa K., Aiba H. (2002). SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells 7 629–638 10.1046/j.1365-2443.2002.00549.x [DOI] [PubMed] [Google Scholar]
- Akerley B. J., Rubin E. J., Novick V. L., Amaya K., Judson N., Mekalanos J. J. (2002). A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 99 966–971 10.1073/pnas.012602299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Kurita D., Takada K., Konno T., Muto A., Himeno H. (2005). Competition between trans-translation and termination or elongation of translation. Nucleic Acids Res. 33 5544–5552 10.1093/nar/gki871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends S., Karzai A. W., Sauer R. T., Wower J., Kraal B. (2001). Simultaneous and functional binding of SmpB and EF-Tu-GTP to the alanyl acceptor arm of tmRNA. J. Mol. Biol. 314 9–21 10.1006/jmbi.2001.5114 [DOI] [PubMed] [Google Scholar]
- Barends S., Wower J., Kraal B. (2000). Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry 39 2652–2658 10.1021/bi992439d [DOI] [PubMed] [Google Scholar]
- Barends S., Zehl M., Bialek S., de Waal E., Traag B. A., Willemse J., et al. (2010). Transfer-messenger RNA controls the translation of cell-cycle and stress proteins in Streptomyces. EMBO Rep. 11 119–125 10.1038/embor.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y., Shibata R., Sekine S., Murayama K., Higashijima K., Hori-Takemoto C., et al. (2007). Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 104 8293–8298 10.1073/pnas.0700402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha G., Stanley R. E., Steitz T. A. (2009). EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 325 966–970 10.1126/science.1175800 [DOI] [PubMed] [Google Scholar]
- Camenares D., Dulebohn D. P., Svetlanov A., Karzai A. W. (2013). Active and accurate trans-translation requires distinct determinants in the C-terminal tail of SmpB protein and the mRNA-like domain of transfer messenger RNA (tmRNA). J. Biol. Chem. 288 30527–30542 10.1074/jbc.M113.503896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadani Y., Ito K., Kutsukake K., Abo T. (2012). ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl-tRNA hydrolysis in Escherichia coli. Mol. Microbiol. 86 37–50 10.1111/j.1365-2958.2012.08190.x [DOI] [PubMed] [Google Scholar]
- Chadani Y., Matsumoto E., Aso H., Wada T., Kutsukake K., Sutou S., et al. (2011a). trans-Translation-mediated tight regulation of the expression of the alternative ribosome-rescue factor ArfA in Escherichia coli. Genes Genet. Syst. 86 151–163 10.1266/ggs.86.151 [DOI] [PubMed] [Google Scholar]
- Chadani Y., Ono K., Kutsukake K., Abo T. (2011b). Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol. Microbiol. 80 772–785 10.1111/j.1365-2958.2011.07607.x [DOI] [PubMed] [Google Scholar]
- Chadani Y., Ono K., Ozawa S., Takahashi Y., Takai K., Nanamiya H., et al. (2010). Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol. Microbiol. 80 772–775 10.1111/j.1365-2958.2011.07607.x [DOI] [PubMed] [Google Scholar]
- Chen C., Deutscher M. P. (2005). Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 280 34393–34396 10.1074/jbc.C500333200 [DOI] [PubMed] [Google Scholar]
- Chen L., Muhlrad D., Hauryliuk V., Cheng Z., Lim M. K., Shyp V., et al. (2010). Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat. Struct. Mol. Biol. 17 1233–1240 10.1038/nsmb.1922 [DOI] [PubMed] [Google Scholar]
- Cheng K., Ivanova N., Scheres S. H., Pavlov M. Y., Carazo J. M., Hebert H., et al. (2010). tmRNA.SmpB complex mimics native aminoacyl-tRNAs in the A site of stalled ribosomes. J. Struct. Biol. 169 342–348 10.1016/j.jsb.2009.10.015 [DOI] [PubMed] [Google Scholar]
- Chiba S., Ito K. (2012). Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Mol. Cell 47 863–872 10.1016/j.molcel.2012.06.034 [DOI] [PubMed] [Google Scholar]
- Chiba S., Lamsa A., Pogliano K. (2009). A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 28 3461–3475 10.1038/emboj.2009.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy J. S., Aung L. L., Karzai A. W. (2007). Lon protease degrades transfer-messenger RNA-tagged proteins. J. Bacteriol. 189 6564–6571 10.1128/JB.00860-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J., Binet E., Bouloc P. (2002). Competition between SsrA tagging and translational termination at weak stop codons in Escherichia coli. Mol. Microbiol. 45 745–754 10.1046/j.1365-2958.2002.03045.x [DOI] [PubMed] [Google Scholar]
- Collier J., Bohn C., Bouloc P. (2004). SsrA tagging of Escherichia coli SecM at its translation arrest sequence. J. Biol. Chem. 279 54193–54201 10.1074/jbc.M314012200 [DOI] [PubMed] [Google Scholar]
- Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M. V. (2013). EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339 85–88 10.1126/science.1229017 [DOI] [PubMed] [Google Scholar]
- Dong G., Nowakowski J., Hoffman D. W. (2002). Structure of small protein B: the protein component of the tmRNA-SmpB system for ribosome rescue. EMBO J. 21 1845–1854 10.1093/emboj/21.7.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B., Hanawa K., Atkins J. F., Himeno H., Muto A., Gesteland R. F., et al. (1998). Presence and location of modified nucleotides in E. coli tmRNA. Structural mimicry with tRNA acceptor branches. EMBO J. 17 3188–3196 10.1093/emboj/17.11.3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B., Himeno H., Muto A., McCutcheon J. P., Atkins J. F., Gesteland R. F. (1997). Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA 3 89–103 [PMC free article] [PubMed] [Google Scholar]
- Feng S., Chen Y., Kamada K., Wang H., Tang K., Wang M., et al. (2013). YoeB-ribosome structure: a canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res. 41 9549–9556 10.1093/nar/gkt742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. M., Levchenko I., Seidel M., Wickner S. H., Sauer R. T., Baker T. A. (2001). Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98 10584–10589 10.1073/pnas.191375298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Hashem Y., Wower I., Lei J., Liao H. Y., Zwieb C., et al. (2010). Visualizing the transfer-messenger RNA as the ribosome resumes translation. EMBO J. 29 3819–3825 10.1038/emboj.2010.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara A., Tomatsu H., Inagaki S., Tadaki T., Ushida C., Himeno H., et al. (2002). Detection of tmRNA-mediated trans-translation products in Bacillus subtilis. Genes Cells 7 343–350 10.1046/j.1365-2443.2002.00523.x [DOI] [PubMed] [Google Scholar]
- Gagnon M. G., Seetharaman S. V., Bulkley D., Steitz T. A. (2012). Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335 1370–1372 10.1126/science.1217443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F., Janssen B. D., Hayes C. S. (2006). Prolyl-tRNAPro in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J. Biol. Chem. 281 34258–34268 10.1074/jbc.M608052200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F., Schaub R. E., Janssen B. D., Hayes C. S. (2011). tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol. Microbiol. 80 1204–1219 10.1111/j.1365-2958.2011.07638.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F., Shoji S., Fredrick K., Hayes C. S. (2009). RNase II is important for A-site mRNA cleavage during ribosome pausing. Mol. Microbiol. 73 882–897 10.1111/j.1365-2958.2009.06813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z., Karzai A. W. (2009). Co-evolution of multipartite interactions between an extended tmRNA tag and a robust Lon protease in Mycoplasma. Mol. Microbiol. 74 1083–1099 10.1111/j.1365-2958.2009.06923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z., Mehta P., Richards J., Karzai A. W. (2011). Non-stop mRNA decay initiates at the ribosome. Mol. Microbiol. 78 1159–1170 10.1111/j.1365-2958.2010.07396.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M., Cruz-Vera L. R., Yanofsky C. (2007). Ribosome recycling factor and release factor 3 action promotes TnaC-peptidyl-tRNA dropoff and relieves ribosome stalling during tryptophan induction of tna operon expression in Escherichia coli. J. Bacteriol. 189 3147–3155 10.1128/JB.01868-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Roche E., Zhou Y., Sauer R. T. (1998). The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging†system. Genes Dev. 12 1338–1347 10.1101/gad.12.9.1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueneau de Novoa P., Williams K. P. (2004). The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 32 D104–D108 10.1093/nar/gkh102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E., Sauer R. T. (2008). Evolution of the ssrA degradation tag in mycoplasma: specificity switch to a different protease. Proc. Natl. Acad. Sci. U.S.A. 105 16113–16118 10.1073/pnas.0808802105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann S., Haebel P. W., Metzinger L., Sutter M., Felden B., Ban N. (2003). Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature 424 5503–5509 10.1038/nature01831 [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K., Bordeau V., Himeno H., Muto A., Felden B. (2001). Importance of the conserved nucleotides around the tRNA-like structure of Escherichia coli transfer-messenger RNA for protein tagging. Nucleic Acids Res. 29 4663–4673 10.1093/nar/29.22.4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K., Takagi M., Inokuchi H., Himeno H., Muto A. (2002). SmpB functions in various steps of trans-translation. Nucleic Acids Res. 30 1620–1629 10.1093/nar/30.7.1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y., Hikawa Y., Tochio N., Kogure H., Inoue M., Koshiba S., et al. (2010). Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality. J. Mol. Biol. 404 260–273 10.1016/j.jmb.2010.09.033 [DOI] [PubMed] [Google Scholar]
- Handa Y., Inaho N., Nameki N. (2011). YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res. 39 1739–1748 10.1093/nar/gkq1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes C. S., Bose B., Sauer R. T. (2002a). Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99 3440–3445 10.1073/pnas.052707199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes C. S., Bose B., Sauer R. T. (2002b). Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 277 33825–33832 10.1074/jbc.M205405200 [DOI] [PubMed] [Google Scholar]
- Hayes C. S., Sauer R. T. (2003). Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell 12 903–911 10.1016/S1097-2765(03)00385-X [DOI] [PubMed] [Google Scholar]
- Herman C., Thévenet D., Bouloc P., Walker G. C, D’Ari R. (1998). Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 12 1348–1355 10.1101/gad.12.9.1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgué-Hamard V., Karimi R., Mora L., MacDougall J., Leboeuf C., Grentzmann G., et al. (1998). Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J. 17 808–816 10.1093/emboj/17.3.808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H. (2010). Cellular mechanisms to rescue the ribosome stalled during translation. Mol. Microbiol. 78 789–791 10.1111/j.1365-2958.2010.07379.x [DOI] [PubMed] [Google Scholar]
- Himeno H., Sato M., Tadaki T., Fukushima M., Ushida C., Muto A. (1997). In vitro trans-translation mediated by alanine-charged 10Sa RNA. J. Mol. Biol. 268 803–808 10.1006/jmbi.1997.1011 [DOI] [PubMed] [Google Scholar]
- Hong S.-J., Lessner F. H., Mahen E. M., Keiler K. C. (2007). Proteomic identification of tmRNA substrates. Proc. Natl. Acad. Sci. U.S.A. 104 17128–17133 10.1073/pnas.0707671104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.-J., Tran Q. A., Keiler K. (2005). Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol. Microbiol. 57 565–575 10.1111/j.1365-2958.2005.04709.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wolfgang M. C., Withey J., Koomey M., Friedman D. I. (2000). Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 19 1098–1107 10.1093/emboj/19.5.1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C. A., Peterson S. N., Gill S. R., Cline R. T., White O., Fraser C. M., et al. (1999). Global transposon mutagenesis and a minimal mycoplasma genome. Science 286 2165–2169 10.1126/science.286.5447.2165 [DOI] [PubMed] [Google Scholar]
- Ivanova N., Lindell M., Pavlov M., Holmberg Schiavone L., Wagner E. G., Ehrenberg M. (2007). Structure probing of tmRNA in distinct stages of trans-translation. RNA 13 713–722 10.1261/rna.451507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N., Pavlov M. Y., Ehrenberg M. (2005). tmRNA-induced release of messenger RNA from stalled ribosomes. J. Mol. Biol. 350 897–905 10.1016/j.jmb.2005.05.033 [DOI] [PubMed] [Google Scholar]
- Ivanova N., Pavlov M. Y., Felden B., Ehrenberg M. (2004). Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 338 33–41 10.1016/j.jmb.2004.02.043 [DOI] [PubMed] [Google Scholar]
- Jacob Y., Seif E., Paquet P.-O., Lang B. F. (2004). Loss of the mRNA-like region in mitochondrial tmRNAs of jakobids. RNA 10 605–614 10.1261/rna.5227904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y., Sharkady S. M., Bhardwaj K., Sanda A. A., Williams K. P. (2005). Function of the SmpB tail in transfer-messenger RNA translation revealed by a nucleus-encoded form. J. Biol. Chem. 280 5503–5509 10.1074/jbc.M409277200 [DOI] [PubMed] [Google Scholar]
- Janssen B. D., Hayes C. S. (2009). Kinetics of paused ribosome recycling in Escherichia coli. J. Mol. Biol. 394 251–267 10.1016/j.jmb.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Schaffitzel C., Bingel-Erlenmeyer R., Ban N., Korber P., Koning R. I., et al. (2009). Recycling of aborted ribosomal 50S subunit-nascent chain-tRNA complexes by the heat shock protein Hsp15. J. Mol. Biol. 386 1357–1367 10.1016/j.jmb.2008.10.079 [DOI] [PubMed] [Google Scholar]
- Karzai A. W., Sauer R. T. (2001). Protein factors associated with the SsrA-SmpB tagging and ribosome rescue complex. Proc. Natl. Acad. Sci. U.S.A. 98 3040–3044 10.1073/pnas.051628298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai A. W., Susskind M. M., Sauer R. T. (1999). SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18 3793–3799 10.1093/emboj/18.13.3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Gillet R., Li W., Gursky R., Frank J. (2006). Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc. Natl. Acad. Sci. U.S.A. 103 16484–16489 10.1073/pnas.0607438103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C. (2008). Biology of trans-translation. Annu. Rev. Microbiol. 62 133–151 10.1146/annurev.micro.62.081307.162948 [DOI] [PubMed] [Google Scholar]
- Keiler K. C., Shapiro L. (2003a). tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. J. Bacteriol. 185 1825–1830 10.1128/JB.185.6.1825-1830.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C., Shapiro L. (2003b). tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J. Bacteriol. 185 573–580 10.1128/JB.185.2.573-580.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C., Shapiro L., Williams K. P. (2000). tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobactor. Proc. Natl. Acad. Sci. U.S.A. 97 7778–7783 10.1073/pnas.97.14.7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K. C., Waller P. R., Sauer R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271 990–993 10.1126/science.271.5251.990 [DOI] [PubMed] [Google Scholar]
- Kogure H., Handa Y., Nagata M., Kanai N., Güntert P., Kubota K., et al. (2014). Identification of residues required for stalled-ribosome rescue in the codon-independent release factor YaeJ. Nucleic Acids Res. 10.1093/nar/gkt1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure H., Hikawa Y., Hagihara M., Tochio N., Koshiba S., Inoue Y., et al. (2012). Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65. Proteins 80 2629–2642 10.1002/prot.24152 [DOI] [PubMed] [Google Scholar]
- Komine Y., Kitabatake M., Inokuchi H. (1996). 10Sa RNA is associated with 70S ribosome particles in Escherichia coli. J. Biochem. 119 463–467 10.1093/oxfordjournals.jbchem.a021264 [DOI] [PubMed] [Google Scholar]
- Komine Y., Kitabatake M., Yokogawa T., Nishikawa K., Inokuchi H. (1994). A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 91 9223–9227 10.1073/pnas.91.20.9223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T., Kurita D., Takada K., Muto A., Himeno H. (2007). A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. RNA 13 1723–1731 10.1261/rna.604907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T., Takahashi T., Kurita D., Muto A., Himeno H. (2004). A minimum structure of aminoglycosides that causes an initiation shift of trans-translation. Nucleic Acids Res. 32 4119–4126 10.1093/nar/gkh750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber P., Stahl J. M., Nierhaus K. H., Bardwell J. C. (2000). Hsp15: a ribosome-associated heat shock protein. EMBO J. 19 741–748 10.1093/emboj/19.4.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita D., Muto A., Himeno H. (2010). Role of the C-terminal tail of SmpB in the early stage of trans-translation. RNA 16 980–990 10.1261/rna.1916610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita D., Muto A., Himeno H. (2012). In vitro trans-translation assay. Methods Mol. Biol. 905 311–325 10.1007/978-1-61779-949-5_20 [DOI] [PubMed] [Google Scholar]
- Kurita D., Sasaki R., Muto A., Himeno H. (2007). Interaction of SmpB with ribosome from directed hydroxyl radical probing. Nucleic Acids Res. 35 7248–7255 10.1093/nar/gkm677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K., Horiguchi N., Aiba H., Inada T. (2009). Analysis of nonstop mRNA translation in the absence of tmRNA in Escherichia coli. Genes Cells 14 739–749 10.1111/j.1365-2443.2009.01304.x [DOI] [PubMed] [Google Scholar]
- Lee S., Ishii M., Tadaki T., Muto A., Himeno H. (2001). Determinants on tmRNA for initiating efficient and precise trans-translation: some mutations upstream of the tag-encoding sequence of Escherichia coli tmRNA shift the initiation point of trans-translation in vitro. RNA 7 999–1012 10.1017/S1355838201010342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessner F. H., Venters B. J., Keiler K. C. (2007). Proteolytic adaptor for transfer-messenger RNA-tagged proteins from alpha-proteobacteria. J. Bacteriol. 189 272–275 10.1128/JB.01387-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yagi M., Morita T., Aiba H. (2008). Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Mol. Microbiol. 68 462–473 10.1111/j.1365-2958.2008.06167.x [DOI] [PubMed] [Google Scholar]
- Li X., Yokota T., Ito K., Nakamura Y., Aiba H. (2007). Reduced action of polypeptide release factors induces mRNA cleavage and tmRNA tagging at stop codons in Escherichia coli. Mol. Microbiol. 63 116–126 10.1111/j.1365-2958.2006.05498.x [DOI] [PubMed] [Google Scholar]
- Liang W., Deutscher M. P. (2010). A novel mechanism for ribonuclease regulation: transfer-messenger RNA (tmRNA) and its associated protein SmpB regulate the stability of RNase R. J. Biol. Chem. 285 29054–29058 10.1074/jbc.C110.168641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Malhotra A., Deutscher M. P. (2011). Acetylation regulates the stability of a bacterial protein: growth stage-dependent modification of RNase R. Mol. Cell 44 160–166 10.1016/j.molcel.2011.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Bhardwaj K., Sharkady S. M., Fish R. I., Driscoll T., Wower J., et al. (2009). Variations on the tmRNA gene. RNA Biol. 6 355–361 10.4161/rna.6.4.9172 [DOI] [PubMed] [Google Scholar]
- Mehta P., Richards J., Karzai A. W. (2006). tmRNA determinants required for facilitating nonstop mRNA decay. RNA 12 2187–2198 10.1261/rna.247706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. R., Buskirk A. R. (2014). An unusual mechanism for EF-Tu activation during tmRNA-mediated ribosome rescue. RNA 20 225–238 10.1261/rna.042226.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. R., Liu Z., Cazier D. J., Gebhard G. M., Herron S. R., Zaher H. S., et al. (2011). The role of SmpB and the ribosomal decoding center in licensing tmRNA entry into stalled ribosomes. RNA 17 1727–1736 10.1261/rna.2821711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munavar H., Zhou Y., Gottesman S. (2005). Analysis of the Escherichia coli Alp phenotype: heat shock induction in ssrA mutants. J. Bacteriol. 187 4739–4751 10.1128/JB.187.14.4739-4751.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Fujihara A., Ito K., Matsuno J., Ushida C., Himeno H. (2000). Requirement of transfer-messenger RNA (tmRNA) for the growth of Bacillus subtilis under stresses. Genes Cells 5 627–636 10.1046/j.1365-2443.2000.00356.x [DOI] [PubMed] [Google Scholar]
- Muto A., Sato M., Tadaki T., Fukushima M., Ushida C., Himeno H. (1996). Structure and function of bacterial 10Sa RNA: trans-translation system. Biochimie 78 985–991 10.1016/S0300-9084(97)86721-1 [DOI] [PubMed] [Google Scholar]
- Muto A., Ushida C., Himeno H. (1998). A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci. 23 25–29 10.1016/S0968-0004(97)01159-6 [DOI] [PubMed] [Google Scholar]
- Muto H., Nakatogawa H., Ito K. (2006). Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol. Cell 22 545–552 10.1016/j.molcel.2006.03.033 [DOI] [PubMed] [Google Scholar]
- Nameki N., Tadaki T., Muto A., Himeno H. (1999a). Amino acid acceptor identity switch of Escherichia coli tmRNA from alanine to histidine in vitro. J. Mol. Biol. 289 1–7 10.1006/jmbi.1999.2754 [DOI] [PubMed] [Google Scholar]
- Nameki N., Felden B., Atkins J. F., Gesteland R. F., Himeno H., Muto A. (1999b). Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E. coli tmRNA. J. Mol. Biol. 286 733–744 10.1006/jmbi.1998.2487 [DOI] [PubMed] [Google Scholar]
- Nameki N., Chattopadhyay P., Himeno H., Muto A., Kawai G. (1999c). An NMR and mutational analysis of an RNA pseudoknot of E. coli tmRNA involved in trans-translation. Nucleic Acids Res. 27 3667–3675 10.1093/nar/27.18.3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameki N., Someya T., Okano S., Suemasa R., Kimoto M., Hanawa-Suetsugu K., et al. (2005). Interaction analysis between tmRNA and SmpB from Thermus thermophilus. J. Biochem. 138 729–739 10.1093/jb/mvi180 [DOI] [PubMed] [Google Scholar]
- Nameki N., Tadaki T., Himeno H., Muto A. (2000). Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett. 470 345–349 10.1016/S0014-5793(00)01349-1 [DOI] [PubMed] [Google Scholar]
- Neubauer C., Gillet R., Kelly A. C., Ramakrishnan V. (2012). Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 335 1366–1369 10.1126/science.1217039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonin-Lecomte S., Germain-Amiot N., Gillet R., Hallier M., Ponchon L., Dardel F., et al. (2009). Ribosome hijacking: a role for small protein B during trans-translation. EMBO Rep. 10 160–165 10.1038/embor.2008.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B. K., Apirion D. (1991). 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Mol. Gen. Genet. 25 737–749 [DOI] [PubMed] [Google Scholar]
- Okan N. A., Bliska J. B., Karzai A. W. (2006). A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2:e6. 10.1371/journal.ppat.0020006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Kutsukake K., Abo T. (2009). Suppression by enhanced RpoE activity of the temperature-sensitive phenotype of a degP ssrA double mutant in Escherichia coli. Genes Genet. Syst. 84 15–24 10.1266/ggs.84.15 [DOI] [PubMed] [Google Scholar]
- Oussenko I. A., Abe T., Ujiie H., Muto A., Bechhofer D. H. (2005). Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 187 2758–2767 10.1128/JB.187.8.2758-2767.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecková P., Felsberg J., Bobek J, Mikulík K. (2007). tmRNA abundance in Streptomyces aureofaciens, S. griseus and S. collinus under stress-inducing conditions. Folia Microbiol. 52 463–470 10.1007/BF02932105 [DOI] [PubMed] [Google Scholar]
- Pech M., Karim Z., Yamamoto H., Kitakawa M., Qin Y., Nierhaus K. H. (2011). Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc. Natl. Acad. Sci. U.S.A. 108 3199–3203 10.1073/pnas.1012994108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., Ehrenberg M. (2003). The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112 131–140 10.1016/S0092-8674(02)01248-5 [DOI] [PubMed] [Google Scholar]
- Pisareva V. P., Skabkin M. A., Hellen C. U., Pestova T. V., Pisarev A. V. (2011). Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30 1804–1817 10.1038/emboj.2011.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Shimizu Y., Ueda T. (2007). Ribosomal protein S1 is not essential for the trans-translation machinery. J. Mol. Biol. 368 845–852 10.1016/j.jmb.2007.02.068 [DOI] [PubMed] [Google Scholar]
- Ramadoss N. S., Zhou X., Keiler K. C. (2013a). tmRNA is essential in Shigella flexneri. PLoS ONE 8:e57537 10.1371/journal.pone.0057537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadoss N. S., Alumasa J. N., Cheng L., Wang Y., Li S., Chambers B. S., et al. (2013b). Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proc. Natl. Acad. Sci. U.S.A. 110 10282–10287 10.1073/pnas.1302816110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath D. J., Yamamoto H., Rother K., Wittek D., Pech M., Mielke T., et al. (2012). The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature 485 526–529 10.1038/nature11006 [DOI] [PubMed] [Google Scholar]
- Ranquet C., Gottesman S. (2007). Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. J. Bacteriol. 189 4872–4879 10.1128/JB.01838-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezzonico E., Lariani S., Barretto C., Cuanoud G., Giliberti G., Delley M., et al. (2007). Global transcriptome analysis of the heat shock response of Bifidobacterium longum. FEMS Microbiol. Lett. 271 136–145 10.1111/j.1574-6968.2007.00704.x [DOI] [PubMed] [Google Scholar]
- Richards J., Mehta P., Karzai A. W. (2006). RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol. 62 1700–1712 10.1111/j.1365-2958.2006.05472.x [DOI] [PubMed] [Google Scholar]
- Richter R., Rorbach J., Pajak A., Smith P. M., Wessels H. J., Huynen M. A., et al. (2010). A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 29 1116–1125 10.1038/emboj.2010.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche E. D., Sauer R. T. (1999). SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 18 4579–4589 10.1093/emboj/18.16.4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche E. D., Sauer R. T. (2001). Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J. Biol. Chem. 276 28509–28515 10.1074/jbc.M103864200 [DOI] [PubMed] [Google Scholar]
- Rudinger-Thirion J., Giegé R., Felden B. (1999). Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA 5 989–992 10.1017/S135583829999101X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkady S. M., Williams K. P. (2004). A third lineage with two-piece tmRNA. Nucleic Acids Res. 32 1–8 10.1093/nar/gkh795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Zhang X., Jiang X., Yuan H., Lee J. S., Barry C. E., III, et al. (2011). Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 333 1630–1632 10.1126/science.1208813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y. (2012). ArfA recruits RF2 into stalled ribosomes. J. Mol. Biol. 423 624–631 10.1016/j.jmb.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Shimizu Y., Ueda T. (2002). The role of SmpB protein in trans-translation. FEBS Lett. 514 74–77 10.1016/S0014-5793(02)02333-5 [DOI] [PubMed] [Google Scholar]
- Shin J.-H., Price C. W. (2007). The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperature. J. Bacteriol. 189 3729–3737 10.1128/JB.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. S., Ahmad R., Sangeetha R., Varshney U. (2008). Recycling of ribosomal complexes stalled at the step of elongation in Escherichia coli. J. Mol. Biol. 380 451–464 10.1016/j.jmb.2008.05.033 [DOI] [PubMed] [Google Scholar]
- Singh N. S., Varshney U. (2004). A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Res. 32 6028–6037 10.1093/nar/gkh924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C. J., Eyler D. E., Green R. (2010). Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330 369–372 10.1126/science.1192430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A., Onodera A., Sugahara J., Kanai A., Yachie N., Tomita M., et al. (2007). Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 19 450–453 10.1126/science.1145718 [DOI] [PubMed] [Google Scholar]
- Someya T., Nameki N., Hosoi H., Suzuki S., Hatanaka H., Fujii M., et al. (2003). Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. FEBS Lett. 535 94–100 10.1016/S0014-5793(02)03880-2 [DOI] [PubMed] [Google Scholar]
- Sunohara T., Jojima K., Yamamoto Y., Inada T., Aiba H. (2002). Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA 10 378–386 10.1261/rna.5169404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara T., Jojima K., Yamamoto Y., Inada T., Aiba H. (2004a). Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA 10 378–386 10.1261/rna.5169404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara T., Jojima K., Tagami H., Inada T., Aiba H. (2004b). Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 279 15368–15375 10.1074/jbc.M312805200 [DOI] [PubMed] [Google Scholar]
- Tadaki T., Fukushima M., Ushida C., Himeno H., Muto A. (1996). Interaction of 10Sa RNA with ribosomes in Escherichia coli. FEBS Lett. 399 223–226 10.1016/S0014-5793(96)01330-0 [DOI] [PubMed] [Google Scholar]
- Takada K., Takemoto C., Kawazoe M., Konno T., Hanawa-Suetsugu K., Lee S., et al. (2007). In vitro trans-translation of Thermus thermophilus: ribosomal protein S1 is not required for the early stage of trans-translation. RNA 13 503–510 10.1261/rna.363207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Konno T., Muto A., Himeno H. (2003). Various effects of paromomycin on tmRNA-directed trans-translation. J. Biol. Chem. 278 27672–27680 10.1074/jbc.M211724200 [DOI] [PubMed] [Google Scholar]
- Thibonnier M., Thiberge J. M, De Reuse H. (2008). Trans-translation in Helicobacter pylori: essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS ONE 3:e3810 10.1371/journal.pone.0003810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu G. F., Reid G. E., Zhang J. G., Moritz R. L., Simpson R. J. (1995). C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem. 270 9322–9326 10.1074/jbc.270.16.9322 [DOI] [PubMed] [Google Scholar]
- Ude S., Lassak J., Starosta A. L., Kraxenberger T., Wilson D. N., Jung K. (2013). Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339 82–85 10.1126/science.1228985 [DOI] [PubMed] [Google Scholar]
- Ueda K., Yamamoto Y., Ogawa K., Abo T., Inokuchi H., Aiba H. (2002). Bacterial SsrA system plays a role in coping with unwanted translational readthrough caused by suppressor tRNAs. Genes Cells 7 509–519 10.1046/j.1365-2443.2002.00537.x [DOI] [PubMed] [Google Scholar]
- Ujiie H., Matsutani T., Tomatsu H., Fujihara A., Ushida C., Miwa Y., et al. (2009). Trans-translation is involved in the CcpA-dependent tagging and degradation of TreP in Bacillus subtilis. J. Biochem. 145 59–66 10.1093/jb/mvn143 [DOI] [PubMed] [Google Scholar]
- Ushida C., Himeno H., Watanabe T., Muto A. (1994). tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res. 22 3392–3396 10.1093/nar/22.16.3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis F., Bron P., Rolland J. P., Thomas D., Felden B., Gillet R. (2010a). Accommodation of tmRNA-SmpB into stalled ribosomes: a cryo-EM study. RNA 16 299–306 10.1261/rna.1757410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis F., Bron P., Giudice E., Rolland J. P., Thomas D., Felden B., et al. (2010b). tmRNA-SmpB: a journey to the centre of the bacterial ribosome. EMBO J. 29 3810–3818 10.1038/emboj.2010.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. P. (2002). Descent of a split RNA. Nucleic Acids Res. 30 2025–2030 10.1093/nar/30.9.2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. P., Martindale K. A., Bartel D. P. (1999). Resuming translation on tmRNA: a unique mode of determining a reading frame. EMBO J. 18 5423–5433 10.1093/emboj/18.19.5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower I. K., Zwieb C., Wower J. (2004). Contributions of pseudoknots and protein SmpB to the structure and function of tmRNA in trans-translation. J. Biol. Chem. 279 54202–54209 10.1074/jbc.M410488200 [DOI] [PubMed] [Google Scholar]
- Wower I. K., Zwieb C. W., Guven S. A., Wower J. (2000). Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J. 19 6612–6621 10.1093/emboj/19.23.6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Sunohara T., Jojima K., Inada T., Aiba H. (2003). SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA 9 408–418 10.1261/rna.2174803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. (2007). RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA 13 1141–1154 10.1261/rna.620507 [DOI] [PMC free article] [PubMed] [Google Scholar]