Abstract
Pancreatic cancer remains the most devastating disease with worst prognosis. There is a pressing need to accelerate the drug discovery process to identify new effective drug candidates against pancreatic cancer. We have developed QSAR models for predicting promiscuous inhibitors using the pharmacological data. Our models achieved maximum Pearson correlation coefficient of 0.86, when evaluated on 10-fold cross-validation. Our models have also successfully validated the drug-to-oncogene relationship and further we used these models to screen FDA approved drugs and tested them in vitro. We have integrated these models in a webserver named as DiPCell, which will be useful for screening and designing novel promiscuous drug molecules. We have also identified the most and least effective drugs for pancreatic cancer cell lines. On the other side, we have identified resistant pancreatic cancer cell lines, which need investigative scanner on them to put light on resistant mechanism in pancreatic cancer.
Pancreatic cancer (PCa) remains the fourth leading cause of cancer-related deaths1, and it is one of the most lethal malignant tumors. An overall 5-year survival rate of less than 6% and 45,220 new cases were reported in United States alone1. The mortality rate of PCa is almost equal to the incidence rate, which demonstrates the aggressiveness and lethal nature of this disease. The contributing factors for this high mortality rate are lack of screening tests for early diagnosis and development of drug resistance in tumor cells2,3,4,5. Over the years, considerable progress has been made in the fight with this disease, and few drugs have been developed to treat this deadly disease. Although gemcitabine is the standard drug of choice6,7,8,9,10; fluorouracil, leucovorin and irinotecan are also used as combination chemotherapy, but existing anti-cancer drugs or therapies are unable to save lives of patients suffering form PCa. One of the major reasons for the inefficiency of existing anti-cancer drugs is acquired drug resistance that is developed due to genetic alterations in various drug targets11,12,13. There is an urgent need to improve pancreatic cancer drug arsenal to combat drug resistance problem and for effective treatment. High-throughput screening of therapeutic molecules from a large pool of chemical compounds is the most suitable way to identify novel anti-cancer molecules. However, it is time and labor consuming effort. In silico methods, which can predict novel inhibitors against pancreatic cancer, will be an attractive alternative approach.
Recently, considerable attention has been paid towards pancreatic cancer drug discovery. In this context, Garnett et. al. have screened 132 anti-cancer drugs on 714 cancer cell lines14 and reported 78070 logIC50 values for different drug and cell line combinations. In another study, Rechard et. al. have demonstrated that cancer cell lines share the same features (i.e. copy number variation, expression abnormality) as the primary tumors15. In 2012, Barretina et. al. clearly demonstrated the correlation between genomic status of primary tumors and cancer cell lines of different lineages16. These studies support the extrapolation of cell line studies to primary tumors and further to clinics. Keeping all these facts into consideration, in the present study, we have developed quantitative structure activity relationship (QSAR) models to predict promiscuous inhibitors against 16 pancreatic cancer cell lines. The pharmacological screening data generated in Genomics of Drug Sensitivity in Cancer (one of the projects in COSMIC) was used to develop models14. QSAR modeling using high-throughput screening data is a powerful technique, which enables the construction of predictive models. These models can be utilized for the in silico screening of libraries of billions of diverse molecules prior to their experimental validation. Here, we have not considered the biological targets of drugs and just tried to demonstrate the potential of chemical descriptors and QSAR to predict anti-cancer activity of unknown molecules. Our QSAR models will complement the pancreatic cancer research by helping in identification of novel inhibitors against pancreatic cancer cell lines. For the advancement of the scientific community, we have integrated these models on a webserver, DiPCell, which is freely accessible at http://crdd.osdd.net/raghava/dipcell/.
Results
Analysis of pharmacological drug profiling
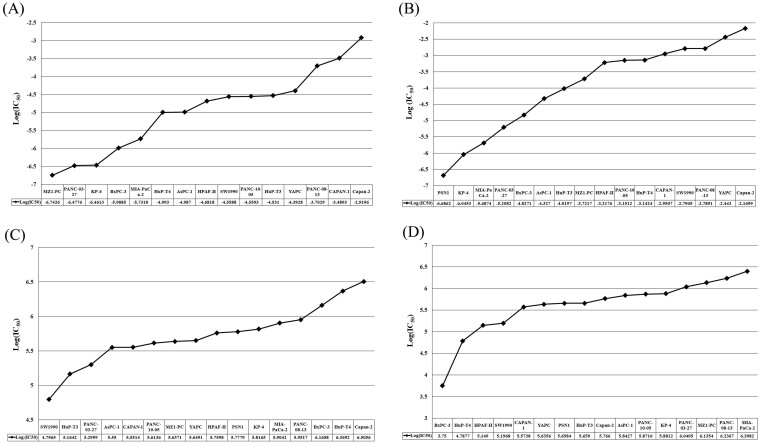
In order to identify the most effective drugs (i.e., killing most of the pancreatic cancer cell lines), we have analyzed the pharmacological profiling of more than 80 drugs on 16 pancreatic cancer cell lines. We found that docetaxel, an inhibitor of microtubule assembly was the most effective as it was effective against 14 out of 16 pancreatic cancer cell lines studied (Figure 1A, Supp. Figure S1A and Supp. Table ST1). Second most effective drug was vinblastine, an inhibitor of DNA topoisomerase I, effective against 11 cell lines having logIC50 values in nanomolar range (Figure 1B, Supp. Figure S1B and Supp. Table ST1). This analysis suggests that these drugs can be used in combination with other drugs against pancreatic cancer. On the other hand, ABT-888 (PARP inhibitor) and LFMA-13 (BTK inhibitor) were the least effective (Figure 1C & 1D, Supp. Figure S1C & D and Supp. Table ST1). Furthermore, clustering of all the anticancer drugs was carried out, and it was observed that most effective drugs were clustered together (Supp. Figure S2). In addition, we found that Capan-2 and YAPC were the most resistant cell lines against most of the anti-cancer drugs (Figure 2A & 2B, Supp. Figure S3A & B and Supp. Table ST2). Behavior of these two cell lines can be subjected to investigate the mechanism of drug resistance in pancreatic cancer. On the other hand, KP-4 and MIA-PaCa-2 were found to be the most sensitive among all the pancreatic cancer cell lines (Figure 2C & 2D, Supp. Figure S3C & D and Supp. Table ST2).
Figure 1. Pharmacological profiling of two most effective anticancer drugs (A) docetaxel and (B) vinblastine, and two least effective anticancer dugs (C) ABT888 and (D) LFMA13 on 16 pancreatic cancer cell lines.
Figure 2. Pharmacological profiling of the most resistant cell lines (A) Capan-2 and (B) YAPC and the most sensitive cell lines (C) KP-4, and (D) MIA-PaCa-2 against 38 anti-cancer drugs.
Performance of QSAR models
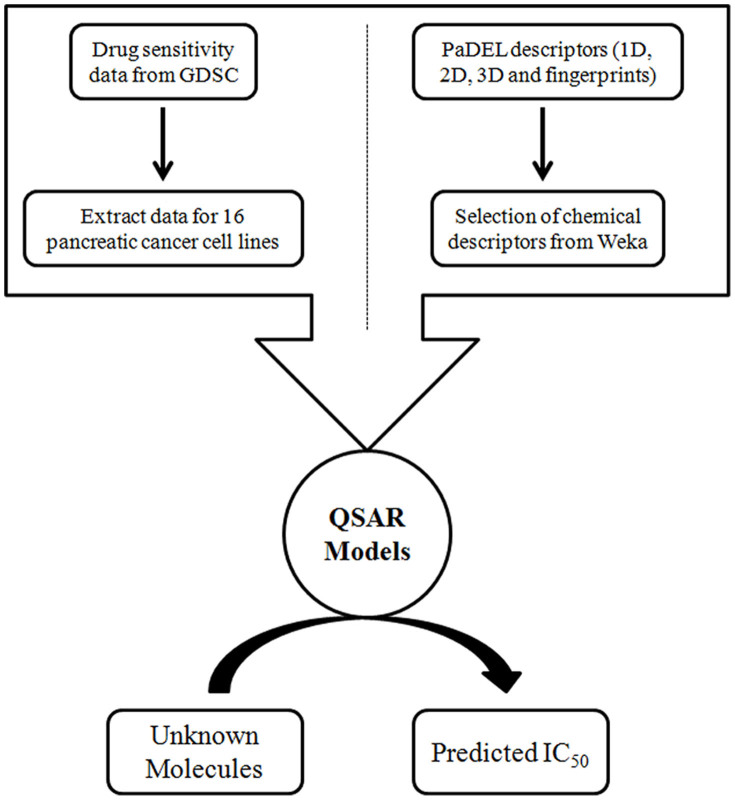
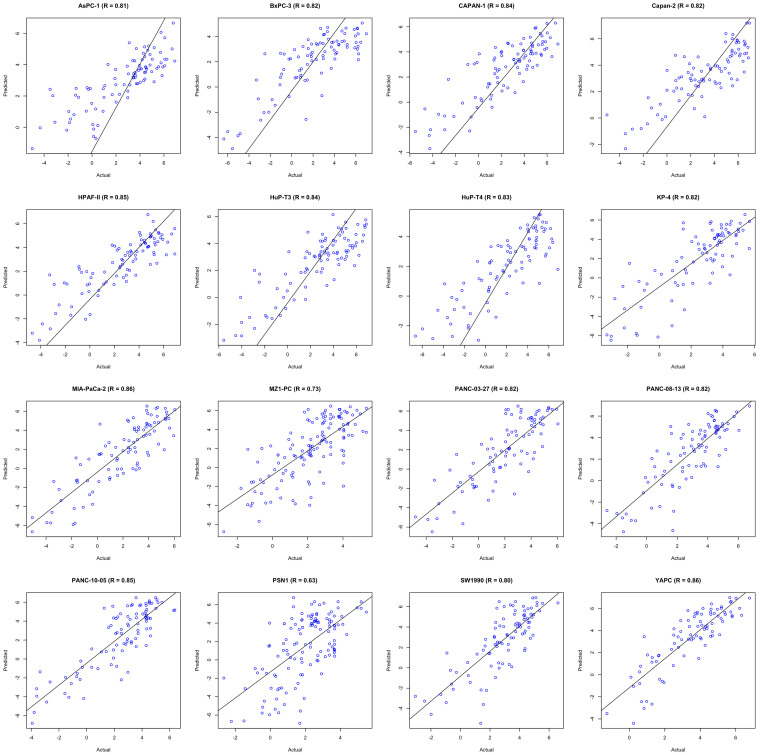
In order to identify the most effective features or descriptors of anticancer drugs, we computed the correlation between chemical features of anti-cancer drugs and their inhibitory activity. We next asked, whether these chemical features have some predictive power to predict anticancer activity of an unknown molecule. To address this issue, we have used the most comprehensive pharmacological screening dataset till now from the GDSC project to develop QSAR models (Figure 3). Performance of QSAR models was evaluated in terms of Pearson correlation coefficient (R), coefficient of determination (R2) and root mean square error (RMSE). Performance of QSAR models was evaluated at two different levels of descriptor selection. At first level, descriptors were selected using CfsSubsetEval module implemented in Weka. At this level, we selected as minimum as 38 descriptors for SW1990 cell line and maximum of 136 descriptors for MZ1-PC cell line (Table 1). We have achieved maximum correlation (R) of 0.89 in case of YAPC cell line with R2 and RMSE values of 0.78 and 1.24 respectively, and minimum correlation was 0.64 in case of PSN1 cell line. Although we achieved a decent correlation for most of the cell lines at this level, but the ratio of number of descriptors and number of drugs is around 1:2 or more (Table 1). For the development of robust QSAR models, this ratio should be around 1:4. So, we further reduced descriptors as much as possible by applying F-stepping technique, which removes each descriptor one by one. At this level, we have achieved maximum correlation (R) of 0.86 in case of MIA-PaCa-2 and YAPC cell lines as shown in Table 1 and minimum correlation was 0.63 in case of PSN1 cell line and maintained the ratio of number of descriptors and number of drugs to 1:4. Figure 4 demonstrates the scatter plot between observed and predicted logIC50 (μM) for different pancreatic cancer cell lines.
Figure 3. Schematic diagram demostrating work flow of DiPCell.
Table 1. Pearson correlation and root mean square error values obtained for each pancreatic cell line by their respective QSAR models.
| S. No. | Cell Line | ND* | D1 | R | R2 | RMSE | D2 | R | R2 | RMSE |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | AsPC-1 | 92 | 45 | 0.84 | 0.70 | 1.6 | 23 | 0.81 | 0.66 | 1.78 |
| 2. | BxPC-3 | 92 | 47 | 0.82 | 0.67 | 1.82 | 19 | 0.82 | 0.67 | 1.83 |
| 3. | CAPAN-1 | 96 | 42 | 0.83 | 0.69 | 1.61 | 21 | 0.84 | 0.71 | 1.58 |
| 4. | Capan-2 | 90 | 54 | 0.86 | 0.73 | 1.47 | 22 | 0.82 | 0.67 | 1.63 |
| 5. | HPAF-II | 95 | 42 | 0.86 | 0.74 | 1.45 | 22 | 0.85 | 0.73 | 1.48 |
| 6. | HuP-T3 | 96 | 54 | 0.85 | 0.73 | 1.57 | 22 | 0.84 | 0.71 | 1.63 |
| 7. | HuP-T4 | 92 | 41 | 0.79 | 0.61 | 2.00 | 24 | 0.83 | 0.69 | 1.81 |
| 8. | KP-4 | 88 | 62 | 0.82 | 0.68 | 1.94 | 22 | 0.82 | 0.67 | 2.04 |
| 9. | MIA-PaCa-2 | 96 | 45 | 0.85 | 0.71 | 1.81 | 23 | 0.86 | 0.73 | 1.76 |
| 10. | MZ1-PC | 132 | 136 | 0.79 | 0.61 | 1.94 | 29 | 0.73 | 0.53 | 2.16 |
| 11. | PANC-03-27 | 91 | 44 | 0.8 | 0.64 | 2.00 | 20 | 0.82 | 0.67 | 1.91 |
| 12. | PANC-08-13 | 92 | 50 | 0.81 | 0.65 | 1.75 | 22 | 0.82 | 0.68 | 1.74 |
| 13. | PANC-10-05 | 95 | 41 | 0.81 | 0.65 | 1.92 | 24 | 0.85 | 0.73 | 1.71 |
| 14. | PSN1 | 129 | 116 | 0.64 | 0.42 | 2.68 | 31 | 0.63 | 0.39 | 2.81 |
| 15. | SW1990 | 94 | 38 | 0.77 | 0.56 | 1.89 | 23 | 0.80 | 0.63 | 1.83 |
| 16. | YAPC | 86 | 65 | 0.89 | 0.79 | 1.24 | 21 | 0.86 | 0.74 | 1.45 |
*ND: Number of drugs used to develop QSAR models; D1: Number of descriptors selected from the CfsSubsetEval algorithm; D2: Number of descriptor selected from CfsSubsetEval followed by F-stepping; R: Pearson Correlation; RMSE: Root Mean Square Error.
Figure 4. Scatter plots between actual and predicted logIC50 values of 16 pancreatic cancer cell lines.
Analysis of descriptors
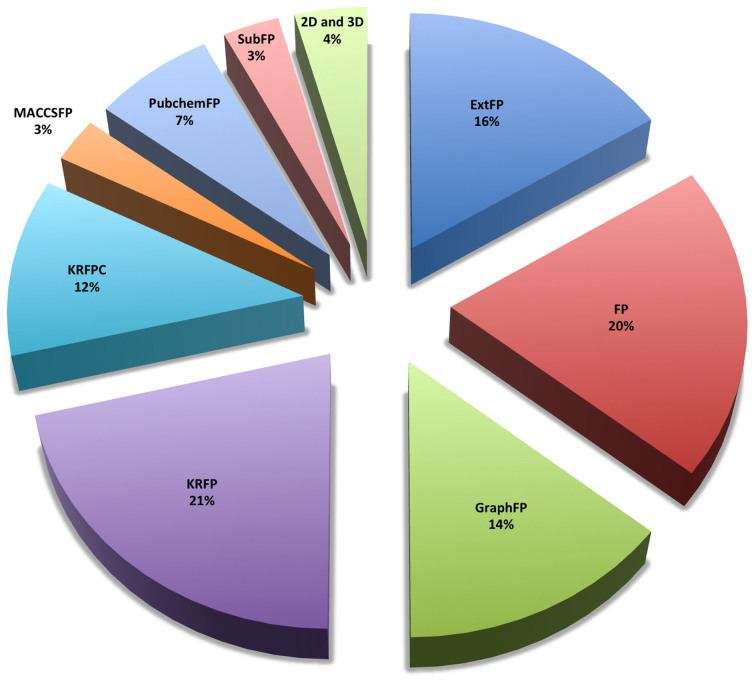
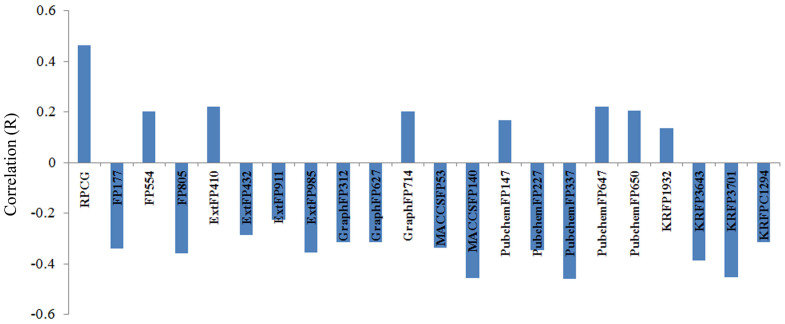
We have analysed all the descriptors used in developing 16 QSAR models and observed that in total 212 descriptors were sufficient enough to predict the effect of anti-cancer drugs on 16 pancreatic cancer cell lines (Figure 5 and Table ST3). While analyzing the properties of these descriptors, we observed that 96% of all the descriptors were binary fingerprints and rest 4% were 2D and 3D descriptors (Figure 5). As shown in Figure 5, KRFPs are the most contributing descriptors (22%) followed by the CDK fingerprints (21%). Further analysis suggested that extended fingerprint 153 (ExtFP153) (describes the ring feature in a drug molecule) and fingerprint (FP1013) showed a negative correlation for 9 and 11 pancreatic cancer cell lines respectively (Supp. Figure S4 and Supp. Table ST4). However, the graph fingerprint 40 (GraphFP40) showed a positive correlation with drug activity (Supp. Figure S4 and Table ST5). Relative positive charge descriptor (RPCG) is the only single 3D descriptor which showed a high positive correlation with the drug activity in Capan-2 cell line (Figure 6). It suggests that relative positive charge plays some role in anti-cancer activity of drugs, and it would be recommendable to have more relative positive charge for better antiproliferative activity. On the other hand, PubChem fingerprint, PubchemFP337, which corresponds to substructure C(~C)(~C)(~C)(~O) showed a negative correlation with the drug activity (Supp. Figure S4 and S5) (‘~' depicts irrespective of bond order). Similarly, activity of anti-cancer drugs for the other cell lines was correlated with different types of descriptors, suggesting that these descriptors play crucial roles in the functioning of these anti-cancer drugs (Supp. Figure S4).
Figure 5. Different classes of descriptors associated with inhibitory activity prediction.
Figure 6. Correration of descriptors (R) with the drug activitiy in Capan-2 cell line.
Validation of drug-to-oncogene relation
From these QSAR models, we tried to recapitulate the drug-to-oncogene associations, which were suggested by the experimental data14. For instance, loss of SMAD4 was associated with sensitivity to EGFR-family inhibitor BIBW299214. First, we divided the 16 pancreatic cancer cell lines into two classes, first one, which is mutated for SMAD4, and second, which is wild type for SMAD4. We developed different QSAR models for wild type and mutated cell lines (BIBW2992 was not used in the training of these models to avoid any biases). Then, we predicted the logIC50 value of BIBW2992 (as an independent molecule) using our QSAR models for each cell line. We got the same association from the predicted logIC50 values as earlier suggested by the experimental data (Figure 7).
Figure 7. Scatter plot showing (A) experiemental and (B) predicted (obtained by QSAR models) LogIC50 values for SMAD4 mutated and wild type pancreatic cancer cell lines.
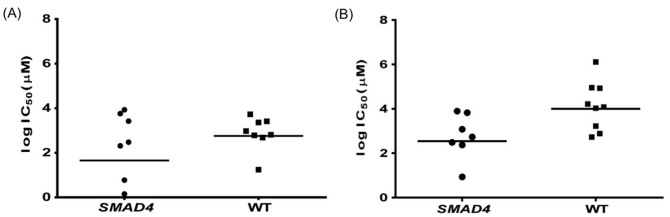
Each dot represents the cell line and horizontal line is the geometric mean. In panel (A), 15 cell lines is presented instead of 16 because for one cell line logIC50 is having a negative value.
Screening of FDA approved drugs
Drug repositioning is the well established concept in the field of drug designing and pharmaco-informatics22,23. In 2012, Debnath and coworkers carried out the high throughput screening of FDA approved drugs against the intestinal parasite Entamoeba histolytica, which is the causative agent of human amebiasis24. They found auranofin, which is a prescribed drug in rheumatoid arthritis is ten times more potent than metronidazole (drug of choice for human amebiasis). This finding and many other earlier such reports advocated the potential of FDA approved drugs for their unknown therapeutic potential in other diseases. To capitalize these findings, we have screened FDA approved drugs by our in silico QSAR models and sorted them according to their predicted IC50 values. We got interesting result, out of top 10 FDA approved drugs (Table 2), 7 are well known anticancer drugs, which uphold the utility of our QSAR models for screening anticancer activity. Remaining 3 drugs, have yet to be characterized for their anticancer activity. Whole rank wise list of FDA approved drugs is available in supplementary material (Table ST5).
Table 2. Rank wise list of predicted anticancer drugs (Top 10).
| S. No. | Drug Bank ID | Name | Therapeutic Use |
|---|---|---|---|
| 1. | DB01248 | Docetaxel | Anticancer |
| 2. | DB01229 | Paclitaxel | Anticancer |
| 3. | DB00541 | Vincristine | Anticancer |
| 4. | DB00570 | Vinblastine | Anticancer |
| 5. | DB06772 | Cabazitaxel | Anticancer |
| 6. | DB00361 | Vinorelbine | Anticancer |
| 7. | DB00954 | Dirithromycin | Antibiotics |
| 8. | DB00864 | Tacrolimus | Immunosuppresent |
| 9. | DB00337 | Pimicrolimus | Immunosuppresent |
| 10. | DB01030 | Topotecan | Anticancer |
Experimental Validation
In the list of top ten predicted anticancer drugs, three drugs (pimicrolimus, tacrolimus and dirithromycin) were not known previsouly for their anticancer activity (Table 2). Therefore, we analysed in vitro antiproliferative effect of these three drugs on two pancreatic cancer cell lines, MIA-PaCa-2 and PANC-1. We have taken paclitaxel as a positive control for the anticancer activity and the same was also present in our predicted list of anticancer drugs. As predicted, all three drugs have shown anti-cancer activity on both the cell lines. Tacrolimus was the most effective drug at higher concentration (above 50 μM) as it has shown ~100% cytotoxicity at 100 μM (Figure 8a and 8b) on both the cell lines. Pimicrolimus has shown more than 60% cytotxicity at 100 μM on both the cell lines (Figure 8a and 8b). These results shows that the tacrolimus have prominent anticancer activity as compared to the other predicted drugs and paclitaxel (positive control) at higher concentration (100 μM) but found to be less effective at lower concentrations. Pimicrolimus was more effctive than tacrolimus below 50 μM concentration range. On the other hand, dirithromycin was less effective even at higher concentration.
Figure 8. Inhibition of cell proliferation by paclitaxel, dirithromycin, pimicrolimus and tacrolimus of pancreatic cancer cell lines:
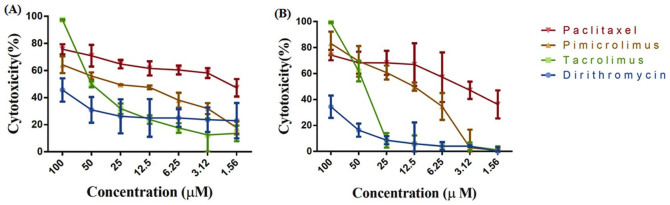
(a). PANC-1 (b). MIA-PaCa-2.
Web Implementation
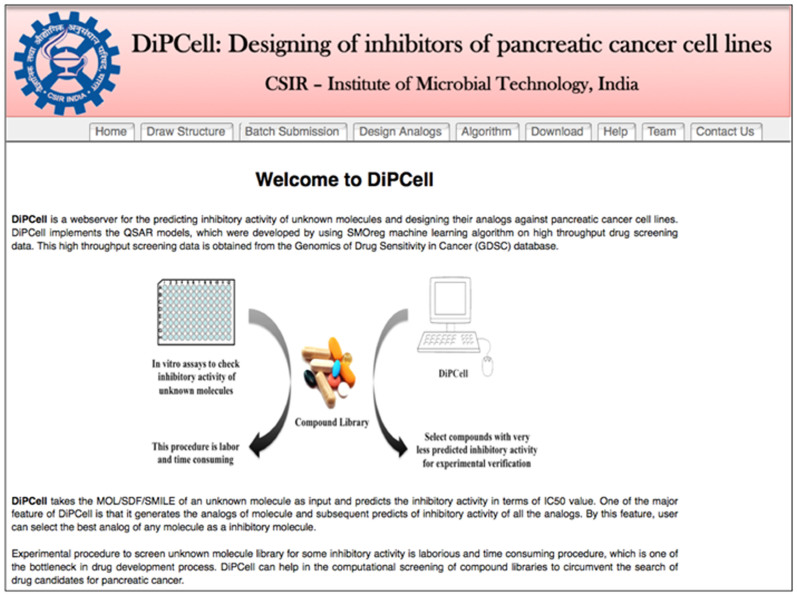
As the results demonstrated that developed QSAR models are quite effective in predicting the inhibitory activity (logIC50) of unknown molecules and in reproducing the drug-to-oncogene association, these QSAR models have been implemented to the user friendly webserver named as DiPCell (Figure 9), where users can predict the inhibitory activity of unknown molecules (or a whole library of chemicals) against 16 pancreatic cancer cell lines in terms of logIC50 value. DiPCell includes following tools:
Figure 9. Web interface showing the home page of DiPCell.
Draw structure
This tool allows users to draw chemical structure of their molecule using Marvin editor. At one time, user can predict drug sensitivity on a maximum of 16 pancreatic cancer cell lines. Since it is very difficult to define the cut-off logIC50 value, which discriminates between sensitive and resistance cancer cell lines hence, an option of logIC50 cut-off value has been provided, which will be defined by the user on the basis of their experimental criteria. After submission, DiPCell returns with logIC50 values against pancreatic cancer cell lines selected by the users along with an option to calculate chemical descriptors of the query molecule (Supp. Figure S6).
Batch submission
This allows users to submit more than one molecule at a time. Users have to choose the cell lines on which they want to test their query molecules along with the cut-off logIC50 values (Supp. Figure S7).
Design analogs
Since analogs of known drug/certain molecule may be more potent than parent molecule. Therefore, it is a common practice to identify a better molecule of a certain existing drug by structural activity relationship (SAR). In DiPCell, we have incorporated the similar kind of module, where user can design analogs and simultaneously predict their drug sensitivity on pancreatic cancer cell lines. User has to provide scaffold structures, building blocks and linkers as input for this module (Supp. Figure S8). This webserver will be useful and can actively contribute in research on pancreatic cancer by helping in discovering the new candidate drug molecules. This web service is freely accessible at http://crdd.osdd.net/raghava/dipcell.
Discussion
Continuous discovery of novel inhibitors against pancreatic cancer will not only improve the current treatment but also provide more options to select suitable drugs for the right subset of patients. Identification of novel drug candidates is not as simple as it looks and the whole process usually takes a long time (~15–20 years) to funnel out a single drug molecule out of billions of compounds. On the other hand, computational screening of billions of molecules to identify/predict drug like compounds based on certain features of well known drug molecules seems to be a potential approach. In the present study, we have developed QSAR models for prediction of inhibitors against pancreatic cancer cell lines to enhance and complement the drug development process. Our results demonstrated that chemical features of drug molecules can be correlated to their activity and thus, can be used to predict activity of unknown molecules. Availability of high throughput drug screening data made it possible to develop such efficient models, and we anticipate that as more and more screening data will be available, the predictive power of these models will increase further. Our models were also able to recapitulate the drug-to-oncogene association, which were revealed by the experimental data. So, it would help to link up the genes as biomarker of drug sensitivity14,25. As we have shown in our results, Capan-2 and YAPC cell lines were resistant against most of the anti-cancer drugs and earlier studies demonstrated that cancer cell lines are like a mirror image of primary tumors in terms of genomic and transcriptional abnormalities15,16 and moreover, the high throughput data as in our case can recapitulate the real conditions up to great extent and help in systematic identification of new anticancer drug candidates. Therefore, we can hypothesize that genomic and transcriptomic studies of these two cell lines can put some light on the drug resistance mechanism in pancreatic cancer. As suggested in the literature, it is not solely the drug, which determined its activity, rather genomics and proteomic signatures of a cell line are also substantial contributors in determining the activity26,27,28,29. We are currently investigating these aspects, and in the future we will integrate these signatures with QSAR models to make them more robust and efficient.
Limitations
Recently, Quackenbash and colleague have shown an interesting comparison between pharmacological data from CCLE and CGP30. They have shown that the pharmacolgical data between these two studies are miserably correlated (Spearman's rank correlation of 0.28). In the light of this comparative study, one can question about the validity of our QSAR models, whether they will accurately predict the anticancer activity or not. We agree with this and certainly it would limit down the spectrum of these QSAR models. But if we carefully look, this is not the limitation of QSAR models, this is the limitation of the pharmacological data available31. This inadequation of the pharmacological data is also reflected in our experimental validation, where we got the anticancer activity in tacrolimus and dirithromycin, but at very high concentration (100 μM). But, we can anticipate as the quality of the data will increase, predictive power of these models will increase more and more. This study is solely based on cell line data, this is also an another constraint, which further narrows down the spectrum of these models. But from somewhere at some point, we have to start and this study is just a beginning of a new arena for drug sensitivity prediction.
Methods
Pharmacological data
In this study, we have used a dataset of 132 anti-cancer drugs and their log transformed IC50 values against 714 cancer cell lines and this data was obtained from the GDSC Website14 (Genomics of Drug Sensitivity, http://www.cancerrxgene.org/translation/Drug, Date of access: 20/11/2012, published in 2012) and CancerDR database17 (CancerDR: Cancer Drug Resistance Database, http://crdd.osdd.net/raghava/cancerdr, Date of access: 07/12/2012, published in 2013). Among the 714 cancer cell lines, 16 were pancreatic cancer cell lines. We extracted the pharmacological screening data of these 16 pancreatic cancer cell lines. LogIC50 values of these drugs vary from −11 to +13.6. Higher logIC50 values are just an extrapolation of the drug-response curve, and they do not have any biological relevance. But, if we reduce this scale to a somewhat narrow range, number of drugs will reduced apparently. Accordingly to make a balance between drugs and logIC50 range, we restricted ourself to −7 to +7 scale of logIC50, so that we can get optimum number of drugs to develop QSAR models and moreover, to avoid any fallacy in machine learning.
Structure of Drugs
To obtain the structure of drugs, we have downloaded the SDF file of molecules available at PubChem and for rest of the drugs, structures were drawn using PubChem editor. These 2D structures were further converted into 3D structures, and their energy was minimized by OpenBabel software18.
Descriptors Calculation
To develop cell line specific QSAR models, we have computed 863 chemical descriptors (1D, 2D, and 3D), which include constitutional, topological, geometric, electrostatic, hydrophobic, etc. using PaDEL software [18]. In addition, we have calculated 10 different classes of binary fingerprints (FP's) available in PaDEL software.
Descriptor Selection
It is a well known fact that all the descriptors are not relevant to the activity, and it is a fundamental requirement to remove irrelevant descriptors to develop robust QSAR models, thus we used feature selection techniques in order to select relevant features/descriptors. We used remove-useless function followed by CfsSubsetEval module with best-fit algorithm implemented in Weka19 for the selection of relevant descriptors. CfsSubsetEval determines the predictive ability of each attribute (chemical descriptor) and the redundancy among the descriptors. It also selects the best set of attributes that are highly correlated with the class for prediction, but at the same time have low inter-correlation. Further, we applied F-stepping, which removes one descriptor at a time to check its correlation with activity.
QSAR Models
We developed individual QSAR models for each of the 16 pancreatic cancer cell lines using SMOreg algorithm in Weka, which uses the sequential minimal optimization algorithm for training a support vector classifier using polynomial or Gaussian kernels for regression problem20. We used the command line version of Weka machine learning tool (version 3.6.6) for implementing SMOreg at RBF kernel19. Chemical descriptors and fingerprints used as input features for the development of QSAR models.
Cross Validation
Cross-validation was carried out to avoid under, and over-fitting of models21. We used 10-fold cross validation technique for building and evaluating our model. In order to implement this cross validation technique, we have randomly divided the original dataset into 10 parts. Nine datasets were used in training and remaining one was used exclusively for testing. This process is repeated, so that each part was tested once. Finally, we have calculated the Pearson correlation coefficient (R), coefficient of determination (R2) and root mean square error (RMSE) as the performance measures.
Reagents and Cell Culture
Paclitaxel and tacrolimus were purchased from Calbiochem. Dirithromycin and pimicrolimus were purchased from Sigma with purity of 95%. Non-radioactive proliferation kit (based on MTS reagent) was purchased form Promega. Human pancreatic cancer cell lines MIA-PaCa-2 and PANC-1 were purchased from American Type Culture Collection (Rockville, MD). Cell lines were maintained in DMEM media supplemented with 10% fetal bovine serum at 37°C in humified atmosphere (5% CO2).
In vitro cytotoxicity assay
First, 1×104 cells in 100 μl of media were plated in 96 well plates and allowed them to grow for 24 hours and treated with paclitaxel, dirithromycin, pimicrolimus and tacrolimus in various concentrations. After 72 hours, 20 μl of MTS reagent (prepared according to the manufacturer's protocol) added to the each well followed by the additional incubation of 2 hours. Absorbance was measured at 490 nm using microplate reader (Tecan).
Author Contributions
R.K. collected the data and created the datasets. R.K., K.C. and D.S. developed QSAR models. R.K. and K.C. created the back end server. R.K., K.C., A.G., and D.S. developed the front end user interface. R.K., A.G. and G.P.S.R. analysed the results and wrote the manuscript. R.K. carried out the experimental validations. R.K. and A.G. prepared figure 1, 2, 3, 5, 7, 8 and 9. D.S. prepared figure 4 and 6. G.P.S.R. conceived and coordinated the project, helped in the interpretation of data, refined the drafted manuscript and gave overall supervision to the project. All of the authors read and approved the final manuscript.
Supplementary Material
Supplementary Information 1
Supplementary Information 2
Acknowledgments
Authors are thankful to funding agencies Council of Scientific and Industrial Research (project Open Source Drug Discovery and GENESIS BSC0121) and Department of Biotechnology (project BTISNET), Govt. of India for financial support.
References
- Am. Cancer Soc., Cancer Facts & Figures. Atlanta Am. Cancer Soc. 2013 64 (2013). [Google Scholar]
- Riker A., Libutti S. K. & Bartlett D. L. Advances in the early detection, diagnosis, and staging of pancreatic cancer. Surg oncol 6, 157–69 (1997). [DOI] [PubMed] [Google Scholar]
- Delhaze M., Jonard P., Gigot J. F., Descamps C. & Dive C. Chronic pancreatitis and pancreatic cancer. An often difficult differential diagnosis. Acta Gastroenterol. Belg. 52, 458–66. [PubMed] [Google Scholar]
- Long J. et al. Overcoming drug resistance in pancreatic cancer. Expert Opin. Ther. Targets 15, 817–28 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh R., Walsh N., Clynes M., O'Connor R. & McDermott R. Challenges of drug resistance in the management of pancreatic cancer. Expert Rev. Anticancer Ther. 10, 1647–61 (2010). [DOI] [PubMed] [Google Scholar]
- Pliarchopoulou K. & Pectasides D. Pancreatic cancer: current and future treatment strategies. Cancer Treat. Rev. 35, 431–6 (2009). [DOI] [PubMed] [Google Scholar]
- Di Marco M. et al. Metastatic pancreatic cancer: is gemcitabine still the best standard treatment? (Review). Oncol. Rep. 23, 1183–92 (2010). [DOI] [PubMed] [Google Scholar]
- Patra C. R. et al. Targeted delivery of gemcitabine to pancreatic adenocarcinoma using cetuximab as a targeting agent. Cancer Res. 68, 1970–8 (2008). [DOI] [PubMed] [Google Scholar]
- Hidalgo M. Pancreatic cancer. N. Engl. J. Med. 362, 1605–17 (2010). [DOI] [PubMed] [Google Scholar]
- Shore S., Raraty M. G. T., Ghaneh P. & Neoptolemos J. P. Review article: chemotherapy for pancreatic cancer. Aliment. Pharmacol. Ther. 18, 1049–69 (2003). [DOI] [PubMed] [Google Scholar]
- Grønbaek K., Hother C. & Jones P. A. Epigenetic changes in cancer. APMIS 115, 1039–59 (2007). [DOI] [PubMed] [Google Scholar]
- Sadikovic B., Al-Romaih K., Squire J. A. & Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr. Genomics 9, 394–408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. Identification of the MEK1(F129L) activating mutation as a potential mechanism of acquired resistance to MEK inhibition in human cancers carrying the B-RafV600E mutation. Cancer Res. 71, 5535–45 (2011). [DOI] [PubMed] [Google Scholar]
- Garnett M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R. M. et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–27 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. et al. CancerDR: cancer drug resistance database. Sci. Rep. 3, 1445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle N. M. et al. Open Babel: An open chemical toolbox. J. Cheminform. 3, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank E., Hall M., Trigg L., Holmes G. & Witten I. H. Data mining in bioinformatics using Weka. Bioinformatics 20, 2479–81 (2004). [DOI] [PubMed] [Google Scholar]
- Cao L. J. et al. Parallel sequential minimal optimization for the training of support vector machines. IEEE Trans. Neural Netw. 17, 1039–49 (2006). [DOI] [PubMed] [Google Scholar]
- Kooperberg C. et al. Logic regression for analysis of the association between genetic variation in the renin-angiotensin system and myocardial infarction or stroke. Am. J. Epidemiol. 165, 334–43 (2007). [DOI] [PubMed] [Google Scholar]
- Ashburn T. T. & Thor K. B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–83 (2004). [DOI] [PubMed] [Google Scholar]
- Chong C. R. & Sullivan D. J. New uses for old drugs. Nature 448, 645–6 (2007). [DOI] [PubMed] [Google Scholar]
- Debnath A. et al. A high-throughput drug screen for Entamoeba histolytica identifies a new lead and target. Nat. Med. 18, 956–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Paredes M. & Esteller M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 17, 330–9 (2011). [DOI] [PubMed] [Google Scholar]
- Chen B.-J. et al. Harnessing gene expression to identify the genetic basis of drug resistance. Mol. Syst. Biol. 5, 310 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. S. et al. Integrative analysis of proteomic signatures, mutations, and drug responsiveness in the NCI 60 cancer cell line set. Mol. Cancer Ther. 9, 257–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold W. C. et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 72, 3499–511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal G. et al. PCMdb: Pancreatic Cancer Methylation Database. Sci. Rep. 4, 4197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B. et al. Inconsistency in large pharmacogenomic studies. Nature 504, 389–393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming J. G., Davis A. M., Muresan S., Haeberlein M. & Chen H. Chemical predictive modelling to improve compound quality. Nat. Rev. Drug Discov. 12, 948–62 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information 1
Supplementary Information 2