Abstract
Objective
To analyse molecular detection of coliforms and shorten the time of PCR.
Methods
Rapid detection of coliforms by amplification of lacZ and uidA genes in a multiplex PCR reaction was designed and performed in comparison with most probably number (MPN) method for 16 artificial and 101 field samples. The molecular method was also conducted on isolated coliforms from positive MPN samples; standard sample for verification of microbial method certificated reference material; isolated strains from certificated reference material and standard bacteria. The PCR and electrophoresis parameters were changed for reducing the operation time.
Results
Results of PCR for lacZ and uidA genes were similar in all of standard, operational and artificial samples and showed the 876 bp and 147 bp bands of lacZ and uidA genes by multiplex PCR. PCR results were confirmed by MPN culture method by sensitivity 86% (95% CI: 0.71-0.93). Also the total execution time, with a successful change of factors, was reduced to less than two and a half hour.
Conclusions
Multiplex PCR method with shortened operation time was used for the simultaneous detection of total coliforms and Escherichia coli in distribution system of Arak city. It's recommended to be used at least as an initial screening test, and then the positive samples could be randomly tested by MPN.
Keywords: Water, Coliforms, PCR, Rapid detection
1. Introduction
Drinking water should be free from known pathogenic micro-organisms and indicator bacteria which is a symptom for fecal contamination of water. Coliforms are the most important indicator bacteria which are considered in the bacteriological examination of water[1]. Determination of coliform as an indicator of water contamination has been explained in 1011 standard (microbiological characteristics of water) and the use of Escherichia coli (E. coli) and other coliform bacteria and those coliforms that are indicator bacteria has been recommended for the daily control of drinking water. Coliform bacteria are used for monitoring the bacteriological safety of water supplies on the basis of the realization that the presence of coliform bacteria in water is an indicator of potential human fecal contamination and therefore the possible presence of enteric pathogens[2].
In general, this standard focuses on three groups of bacteria including coliform, thermotolerant coliform and E. coli[3]. E. coli is used as an indicator of fecal contamination of water.
Detection of indicator bacteria is one of the best ways to evaluate the effectiveness of water disinfection methods. The most important indicator bacteria, in terms of their importance, include E. coli, coliforms and other thermotolerant coliforms. The presence of these bacteria in the water is an indicator of insufficient disinfection process and also recent and frequent contamination of water with human and animal feces[4]. Thermo tolerant coliforms, except E. coli, can enter the drinking water through water contaminated by industrial wastewaters and under deterioration soil and water. Conventional methods for the detection of microbial contamination of water include methods which are based on culture of water sample and diagnosis of β-galactosidase (using ortho-nitrophenyl-β-D-galactopyranoside) which is cumbersome, expensive and often with personnel error in routine use[5].
Culture-based methods have limitations such as long incubation period (48 to 72 h), possibility of contamination with other bacteria, limited ability to detect low-growing bacteria and the inability to detect “viable but non-cultivable bacteria” (VBNC) such as those stressed by chemicals in the water which has recently been considered in Iran and lack of specificity for detection of true fecal coliforms[3].
PCR has been recommended as a specific and reliable method for the detection of coliforms in drinking water[6]. PCR-based molecular methods have recently been considered due to their high sensitivity and specificity, quickly achieving to the relevant responses and reducing the workload of experts, especially in those centers with a high number of samples per day. These methods have great features due to the ability of rapid detection and being highly specific. In these methods, DNA of target bacteria is searched using specific primers. In PCR, amplification of lacZ gene (β-galactosidase gene) to detect total coliforms and uidA gene (β-glucuronidase gene) to detect E. coli are specifically used[3]. Other genes such as dct A, dcuB, frdA, dcuS and dcuR are also provided for the precise and rapid study of E. coli[7],[8].
2. Materials and methods
2.1. Samples
Two types of samples were used in this study. Samples for evaluation of the method and city samples consist of distribution network supply wells. About 14 samples were used as control.
2.1.1. Samples for evaluation of the method
a-1) E. coli DH5α strain was used to evaluate the study conducted for the detection of E. coli using specific primers.
a-2) CRM04 and CRM07: CRM04 (QCMIC-001-04) contains 83.00±CFU/100 mL of coliform bacteria except E. coli, and CRM007 (QCMIC-007) contains (100.00±2.00) CFU/mL of coliform bacteria and E. coli.
a-3) Synthetic samples: 10 samples containing isolated bacteria in 250 mL distilled water.
a-4) Isolated bacteria from contaminated water: Three bacterial strains were isolated from positive most probable number (MPN) samples and then purified. They were identified using microbiological techniques (up to the level of identification of species). Therefore, mediums such as nutrient agar, EMB, IMVIC phenylalanine and urea were used in addition to Gram stain.
2.1.2. Composition of field samples
b-1) Samples of city water distribution network: 36 samples were collected from Arak city water.
b-2) Wells that supply drinking water to the city of Arak: 41 samples were collected from Arak city drinking water.
b-3) Control samples: 24 purified colonies as positive control and distilled water as negative control.
2.2. Sample preparation
In this analysis, 150 mL samples were passed through a filter with a diameter of 0.42 micron by the help of a vacuum pump. To avoid possible contamination, it was conducted as class 2 laminar flow.
Separation of microbes from the filter paper was carried out in two ways:
A - Washing with distilled water.
B - Washing with diethylpyrocarbonate (DEPC).
Filter paper was placed in 50 cc Iso glass and about 10 cc of 0.1% solution of autoclaved DEPC was added to the Iso glass. Filter was properly washed three times with the help of sampler in the Iso glass wall through immersion and extreme turbulence. Then the solution resulted from washing were transferred into centrifuge Falcon tubes. Then, the solution was centrifuged for 10 min with 14 000 r/min.
Supernatant was removed carefully with a sampler and about 10 cc of autoclaved saline was added to the deposits. Then it was centrifuged after application of vortex and turbulence. Thus, DEPC was washed from the deposits. About 300 micro liters of the final deposits were transferred to a microtube 1.5 and finally about 50 micro liters of the final deposit was transferred as a bacterial mass to a microtube 1.5, after it was centrifuged at 14 000 r/min for 2 min and at the next stage, it was analyzed by PCR test. Moreover, a number of samples were comparatively tested with or without DEPC to evaluate the necessity of its application.
From 250 mL sample in the sample container, 100 cc was used for doing MPN test, about 30 cc was used for doing turbidity test and the rest was used for doing molecular filtrations.
In this study, three types of samples were studied:
1 - Wells and water distribution system samples: MPN culture test were first performed as the gold standard tests, then the samples were filtered and analyzed by PCR test.
2 - Standard bacteria: E. coli and CRM.
3 - Synthetic samples: In order to evaluate PCR, synthetic samples were used as positive examples. These samples were positive BGB (their microbial contamination with coliform was confirmed) and they are kept and expanded for setting up PCR. After isolation of bacteria, purified bacteria were added to the negative MPN water sample. In this study, all water samples sent to the laboratory culture were kept in a refrigerator for 24 h after being cultured in a lauryl tryptose broth for doing MPN.
2.3. PCR method
In this study, culture and PCR methods were compared to detect total coliform (Citrobacter, Klebsiella, E. coli and Enterobacter) in water samples. Blast survey indicated that the LacZ gene as the β-galactosidase gene encoding is found in all these bacteria and they are relatively synonymous. Blast revealed that the gene existed in mentioned coliform bacteria.
Furthermore, uidA gene which is a coliform-specific gene can specifically be used to prove the presence of E. coli in water samples. Therefore, forward and reverse primers of uidA. (UAL: TGGTAATTACCGACGAAAACGG, UAR: ACGCGTGGTTACAGTCTTGCG) and lacZ (LZL: ATGAAAGCTGGCTACAGGAAGGCC, LZR: CACCATGCCGTGGGTTTCAATATT)
Genes were selected to detect the bacteria in water samples[3]. As can be seen in MEGA 4.0 software an 874-bp fragment was selected from nucleotides 392 to 1266 of lacZ gene by specified primers.
In order to perform PCR test, methods of isolating DNA from purified bacteria and filtered water samples were first used in this study. Then, with the aim of accelerating and simplifying the test procedure in the routine implementation, mass PCR was performed. In mass PCR method, cell mass obtained from water samples was used directly after filtration and concentration. Final volume of PCR was considered as 25 mL, containing 20 mL of all PCR components together with 5 mL of microbial biomass.
To avoid any errors, PCR reaction for pure colonies of isolated bacteria were conducted in the same method. Colonies dissolved in 250 mL distilled water, and filtrated and concentrated biomass used in PCR reaction.
2.4. Multiplex PCR
In this study, samples identification through detection of lacZ and uidA genes using PCR simultaneously (multiplex PCR) was approved. Primers listed in Table 1 are used to amplify 876 bp and 147 bp fragments of lacZ and uidA genes, respectively. Amplification reaction in a final volume of 25 mL contains 5 mL of diluted bacteria or purified DNA; and 2.5 micro liters buffer containing 10 Xmg, 1 mL dNTPs, 1.5 units of Taq enzyme and 2 mL of both primers were added to it. The PCR reaction steps were as follows.
Table 1. Corrected programs of PCR.
| Corrected program | PCR program | Target gene | Bacteria |
| 94-40 s | 95-10 min | lacZ | Coliform bacteria |
| 94-1 min | 55-1 min | ||
| 55-40 s | 72-1 min | ||
| 72-30 s | 72-2 min | ||
| Cycles 23 | Cycle 25 | ||
| 94-30 s | 95-10 min | uidA | E. coli |
| 94-45 s | |||
| 55-30 s | 62-45 s | ||
| 72-45 s | |||
| 72-30 s | 72-2 min | ||
| Cycles 23 | Cycles 30 |
Initial denaturating temperature: 94 °C for 9 min and then 23 thermal cycles for amplification in PCR reaction as: 94 °C for 1 min, 55 °C for 1 min and 72 °C for 50 s and final amplification cycle (extension) at 72 °C for 2 min. Corrected programs of PCR are shown in Table 1.
3. Results
In this study, DNA isolation step was removed to reduce the steps of PRC, accelerate the reaction, and most importantly to reduce personnel error; and then colony PCR method was performed successfully.
3.1. Standard strains
A) PCR performed and presented 876 bp bands in electrophoresis that shows the correct designing of this study.
B) CRMs: PCR performed on CRM 04 for searching lacZ gene showed a 876 bp band which in turn shows the presence of coliform (except E. coli bacteria) and the correct design of this study.
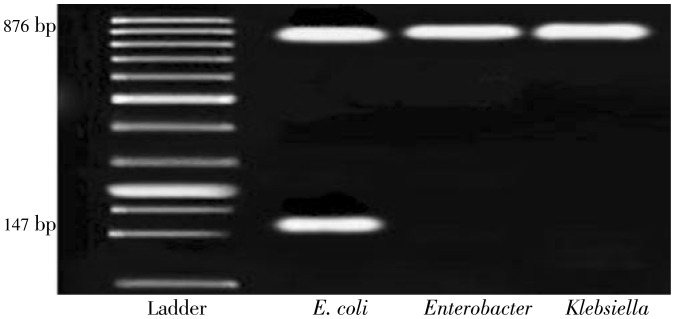
Moreover, 147 bp band obtained from PCR is used to study and indicate the existence of uidA gene and also the presence of E. coli bacteria in CRM07. Also, microbial analysis of CRM007 CRM 04 identified bacteria from the Coliform and E. coli family, the pure culture of which was used for doing PCR. Performing PCR on all the bacteria separately, revealed 876 bp and 147 bp bands which demonstrates the applicability of the proposed method for the detection of drinking water contamination (Figure 1). Bacteria that cause water contamination which are the members of total coliform group were purified and isolated from samples of contaminated drinking water and samples with positive MPN; and detection tests were led to the identification of four bacterial species of E. coli, Klebsiella, pneumonia, Enterobacter and Pseudomonas aeruginosa.
Figure 1. PCR and multiplex PCR amplification of LacZ and gene uidA.
Lane 2-multiplex PCR amplification of LacZ and uidA gene from contaminated water with coliformm bacteria (E.coli); Lane 1 ladder, lane 3-4 PCR LacZ amplification from contaminated water with coliformm bacteria (Enterobacter and Klebsiella ).
Results of PCR for lacZ and uidA genes on these samples confirmed the occurrence of 876 bp and 147 bp bands.
3.2. Results of evaluating the water samples
The study was conducted on 77 samples of waters. Of the samples sent to the laboratory, 41 samples were taken from the wells, 36 samples were taken from Arak city water. About 12 samples were positive control samples and 12 samples were also considered as negative control. Results of performing MPN and PCR on all the samples are shown in Table 2 and Figure 1.
Table 2. Results of MPN and PCR microbial tests on samples taken from different parts of drinking water production and distribution networks of Arak city in the winter of 2011.
| Methods | Quantity | MPN |
PCR |
||
| Positive | Negative | Positive | Negative | ||
| Samples of city water distribution network | 36 | 0 | 36 | 5 | 31 |
| Wells that supply drinking water to the city of Arak | 41 | 0 | 41 | 0 | 41 |
| Control samples | 24 | Positive control samples: 12 | |||
| Negative control samples: 12 | |||||
PCR: Polymerase chain reaction; MPN: Most probable number of coliform.
As it is obvious in Table 2, of the total samples sent to the laboratory, PCR results of 5 samples were positive, whereas the MPN results have been reported as negative. The results of doing PCR and MPN tests on other samples of the water system were similar with each other and both tests were negative.
About the water samples taken from the wells sent to the laboratory, following results were reported. Of 41 taken samples, two samples had positive results in both the MPN and PCR tests and about the rest of the sample taken from wells, the results of both tests were reported negative which indicates the compatibility of the both tests.
4. Discussion
In this study, the lacZ gene was successfully used as a target molecule for the detection of coliform group bacteria. The indicator of the existence of total coliform in the samples was determined after observing 876 bp gene fragment. Moreover, the uidA gene which is specifically found in E. coli 876 bp (among coliforms) was used as a target molecule for the detection of E. coli bacteria. Then a gene fragment around 174 bp was obtained using designed primers[9].
Molecular techniques are accurate, rapid and sensitive methods for the study of specific pathogenic bacteria. These tools can be used to accurately analyze the drinking water performance of the elimination of pathogens in drinking water and treatment of water used for drinking[10].
Culture methods for the detection of coliforms have limitations such as long incubation period, interactions with other microorganisms, lack of precision and required sensitivity and poor identification of VBNC bacteria. As an accurate and rapid method for the detection of coliforms, molecular methods have been proposed[11].
PCR can detect coliform bacteria using lacZ gene (gene β-galacotozidase) and E. coli bacteria using uidA gene (β-glucuronidase gene)[12]. In the present study, common ways to identify coliforms are compared with the new molecular methods and the sensitivity and precision of these methods is evaluated in practical application. In this study, different items such as molecular method for reducing the volume of routine operations in the Iran's water bacteriology laboratory, reduction in the costs of samples analysis, increase in the accuracy and possible coverage of VBNC bacteria in the water by molecular methods are presented. Plan targeting is based on PCR method in the initial screening of all samples accepted per day. As high percentage of sample accepted each day have negative test results, initial screening by PCR, removal of samples having negative test results and focus of tests on positive samples cause reduction in the consumption of a high volume of medium and involvement of the expert in the creation and removal of media[13].
To prove the correctness of the performance of the two lacZ and uidA genes, at first standard strains and then reference strain (CRM) were studied and the occurrence of bands in electrophoresis was confirmed. The MPN was used as the gold standard to minimize any error in interpretation of results. To further confirm the results, bacteria isolated in MPN test were identified up to the extent of species and then purified and identified bacteria were successfully undergone molecular evaluation with the help of these two genes. Given that the entire study was planned with the aim of making routine tests in order to perform operation in the Iran's water microbiology laboratories, the results of this study prove the operability of the usage of molecular methods in routine applications for the microbial assessment of drinking water[14].
Filtration and sample preparation method was designed based on samples accepted per day. Of the advantages of PCR system, sensitivity [detection of target bacteria in minimum concentration, specificity of this method for target microorganisms, rate of doing test from the time of sample collection until analysis completion (less than 4 h)], and the ability to simultaneously detect multiple bacterial (including general specific species and a number of target specific pathogens) can be mentioned. With regard to the emerging problem of living bacteria but not being able to be cultured (VBNC) and their role in public health, the results of this study indicated that the provided molecular methods may also be able to detect VBNC bacteria[10]. On the other hand, from the perspective of economic justification, according to the conducted studies, costs of culture methods are higher than the costs of PCR method.
In the PCR method, preparation of the medium for culturing was removed and the device depreciation costs were reduced. Time spent by experts and subsequently personnel error are also minimized. The mentioned PCR method is able to cover three steps of MPN test in the shortest time and with high sensitivity (detecting total coliforms, fecal coliforms and E. coli)[11],[15]. But, in the first operational stage, PCR can be used for the primer screening of the accepted water samples per day. Samples identified as having positive test results by PCR within a few hours, are entered into the stage of cultivation and counting. In this project, the execution time of PRC was reduced with the aim of reducing the operation time. For this reason, the program of amplification was changed with the time reduction so that the replication time fell from 2 h and 15 min to an hour.
Bands resulted from this method have such sizes that remove the need for the use of polyacrylamide for electrophoresis. Therefore, with the increase in the voltage during the use of agarose gel, electrophoresis time is also decreased. When using in vitro cultivation routine methods, it is probable that in the most cases the PCR test shows false negative results due to the low number of pathogenic bacteria in water (less than detectable by culture) or the loss of chromosomes of bacteria at different stages of purification, in addition to being energy consuming of routine methods of in vitro cultivation[16]. In molecular methods, detection scope is much less than medium and bacteria are detected even in small number. On the other hand, the only existing way to detect VBNC is molecular methods. In this study, with the aim of reducing the false negative responses, samples filtration was used based on the method proposed by WHO[6],[17].
Also simultaneous detection of total coliform bacteria for E. coli in drinking water has been carried out in this study using lacZ and uidA genes. Detection using agarose gel electrophoresis has created a band of 876 bp for the lacZ gene for the all of coliform bacteria and a band of 147 bp for uidA gene and a band of 876 bp for lacZ gene for all species of E. coli[10],[18].
In this study, rapid method for the simultaneous detection of total coliforms and E. coli in drinking water was designed using PCR method and tested in the routine work. Validity of the results and concurrent comparison between them and conventional method of cultivation, application of methods on standard samples, bacteria isolated from positive MPN and CRM samples in different wells, water reservoir and system were evaluated and confirmed. Given the volume of water incoming to wastewater laboratories, it was tried to reduce the test execution time to a possible minimum level. Thus, given the particular sensitivity of the public health, multiplex PCR method can be used at least as an initial screening test. Thus, the samples showing positive results in this test can be randomly tested for MPN.
Acknowledgments
The authors would particularly like to thank all colleagues at Tuberculosis and Pediatric Infectious Research Center (TPI) of Arak University of Medical Sciences for their assistance. Supported by the Ministry of Power of I.R.Iran. Grant No. 201.
Comments
Background
With the increasing of world population and it's thickness in built-up area, drinking water is becoming a resource most valuable. Outbreaks, caused by infected water, can be a serious problem of health public. To reduce risk of infection, it is important to use reliable, inexpensive, rapid and robust protocols to analyze a lot of samples in little time.
Research frontiers
This paper evaluates a method for rapid analysis of drinking water for human use. To be precise, the samples came from distribution network supply wells of the city of Arak. The results obtained by multiplex PCR are compared with standard cultural methods. The study underlined the detection of sentinel microorganisms that may indicate the presence of faecal contamination in water sources.
Related reports
This work is interesting for rapidity of assay and for the possible epidemiological implication. It does not contrast with other reports, e.g. Kuo et al. (2010) and Hiraka et al. (2009). It should be useful to know the detection limits of these target genes. These values are depending by selected primers.
Innovations and breakthroughs
The present study has shown that a accurate monitoring should not only be based on standard cultural method but it should be implemented with other tests. Rapid laboratory investigations tools are indispensable for an accurate monitoring, so assay proposed is very interesting.
Applications
The maintenance of sewer and water networks in cities is very expensive. Moreover, not all urban centers have sewage systems. This creates problems, which can be monitored and programmed only in a systemic way. Using methods more sensitive and rapid should be of help in these circumstances.
Peer review
The authors scrupulously highlight any potential of molecular biology techniques and they try to focus the limits on the standard cultural methods. For example, long incubation period, limited ability to detect low-growing bacteria and the inability to detect “viable but non-cultivable bacteria” etc. In the whole the study is interesting.
Footnotes
Foundation Project: Supported by the Ministry of Power of I.R.Iran (Grant No. 201).
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.World Health Organization . Guidelines for drinking-water quality, fourth edition. Geneva: World Health Organization; 2011. [Online] Available from: http://www.who.int/water_sanitation_health/publications/2011/dwq_guidelines/en/. [Accessed on 21st November, 2013] [Google Scholar]
- 2.Bej AK, Steffan RJ, DiCesare J, Haff L, Atlas RM. Detection of coliform bacteria in water by polymerase chain reaction and gene probes. Appl Environ Microbiol. 1990;56(2):307–314. doi: 10.1128/aem.56.2.307-314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tantawiwat S, Tansuphasiri U, Wongwit W, Wongchotigul V, Kitayaporn D. Development of multiplex PCR for the detection of total coliform bacteria for Escherichia coli and Clostridium perfringens in drinking water. Southeast Asian J Trop Med Public Health. 2005;36(1):162–169. [PubMed] [Google Scholar]
- 4.Rodríguez DC, Pino N, Peñuela G. Microbiological quality indicators in waters of dairy farms: detection of pathogens by PCR in real time. Sci Total Environ. 2012;427-428:314–318. doi: 10.1016/j.scitotenv.2012.03.052. [DOI] [PubMed] [Google Scholar]
- 5.Tharannum S, Sunitha S, Nithya J, Chandini M, Vanitha J, Manjula TS, et al. Molecular confirmation of the presence of coliforms in drinking water using polymerase chain reaction. Kathmandu Univ J Sci Eng Technol. 2009;5(2):130–136. [Google Scholar]
- 6.Clifford RJ, Milillo M, Prestwood J, Quintero R, Zurawski DV, Kwak YI, et al. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One. 2012;7(11):e48558. doi: 10.1371/journal.pone.0048558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abo-Amer AE, Soltan el-SM, Abu-Gharbia MA. Molecular approach and bacterial quality of drinking water of urban and rural communities in Egypt. Acta Microbiol Immunol Hung. 2008;55(3):311–326. doi: 10.1556/AMicr.55.2008.3.3. [DOI] [PubMed] [Google Scholar]
- 8.Mannapperuma WM, Abayasekara CL, Herath GB, Werellagama DR, Tanski HH. Comparison of bacteriological methods for detecting and enumerating total coliforms and Escherichia coli in water. Res J Microbiol. 2011;6:851–861. [Google Scholar]
- 9.Girones R, Ferrús MA, Alonso JL, Rodriguez-Manzano J, Calgua B, Corrêa Ade A, et al. Molecular detection of pathogens in water--the pros and cons of molecular techniques. Water Res. 2010;44(15):4325–4339. doi: 10.1016/j.watres.2010.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Chung JW, Foppen JW, Lens PN. Development of low cost two-step reverse transcription-quantitative polymerase chain reaction assays for rotavirus detection in foul surface water drains. Food Environ Virol. 2013;5(2):126–133. doi: 10.1007/s12560-013-9111-7. [DOI] [PubMed] [Google Scholar]
- 11.Park SH, Hanning I, Jarquin R, Moore P, Donoghue DJ, Donoghue AM, et al. Multiplex PCR assay for the detection and quantification of Campylobacter spp., Escherichia coli O157:H7, and Salmonella serotypes in water samples. FEMS Microbiol Lett. 2011;316(1):7–15. doi: 10.1111/j.1574-6968.2010.02188.x. [DOI] [PubMed] [Google Scholar]
- 12.EL-Jakee J, Moussa EI, Mohamed KF, Mohamed G. Using molecular techniques for characterization of Escherichia coli isolated from water sources in Egypt. Global Veterinaria. 2009;3(5):354–362. [Google Scholar]
- 13.Fode-Vaughan KA, Maki JS, Benson JA, Collins ML. Direct PCR detection of Escherichia coli O157:H7. Lett Appl Microbiol. 2003;37(3):239–243. doi: 10.1046/j.1472-765x.2003.01386.x. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization . Guidelines for drinking-water quality. Geneva: World Health Organization; 2010. [Online] Available from: http://www.who.int/water_sanitation_health/dwq/guidelines/en/. [Accessed on 21st December, 2013] [Google Scholar]
- 15.Abtahi H, Ghannadzadeh M, Salmanian AH, Rad EG, Karimi M, Molaei N. Improvement of PCR in detection of coliform in water pollution. J Arak Univ Med Sci. 2008;11(3):1–7. [Google Scholar]
- 16.Heijnen L, Medema G. Quantitative detection of E. coli, E. coli O157 and other shiga toxin producing E. coli in water samples using a culture method combined with real-time PCR. J Water Health. 2006;4(4):487–498. [PubMed] [Google Scholar]
- 17.Kuo JT, Cheng CY, Huang HH, Tsao CF, Chung YC. A rapid method for the detection of representative coliforms in water samples: polymerase chain reaction-enzyme-linked immunosorbent assay (PCR-ELISA) J Ind Microbiol Biotechnol. 2010;37(3):237–244. doi: 10.1007/s10295-009-0666-0. [DOI] [PubMed] [Google Scholar]
- 18.Hidaka A, Hokyo T, Arikawa K, Fujihara S, Ogasawara J, Hase A, et al. Multiplex real-time PCR for exhaustive detection of diarrhoeagenic Escherichia coli. J Appl Microbiol. 2009;106(2):410–420. doi: 10.1111/j.1365-2672.2008.04043.x. [DOI] [PubMed] [Google Scholar]