Abstract
Myeloid cells that orchestrate malignant progression in the tumor microenvironment offer targets for a generalized strategy to attack solid tumors. Through an analysis of tumor microenvironments, we explored an experimental model of lung cancer that uncovered a network of Dll4/Notch/TGF-β1 signals that links myeloid cells to cancer progression. Myeloid cells attracted to the tumor microenvironment by the tumor-derived cytokines CCL2 and M-CSF expressed increased levels of the Notch ligand Dll4, thereby activating Notch signaling in the tumor cells and amplifying intrinsic Notch activation there. Heightened Dll4/Notch signaling in tumor cells magnified TGF-β-induced pSMAD2/3 signaling and was required to sustain TGF-β-induced tumor cell growth. Conversely, Notch blockade reduced TGF-β signaling and limited lung carcinoma tumor progression. Corroborating these findings, interrogating RNAseq results from tumor and adjacent normal tissue in clinical specimens of human head and neck squamous carcinoma we found evidence that TGF-β/Notch crosstalk contributed to progression. In summary, the myeloid cell-carcinoma signaling network we describe uncovers novel mechanistic links between the tumor microenvironment and tumor growth, highlighting new opportunities to target tumors where this network is active.
Keywords: tumor microenvironment, myeloid cells, Dll4, Notch, TGF-β
Introduction
The tumor microenvironment is increasingly recognized as an enabling contributor to tumor progression (1, 2), and strategies that target the tumor microenvironment are effective at reducing tumor growth, despite persistence of genetically modified tumor cells (3–5).
The vasculature is a component of the tumor microenvironment that contributes to tumor growth through angiogenesis. Inhibition of angiogenesis by neutralization of VEGF is effective at reducing progression of certain tumors despite having little effect on most tumor cells (5). “Inflammatory” cells, particularly “M2-type” myeloid cells (2) and stromal fibroblast-like cells/cancer-associated fibroblastic cells (CAFs) (1) are other pro-tumorigenic components of the cancer microenvironment. Through integrin-mediated adhesion signaling and other mechanisms myeloid cells promote cancer cell survival (6). Acting directly or through effector molecules, including TGF-β, fibroblast growth factors (FGFs) and epidermal growth factors, cancer-associated myeloid and fibroblastic cells supply mitogenic signals to tumors (1). By secreting VEGF, bFGF, platelet-derived growth factor, placental growth factor and Bv8, myeloid cells promote tumor angiogenesis (5, 7). By producing various proteases, myeloid cells induce the release VEGF and other mitogenic factors sequestered in the extracellular matrix and disrupt tissue integrity by cleaving homotypic and heterotypic cell adhesion molecules (8).
Pressing motivation to abrogate myeloid-derived pro-tumorigenic signals has produced some encouraging results. Blocking macrophage recruitment with antagonists of colony stimulating factor-1 receptor reduced mammary tumor progression and increased mice survival (4). TGF-β targeted drugs are currently in clinical trials after showing encouraging anti-cancer activity in preclinical studies (9, 10). Matrix metallo proteinases inhibitors, which showed promising anti-tumor activities in mouse but not in human cancer trials, are being re-evaluated in light of emerging new understanding (11).
Despite these advances, the complexities of cell composition of tumor microenvironments and tendency to adaptive change present current obstacles. To overcome some of these difficulties, we have queried the “simpler” tumor microenvironment of Gfi1 (growth factor independence-1)-null mice, which lack mature granulocytes and harbor functionally impaired myeloid cells (12, 13) to identify principal mechanisms that sustain reciprocal communications between tumor cells and cells of the tumor microenvironment. By focusing on Notch signaling, a crucial regulator of cell fate decisions activated by cell-to-cell interaction between Notch ligand (Dll1, Dll3, Dll4, Jagged1 and Jagged2) and Notch receptor (Notch1-Notch4)-expressing cells (14–16), we now uncovered a novel network of Dll4/Notch/TGF-β signals linking myeloid cells and cancer cells that drives lung carcinoma tumor progression. This network provides a mechanistic link between tumor-infiltrating myeloid cells and tumor cells with opportunities for intervention.
Materials and Methods
Cell culture and in vitro treatments
The EL4, LLC1 and B16F10 murine cell lines (from ATCC; authentication confirmed by ATCC through depositor’s analysis of representative cultures from the master seed stock), were propagated in the laboratory for fewer than 6 months in culture medium (RPMI or DMEM with 1% BSA, 2mM L-glutamine, 100IU/ml penicillin, 100µg/ml streptomycin, 50µM 2-ME, and 1mM sodium pyruvate; proliferation was measured by 3H thymidine incorporation (17). Recombinant human TGF-β1 (R&D Systems), DAPT (N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine tert-butyl ester; Sigma-Aldrich), DBZ ((2S)-2-[2-(3,5-difluorophenyl)-acetylamino]-N-(5-methyl-6-oxo-6,7-dihydro-5H-dibenzo[b,d]azepin-7-yl)-propionamide; Millipore) and appropriate diluent controls were added to culture; recombinant mouse His tag-Dll4 (R&D Systems) and His control (Millipore) were immobilized (18hr at 4°C) to culture vessels prior to cell addition.
Animal Studies
Animal experiments were approved by the NCI-Bethesda Institutional Animal Care and Use Committee and conducted according to protocol. Gfi1+/+, +/− and −/− male and female mice (18) were used between 4–8 weeks of age. Mouse tumor cell lines were implanted (10x106) subcutaneously in the left abdominal quadrant. DAPT (10mg/kg, i.p. 5 days/week) or diluent control treatment was initiated 24 hr after tumor-cell injection. Tumors were removed in toto after 12–16 days. Spleens and bone marrows were obtained from tumor-bearing and control mice.
Flow Cytometric Analysis and Cell Sorting
Single-cell suspensions from bone marrow, spleen and tumor tissues were incubated with mouse Fc block CD16/32 antibody (2.4G2 BD Biosciences) for 20 minutes at 4°C in PBS containing 2%BSA (PBS/BSA) to reduce nonspecific antibody binding. After washing in PBS/BSA cells were incubated with control Ig or fluorophore-conjugated antibodies in PBS with 1%BSA and 2mM EDTA. Cell sorting and data collection were performed on a FACSVantage SE or FACSAria (BD Biosciences); data analysis used Flowjo software. Details on antibodies are found in Supplemental Experimental Procedures.
Immunohistochemistry and Immunoblotting
Tissues were fixed with 2% or 4% paraformaldehyde (PFA) overnight or 4hr at 4°C (19). Tissue immunostaining and quantification was performed as described previously (19). Protein extracts prepared as described (19) were run through 4–12% bis-Tris gels (Invitrogen) or 10–20% polyacrylamide gels (Novex), transferred to protran BA83 cellulosenitrate membranes (Whatman) and stained with the primary and secondary antibodies as detailed in Supplemental Experimental Procedures.
Bioinformatics and Statistical Analysis
All bioinformatic analyses were conducted on the publically available gene expression data (normalized values from Illumina RNAseq version 2, level 3) from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/). The data was downloaded from TCGA matrix and was evaluated by box plot analysis and Mann-Whitey U-test using the R system (2.14.1) for statistical computation and graphics. In all other experiments group differences were analyzed by using two-tailed Student’s t test with equal variance assumption and Fisher’s exact test (Microsoft Excel). P values ≤0.05 were considered significant.
Results
Host-dependency of LLC1 carcinoma and EL4 T-cell lymphoma progression
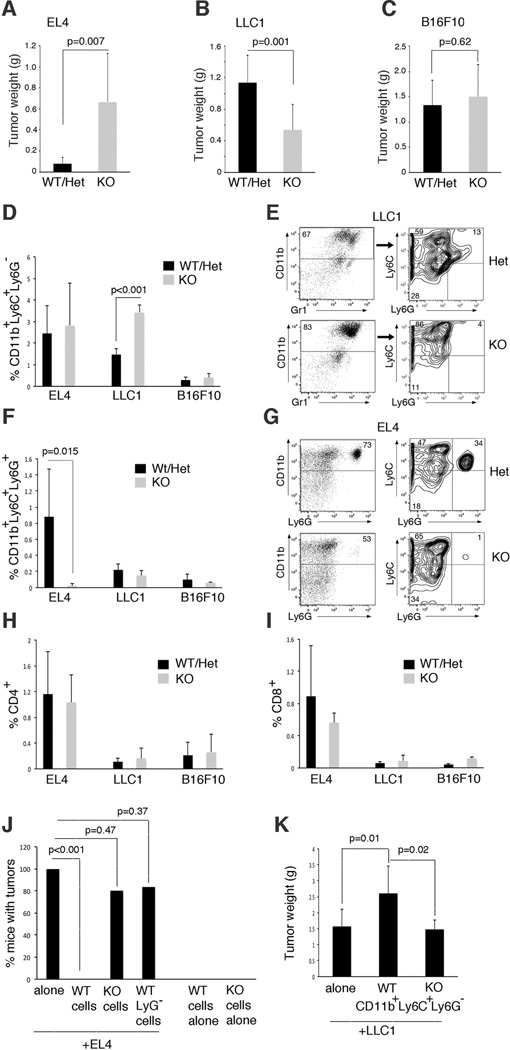
To explore contributions of the tumor microenvironment to tumor progression, we utilized Gfi1-null mice that lack mature granulocytes and have functionally defective monocytes, while displaying a mostly intact lymphoid system (12, 13, 18). Gfi1-heterozygote mice are indistinguishable from wild type (12, 13). By analysis of syngeneic subcutaneous transplant systems, we evaluated tumor growth induced by cell lines representative of T-cell lymphoma (EL4); lung carcinoma (LLC1), and melanoma (B16F10) (Figure 1 A–C; Supplementary (S) Figure S1). EL4 cells generated tumors that grew more aggressively (Figure 1A, Figure S1) in Gfi1-null (KO) mice compared to Gfi1+/+ (wild type WT) or Gfi1+/− heterozygous (Het) mice. By contrast, LLC1 cells generated tumors that grew more aggressively (Figure 1B, Figure S1) in Gfi1-WT/Het mice compared to Gfi1 KO. B16F10 cells generated tumors that grew similarly in Gfi1-WT/Het and KO mice (Figure 1C, Figure S1). We concluded that EL4 and LLC1 tumor progression is significantly affected by host factors.
Figure 1.
The Gfi1-null microenvironment regulates tumor progression. (A–C) Tumor weight from control (WT Gfi1+/+ or het Gfi1+/−) and Gfi1-null (KO Gfi1−/−) mice analyzed 12–15 days post subcutaneous injection of EL4, LLC1 and B16F10 tumor lines. Data are averages ± SD from individual experiments, each representative of 3 performed; (A) EL4 tumors WT/Het n=12; KO n=10; (B) LLC1 tumors WT/Het n=15, KO n=12; (C) B16F10 tumors WT/Het n=12, KO n=10; p values from Student’s t test. (D–G) Monocytes and granulocytes infiltrate tumors from control and Gfi1-null mice. In the bar graphs (D,F), flow cytometry data are expressed as average percentage of total cells from tumor ± SD; EL4: n=5; LLC1: n=6; B16F10 n=3. In the representative flow cytometry plots (E,G), the numerical values are expressed as percentages of total CD11b+ leukocytes in the tumor; p values from Student’s t test. (H,I) Distribution of CD4+ and CD8+ lymphocytes in tumors. The data are expressed as average percentage of total cells from tumor ± SD; EL4: n=5; LLC1: n=6; B16F10 n=3; p values from Student’s t test. (J) Frequency of tumor development in WT mice injected with EL4 cells alone or with splenocytes unfractionated (WT or KO) or depleted of Ly6G+ cells (WT). Splenocytes were from EL4-bearing mice. EL4 alone, EL4+WT cells, EL4+KO cells: n=10; EL4+WT LyG− cells: n=6; WT or KO cells alone: n=3. Data indicate the % mice injected that developed tumors over 14–16 days; p values from Fisher’s exact test. (K) Tumor weight in WT mice injected with LLC1 cell alone or with WT monocytes sorted from bone marrow of LLC1-bearing WT or KO mice; n=6/group. Data are averaged ± SD; p values from Student’s t test.
We hypothesized that the Gfi1 and WT tumor microenvironment differed in EL4 and LLC1 tumors, but not in B16F10 tumors. Since neutrophils, a source of the pro-angiogenic Bv8 factor (7), are absent in Gfi1-null mice, we examined tumor vascularization. We found that vascularization of EL4 and LLC1 tumors from WT/Het and Gfi1-null mice was quantitatively and morphologically similar, as assessed by CD31 immunostaining (Figure S2A,B). A comprehensive analysis of major cell types revealed a significantly greater infiltration of CD11b+Ly6C+Ly6G− cells in LLC1 tumors from Gfi1-null mice compared to control, whereas this population was similarly represented in EL4 tumors from Gf1-null and WT hosts, and was rare in B16F10 tumor tissues (Figure 1D,E). By contrast, the CD11b+Ly6C+Ly6G+ cells were significantly more abundant in EL4 tumors from WT compared to Gfi1-null mice; this population was virtually absent in LLC1 and B16 tumor tissues from WT and Gfi1-null hosts (Figure 1F,G). CD4+ and CD8+ T lymphocytes (Figure 1H,I) were similarly represented in EL4, LLC1 and B16F10 tumor tissues from WT and Gfi1-null mice. CD25+ T cells; CD4+FOXP3+ T cells, B220+ B (and other) cells; CD11c+B220+ dendritic cell populations; SMA+ myofibroblasts; CD49b NK cells and CD11b+F4/80 macrophages were also similarly represented in EL4, LLC1 and B16F10 tumor tissues from WT and Gfi1-null mice (not shown).
Reflecting the Gfi1-null phenotype, spleens (Figure S2 C,D) and bone marrows (Figure S2 D,E) from Gfi1-null mice (control and tumor-bearing) showed an increase of CD11b+Ly6C+Ly6G− cells and a decrease of CD11b+Ly6C+Ly6G+ cells compared to WT mice. Spleens of EL4 tumor-bearing WT mice displayed a significant increase of granulocytes compared to naïve spleens, and spleens of LLC1 tumor-bearing WT mice displayed a significant increase of monocytes compared to naïve spleens (Figure S2C). Given the predominant differences in infiltrates distinguishing EL4 and LLC1 tumors growing in the Gfi1 and WT hosts and changes developing in the spleens of tumor-bearing mice, we focused on the role of CD11b+Ly6C+Ly6G− and CD11b+Ly6C+Ly6G+ cells in these models.
Adoptive transfer experiments supported a tumor-inhibitory activity of WT granulocytes in the EL4 system, because depletion of Ly6G+ cells reduced the anti-tumor activity of WT splenocytes (Figure 1J), and a tumor-promoting function of monocytes in the LLC1 tumor model, because CD11b+Ly6C+Ly6G− cells enhanced LLC1 tumor growth whereas this cell population from Gfi1-null mice did not (Figure 1K).
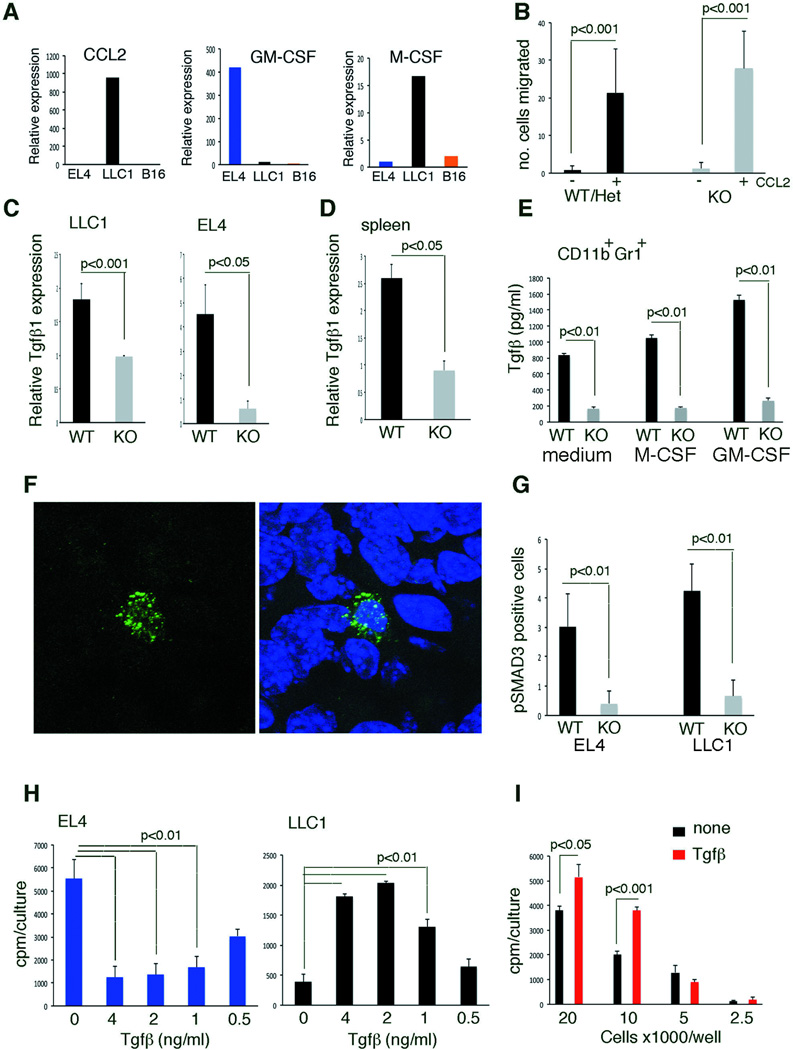
LLC1 cells express CCL2/MCP1 mRNA (Figure 2A) (20) and protein present in LLC1 culture supernatant (2.8 ng/ml). Control and Gfi1-null CD11b+Gr1+ cells similarly migrated to recombinant CCL2 (Figure 2B), suggesting that LLC1-derived CCL2 may recruit myeloid cells to the tumor. LLC1 cells also express M-CSF/CSF1 mRNA (Figure 2A) but protein was undetected in culture supernatant. EL4 cells express GM-CSF/CSF2 mRNA (Figure 2A) and protein detected in the culture supernatant at 21pg/ml, suggesting that it may recruit granulocytes to EL4 tumors (21).
Figure 2.
TGF-β inhibits EL4 cell proliferation and density-dependently increases LLC1 cell proliferation. (A) Relative cytokine mRNA expression in cell lines; results are from qPCR (averages from duplicate measurements). (B) Mouse CCL2 induces migration of bone marrow CD11b+Gr1+ cells. Data are averages ± SD; n=3 experiments. (C) Relative TGF-β1 expression in CD11b+Gr1+ cells sorted from tumor cell suspensions. Data from qPCR are averages ± SD; n=5–6 tumors/group. (D) Relative TGF-β1 expression in naïve splenocytes; qPCR data are means ± SD; n=5. (E) TGF-β in supernatant of bone marrow CD11b+Gr1+ cells; M-CSF (20 ng/ml), GM-CSF (40 ng/ml); ELISA results are means ± SD; triplicate cultures. (F) Representative images of pSMAD3-expressing cells in LLC1 tumor; nuclei are visualized with DAPI. (G) Quantification of pSMAD3-expressing cells in EL4 and LLC1 tumors. Data are averages ± SD cells/field (40x); 5 fields/tumor; n=5 tumors/group. (H) Proliferation of EL4 and LLC1 cells to TGF-β1. Representative results (of 3–5 experiments) are means ± SD (triplicate cultures). (I) LLC1 proliferation to TGF-β is cell density dependent. Representative results (of 3 experiments) are means ± SD of (triplicate cultures). All p values are from Student’s t test.
Identification of myeloid cell-derived signals that modulate tumor cell growth
Results from adoptive transfer experiments suggested that myeloid cell types recruited by EL4 and LLC1 tumor cells might produce paracrine signals that modulate cancer cell growth/survival. To identify such signals, we profiled gene expression in EL4, LLC1 and B16F10 tumors from WT and Gfi1-deficient mice. To distinguish signals derived from the tumor microenvironment from others derived from the tumor cells, we profiled in parallel EL4, LLC1 and B16F10 cell lines from culture. Focusing on genes previously linked to modulation of tumor growth, we measured 57 mRNAs in 10 EL4 tumors (5 each from WT and Gfi1-null mice) and in 15 LLC1 tumors (10 from WT; 5 from Gfi1-null mice) from 2–4 different experiments (Table 1 and Table S1). From this pool, we identified 10 mRNAs expressed at significantly different levels in EL4 and/or LLC1 tumors arising in WT as opposed to Gfi1-null mice (Table 1). Extending analysis of these 10 mRNAs to B16F10 tumors, we found no expression difference between tumors from WT and Gfi1-null mice (Table 1).
Table 1.
Differentially expressed genes in tumors from Gfi1-null and control mice
| Gene | Relative Expressiona | P valueb | Relative Expression | P value | Relative Expression | P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EL4 | WT | KO | LLC1 | WT | KO | B16 | WT | KO | ||||
| Tgif1 | 1 | 0.5(0.4) | 0.9(0.8) | 0.08 | 1 | 5.1(0.2) | 3.1(0.1) | 0.02 | 1 | 2.1(0.4) | 2.5(0.2) | 0.2 |
| Tgif2 | 1 | 4.5 (1.5) | 0.6 (0.2) | 0.02 | 1 | 2.1(0.2) | 2.2(0.1) | 0.6 | 1 | 3.0(0.5) | 3.3(0.2) | 0.6 |
| Dll4 | 1 | 13.8(10.6) | 2.3(2.6) | 0.2 | 1 | 57.2(1.9) | 5.0(1.2) | 0.001 | 1 | 1.8(0.7) | 1.7(1.1) | 0.6 |
| Hey1 | 1 | 30.2(21.0) | 12.2(6.9) | 0.2 | 1 | 15.1(0.7) | 16.4(1.0) | 0.5 | 1 | 2.4(0.3) | 2.9(0.2) | 0.3 |
| Hey2 | 1 | 2.7(1.0) | 1.3(0.8) | 0.06 | 1 | 10.7(0.4) | 1.3(1.0) | 0.05 | 1 | 2.8(2.7) | 3.1(1.7) | 0.9 |
| S100a9 | ndc | 3.6(0.5) | 0.9(0.4) | 0.003 | nd | 1.1(0.1) | 44.9(20.9) | 0.001 | nd | 2.4(2.5) | 0.3(0.3) | 0.08 |
| Lcn2 | nd | 2.7(2.2) | 0.5(0.3) | 0.2 | nd | 1.1(0.1) | 58.9(2.6) | 0.001 | nd | 2.7(4.0) | 1.5(1.8) | 0.2 |
| Il4 | 1 | 1.8(0.4) | 12.3(1.5) | 0.01 | nd | nd | nd | nd | nd | nd | ||
| Il18 | 1 | 3.9(0.2) | 12.3(0.3) | 0.01 | 1 | 1.5(1.3) | 4.7(1.7) | 0.03 | 1 | 5.7(0.2) | 5.9(0.4) | 0.8 |
| Cxcr4 | 1 | 0.22(0.3) | 0.86(0.3) | 0.003 | nd | nd | nd | 1 | 5.7(0.2) | 5.9(0.4) | 0.8 | |
Results from qPCR analysis of RNAs from cell lines (EL4, LLC1, B16) and tumor tissues from WT/Het or Gfi1-null (KO) mice injected with one of these cell lines. Data from tumors in WT/Het and KO mice are expressed as relative mRNA levels (±SD) compared to the injected cell line when expression was detected, or compared to each other when not detected in the injected cell line.
P values were determined by Student’s t-test; n=4–10 tumor tissues per group
not detected
All but one (Cxcr4) of the 10 candidate genes fulfilled the criteria of being likely host induced in that expression was higher in the EL4 or LLC1 tumors compared to the tumor cell line. For the remainder, we looked for myeloid-derived genes preferentially expressed in the WT host that might be linked to suppression of EL4 and stimulation of LLC1 tumor growth. Tgif1 (encodes TGF-β1) and Tgif2 (encodes TGF-β2) fulfilled these criteria (Table 1). TGF-β1/2 is a monocyte and neutrophil product that can inhibit and stimulate cell growth dependent on context: it is a tumor suppressor in early tumor development, but a tumor enhancer in more advanced tumors (3). We confirmed that TGF-β1 mRNA is expressed at higher levels in the LLC1 tumor microenvironment of WT compared to Gfi1-null mice by sorting the CD11b+Gr1+ cells (Figure 2C). Similarly, we confirmed that TGF-β2 mRNA is expressed at significantly higher levels in the EL4 tumor microenvironment of WT mice compared to Gfi1-null mice (not shown). Naïve spleens from WT mice constitutively express higher levels of TGF-β1 (Figure 2D) and TGF-β2 (not shown) mRNA compared to Gfi1-null mice, and naïve CD11b+Gr1+ from WT bone marrow secrete higher levels of TGF-β1 compared to Gfi1-null bone marrow, both constitutively and after activation with M-CSF/CSF1 or GM-CSF/CSF2 (Figure 2E). We also found that levels of the TGF-β signaling mediator pSMAD3 were higher in tumors arising in WT mice than in Gfi1-null mice (Figure 2F,G), indicative of greater TGF-β activity in vivo. We tested the effects of TGF-β on tumor cell growth. TGF-β1 significantly and dose-dependently reduced EL4 proliferation but enhanced LLC1 proliferation (Figure 2H). Increased LLC1 cell proliferation by TGF-β1 is cell density-dependent, suggesting a requirement for cell-cell interaction (Figure 2I); no such cell dose-dependency was observed with EL4 cells (not shown).
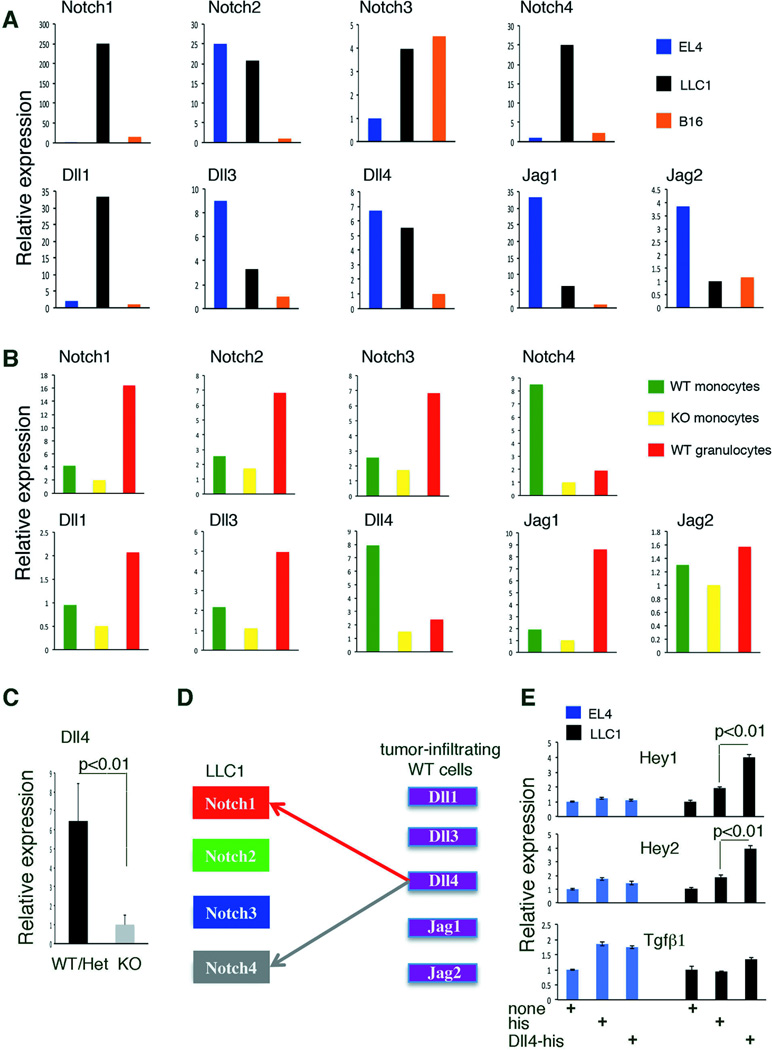
To investigate this cell dose-dependency of LLC1 proliferation to TGF-β, we examined the potential involvement of Notch signaling, because it is induced by cell-to-cell interaction and can establish cooperative crosstalk with TGF-β signaling in other systems (14–16). We found that the Notch ligand Dll4 and the Notch signaling mediator Hey2 were expressed at higher levels in LLC1 tumors from WT compared to Gfi1-KO (Table 1). To identify the cell sources of the differentially expressed Dll4 and Hey2, we characterized Notch receptors/ligands expression in tumor cells and myeloid cells. LLC1 cells expresses Notch1 and Notch4 receptors mRNAs at higher levels than the EL4 and B16F10 cells (Figure 3A); primary WT monocytes sorted from bone marrow express higher levels of Dll4 mRNA (Notch1 and Notch4 ligand) than Gfi1-null monocytes (Figure 3B). LLC1 cells also express endogenous Dll1 mRNA (Notch1/2/3 ligand) and Dll4 mRNA (Figure 3A). We sorted WT and Gfi1-null CD11b+Ly6C+Ly6G− cells from LLC1 tumor cell suspensions to measure expression levels of Notch1, Notch4, Dll4, Hey1 and Hey2 (Figure S3A). LLC1 tumor-infiltrating CD11b+Ly6C+Ly6G− cells from WT mice expressed more Dll4 than this population sorted from Gfi1-null mice (Figure 3C), and more than LLC1 cells from culture (Figure S3B). Notch1 and Notch4 were expressed in LLC1 tumor-infiltrating CD11b+Ly6C+Ly6G− cells from WT mice and Gfi1-null mice (Figure S3B) at somewhat lower levels than found in LLC1 cells (Figure S3B). Since Hey1 and Hey2 mRNAs were at the limit of detection in tumor-infiltrating CD11b+Ly6C+Ly6G− cells from WT and Gfi1-null mice (not shown), we concluded that Hey2 mRNA detected at higher levels in LLC1 tumor tissues from WT as opposed to KO mice was likely tumor-cell derived. Collectively, these results suggested a model in which Dll4-expressing tumor-infiltrating WT myeloid cells, stimulate Notch1 and Notch4 signaling in LLC1 cells inducing Hey2 expression (Figure 3D and Figure S3C). Supporting this model, immobilized Dll4-his specifically induced Hey1 and Hey2 expression in LLC1 cells, but not the expression of TGF-β (Figure 3E). By contrast, Dll4 did not induce Hey1 and Hey2 expression in EL4 cells, which express Notch1 and Notch4 at considerably lower levels than LLC1 cells (Figure 3E).
Figure 3.
Tumor-infiltrating myeloid cells express Dll4, which activates Notch signaling in LLC1 tumor cells that express Notch1 and Notch4. (A,B) Notch receptors and ligands expression in tumor cell lines (A) and in CD11b+Ly6C+Ly6G−/CD11b+Ly6C+Ly6G+ populations (B). Data from qPCR are averages from triplicates measurements (variation <12%); the sorted populations are pools from 3 mice per group. (C) Dll4 expression in CD11b+Gr1+ cells infiltrating LLC1 tumors. Data are averages ± SD; n=3 mice/group. (D) A model for tumor/tumor-infiltrating myeloid cell interaction. (E) Hey1 and Hey 2 expression in tumor cells activated with immobilized Dll4-his; controls: uncoated and His-coated wells. Data from qPCR are averages ± SD; n=4–5. P values are from Student’s t test.
Crosstalk between Notch and TGF-β signaling regulates carcinoma cell growth
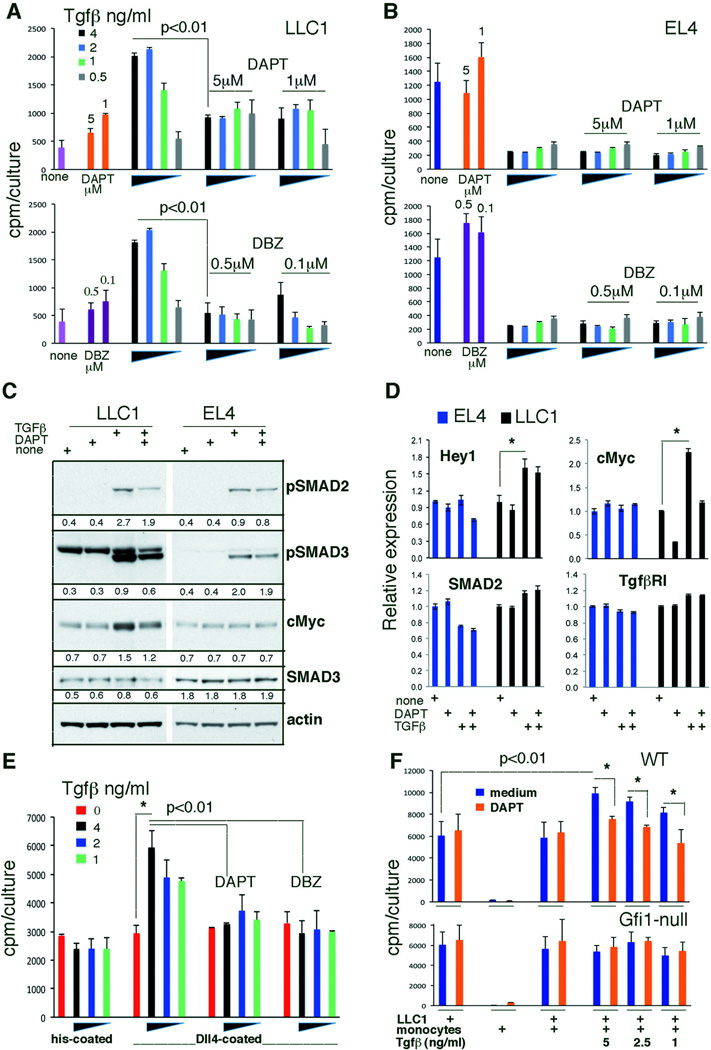
Next, we examined whether Notch/Hey2 signaling in LLC1 cells, attributable at least in part to activation by WT myeloid cell-derived Dll4, modulates TGF-β signaling and function in LLC1 cells. The Notch signaling inhibitors DAPT and DBZ reduced TGF-β-induced LLC1 proliferation (Figure 4A) but minimally affected TGF-β-induced repression of EL4 proliferation (Figure 4B). These results support a functional requirement for Notch signaling in sustaining TGF-β-induced LLC1 growth stimulation.
Figure 4.
Dll4/Notch and TGF-β signaling cooperate to promote LLC1 cell growth. (A,B) LLC1 (A) and EL4 (B) proliferation to TGF-β1 with or without the inhibitors DAPT or DBZ; representative of 5 experiments. Data are cpm averages ± SD of triplicate cultures. (C) Modulation of SMADs phosphorylation and cMyc protein levels in tumor cells by TGF-β1 with or without DAPT. Representative results; band intensity ratios relative to actin are shown. (D) Relative gene expression in tumor cells cultured with TGF-β1 (2ng/ml/48 hr) alone or with DAPT (1µM). Data are averages ± SD; n=4–5 replicates; *p<0.01. (E) Representative data (4 experiments) of TGF-β1-induced LLC1 cell proliferation with or without DAPT (1µM) or DBZ (0.1µM); wells coated with Dll4-histidine or control histidine. Data are cpm averages ± SD of triplicate cultures. *p<0.01. (F) Representative results (3 experiments) of TGF-β1 (0–5ng/ml)-induced LLC1 cell proliferation with or without addition of WT or KO CD11b+Ly6C+Ly6G− cells (ratio 10:1); DAPT 1µM. P values are from Student’s t-test.
DAPT and DBZ block early steps in the Notch signaling cascade by preventing the gamma-secretase-dependent proteolytic cleavage of Notch intracellular domain. To investigate points of potential integration of Notch and TGF-β signaling, we focused on SMADs phosphorylation induced by TGF-β binding to typeI and typeII serine-threonine kinase receptors. SMAD2 and SMAD3 receptor-regulated SMADs are recognized by Type I TGF-β receptors, which are expressed by EL4 and LLC1 cells (see below). We found that TGF-β similarly induces the phosphorylation of SMAD2 and SMAD3 in LLC1 and EL4 cells, despite the different biological outcomes (Figure 4C). However, the Notch inhibitor DAPT reduces this phosphorylation in LLC1, but not in EL4 cells (Figure 4C), indicating that TGF-β-induced SMADs phosphorylation is dependent, in part, upon Notch signaling in LLC1, not EL4, cells. This crosstalk of TGF-β and Notch signaling pathways in LLC1 cells is consistent with the previously recognized binding of the active Notch intracellular domain to SMAD2/3 (14, 15). Based on experiments showing that active Notch promotes cMyc transcription (22) whereas TGF-β inhibits cell cycle progression by transcriptional and non-transcriptional cMyc repression in many cell types (23, 24), we examined cMyc expression. We found that cMyc levels increase in LLC1 cells after TGF-β activation and that the Notch inhibitor DAPT reduces this effect, whereas cMyc levels are unaffected by TGF-β and/or DAPT in EL4 cells (Figure 4C). This provides additional evidence for cooperative crosstalk between TGF-β and Notch signaling resulting in increased cMyc expression in LLC1 cells. Strengthening this evidence, we found that TGF-β induces cMyc mRNA expression in LLC1 cells, which is reduced by DAPT, and that TGF-β promotes expression of the Notch target gene Hey1 (and Hey2, not shown) while minimally affecting expression of SMAD2, SMAD3 (not shown) and TbRI (Figure 4D). TGF-β did not change expression levels of Hey1 or cMyc in EL4 cells (Figure 4D).
Since Hey2 mRNA levels are 8–11-fold higher in LLC1 tumors from WT mice compared to LLC1 tumors from Gfi1-null mice and to LLC1 cells from culture (Table 1), we hypothesized that this might be due to a contribution by WT tumor-infiltrating monocytes that express Dll4 at higher levels than Gfi1-null monocytes (Figure 3C, Figure S3B). We therefore investigated whether increased Hey2 levels in LLC1 tumors are attributable to Dll4-expressing WT monocytes infiltrating the tumor, and whether this increased Notch signaling accelerates LLC1 tumor cell growth. To mimic the monocyte effect, we first used immobilized Dll4 to modulate Notch signaling intensity in LLC1 cells seeded at low density to reduce cell-cell contact expected to activate Notch through membrane-anchored Dll1 and Dll4 ligands. When activated with Dll4-his, low-density LLC1 cells responded to TGF-β with increased proliferation, which was absent from control cultures lacking Dll4-his (Figure 4E). DAPT and DBZ both reduced this proliferation (Figure 4E), providing evidence of functional cooperation between TGF-β and Notch signaling in promoting LLC1 proliferation. We then tested WT monocytes, which express Dll4. TGF-β enhanced the proliferation of low-density LLC1 cells co-cultured with bone marrow CD11b+Ly6C+Ly6G− cells from WT, but not Gfi1-null mice. Hey1 mRNA levels were significantly (P<0.05) higher in co-cultures of LLC1 cells with WT monocytes compared to co-cultures of LLC1 cells with KO monocytes, confirming that WT monocytes activate Notch in LLC1 cells better than KO monocytes. DAPT reduced this stimulatory effect of WT monocytes (Figure 4F). Collectively, these results show that TGF-β enhances LLC1 tumor cell growth in the presence of Notch signaling from either LLC1 cell-cell contact and/or through interaction with myeloid cells expressing Dll4.
Contribution of Notch signaling to carcinoma progression
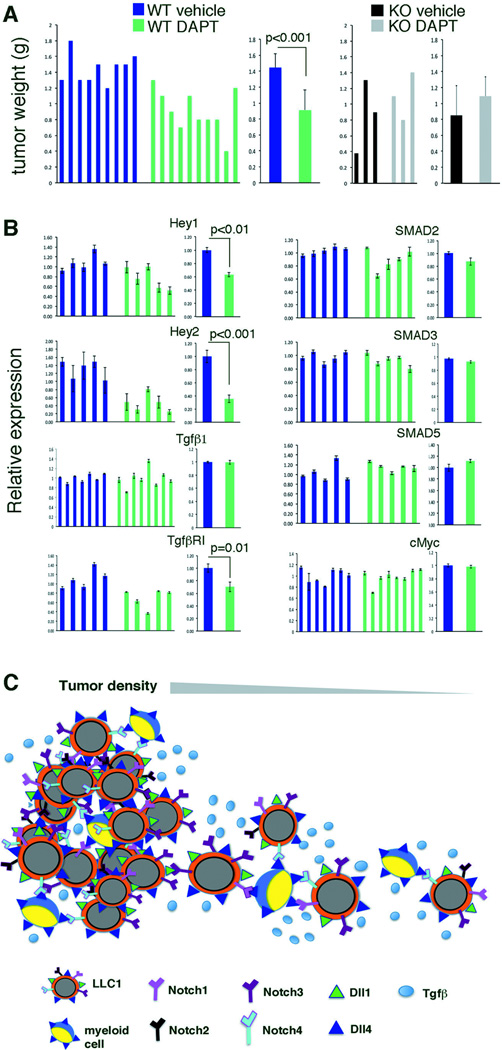
Next, we investigated whether Notch signaling contributes to increased LLC1 tumor progression. WT mice bearing LLC1 tumors showed a significant reduction in tumor progression when treated with the Notch inhibitor DAPT, whereas KO mice did not (Figure 5A, Figure S4). Treatment with DAPT reduced Hey1 and Hey2 mRNA expression in the tumor tissue, indicative of Notch signaling inhibition in vivo (Figure 5B). Instead, mRNA levels of SMAD2, SMAD3 and TGF-β were similar in untreated and DAPT-treated tumors (Figure 5B). Although this was not predicted by the results of short-term experiments in vitrowe found that TGF-βR1 mRNA levels were reduced in DAPT-treated tumors and that cMyc mRNA levels were similar in DAPT-treated mice compared to controls (Figure 5B). This could be explained by observations linking Notch and its downstream mediator RPB-jk to the regulation of TGF-β receptors expression (25), and the complexities of Myc transcriptional regulation by many signaling pathways besides Notch and TGF-β (26). Reduced tumor growth with DAPT treatment could not be attributed to reduced accumulation of pro-tumorigenic myeloid cells, since similar infiltration of CD11b+Ly6C+ cells was present in LLC1 tumors treated or not treated with DAPT (not shown). Overall, these results support a model in which WT myeloid cells recruited to the tumor accelerate tumor progression by secreting mitogenic TGF-β and enhancing TGF-β signaling in the tumor cells via expression of the ligand Dll4, which activates Notch1 and Notch4 (Figure 5C). We envision that the contribution of myeloid cells is most critical at the invasive edge of the tumor where myeloid cells accumulate (27), providing an opportunity for myeloid-tumor cell interactions resulting in Notch activation. Tumor cells are mobile at the tumor invasive edge and tumor cell density is reduced (28), likely limiting Notch signaling from tumor-intrinsic cell-to-cell interactions (Figure 5C).
Figure 5.
Notch signaling promotes LLC1 tumor progression. (A) Groups of littermate mice were treated i.p. every other day (7 total injections) with the Notch inhibitor DAPT (10mg/kg) or vehicle alone, beginning 24 hours after inoculation of LLC1 cells. Tumors were removed 16 days after LLC1 inoculation. Individual tumor weights and average weight ± SD. (B) Relative gene expression in LLC1 tumors after treatment with vehicle alone or DAPT. Data are from individual tumors and group averages ± SD. (C) Model showing the contribution of myeloid cells to Notch signaling and TGF-β function in tumor cells. Where the tumor is dense, tumor intrinsic cell-to-cell interactions are most critical inducers of Notch signaling: tumor cells express the Notch ligands Dll1 and Dll4 and all Notch receptors. At the infiltrating edge of the tumor, where tumor density is low, Dll4-expressing tumor-infiltrating myeloid cells activate Notch1 and/or Notch4 signaling in the tumor cells. Notch signaling enhances TGF-β signaling and growth stimulation in the tumor cells. All p values are from Student’s t test.
Prompted by the current findings showing a role of tumor-infiltrating monocytes in enhancing TGF-β/Notch signaling and LLC1 tumor progression, we sought evidence for this link in clinical samples. We queried The Cancer Genome Atlas (TGCA), focusing on head and neck squamous cell carcinoma, which resembles subcutaneous LLC1 in having significant monocyte infiltration correlating with increased tumor aggressiveness and reduced survival, increased expression CCL2, deregulated TGF-β and Notch signaling components and evidence of active TGF-β signaling (29–32). We also examined lung squamous cell carcinoma based on the lung carcinoma derivation of LLC, albeit LLC1 is a cloned line adapted to culture. Remarkably, head and neck squamous cell carcinomas display significantly greater expression of Dll4, Notch4, Hey1, TGF-β1 and TGF-βRI, compared to normal tissue, much alike LLC1 tumors in WT mice (Figure 6A). By contrast, lung squamous cell carcinoma display significantly lower expression of Dll4, Notch4, TGF-β1, TGF-β2, TGF-βR1 and TGF-βR2 compared to normal tissue (Figure 6A). This discordant pattern of gene expression is strengthened by comparing ratios of tumor/normal control from paired samples of individual patients (Figure 6B). The gene expression signature emerging from this analysis highlights the potential for activation of TGF-β/TGF-βR1 and Dll4/Notch4/Hey1 signaling in head and neck squamous cell carcinoma and contribution to tumor progression. Additional studies are planned to measure TGF-β/TGF-βR1 and Notch signaling activity in head and neck squamous cell tumors and correlate signaling levels with disease outcome.
Figure 6.
Analysis of gene expression in human head and neck squamous cell carcinomas reveals similarities to gene expression in mouse LLC1 tumors. (A) Gene expression in head and neck normal tissue (n=37) and squamous carcinoma (n=303); and in lung normal tissue (n=50) and squamous carcinoma (n=369). C=control tissue; T=tumor. Results from RNAseq (The Cancer Genome Atlas) are expressed as transcript expression levels; data distribution is displayed as box plots (the box limits first and third quartiles; band inside the box is median value; the vertical dotted lines indicate variability; outlier values shown as dots. Statistical significance of group differences are from Mann-Whitney U-test. (B) Distinct patterns of gene expression in head and neck and lung squamous cell carcinomas. Results depict ratios of gene expression in tumor tissue and corresponding normal tissue in paired patient samples with head and neck (blue bars; n=35) or lung (red bars; n=50) squamous cell carcinomas.
Discussion
One of the emerging challenges to the successful treatment of cancer is the tumor microenvironment that variously contributes to cancer progression through reciprocal communication with the tumor cells. In addressing this challenge, our work unveils the Notch and TGF-β signaling pathways as functional partners in a network of cancer cells and tumor-infiltrating CD11b+Ly6C+Ly6G− cells that promotes cancer cell growth and tumor progression. This new understanding offers an opportunity for the targeting of tumors where TGF-β and Notch signaling are linked pro-tumorigenic partners. Head and neck squamous cell carcinoma may represent such setting as we find that this tumor generally displays increased expression of TGF-β and its receptors, in conjunction with evidence of Notch signaling activity.
TGF-β, a product of myeloid-lineage cells in many tumor microenvironments, plays a well-recognized role in tumor progression and resistance to treatment (33, 34). TGF-β has cytostatic and pro-apoptotic effects for normal cells and pre-malignant lesions (33). In advanced cancer, TGF-β promotes tumor progression (33). Such functional change is attributed to inactivating mutations of TGF-ΒR2 and SMAD4 preventing TGF-β tumor-suppressive signaling in gastrointestinal and pancreatic tumors (33, 34). In many other cancer types, including the LLC1 tumor model in our study, TGF-β receptors and SMAD signaling are intact (33). It remains a puzzle how TGF-β can exert contextually different functions. Here we show that crosstalk between the Notch and TGF-β pathways is critical to TGF-β signaling and function as a tumor enhancer in the LLC1 model. Blocking Notch signaling significantly reduces SMADs phosphorylation and cell growth induced by TGF-β. It also slows LLC1 tumor progression, which is accelerated by TGF-β-producing tumor-infiltrating myeloid cells.
TGF-β and Notch signaling converge in regulating a number of developmental processes (35). Notch signaling modulates expression of BMP family members (36) and TGF-β target genes (37). Conversely, TGF-β induces expression of the Notch ligand Jagged1 and target gene Hey1 (38). Interestingly, Notch signal transducers can physically interact with components of the TGF-β signaling pathway contributing more directly to TGF-β and BMP function (14, 39, 15, 16). The present work shows that cross talk between the TGF-β and Notch signaling pathways converge on SMAD2/3 activation, which sustains TGF-β tumor-promoting function.
Our current findings further identify a previously unrecognized role of tumor-infiltrating CD11b+Ly6C+Ly6G− cells as activators of Notch signaling in tumor cells enabling paracrine TGF-β growth stimulation. CD11b+Ly6C+Ly6G− myeloid cells constitute a functionally heterogeneous population (2, 40), which includes tumor-promoting myeloid-derived suppressor cells (MDSCs) with T-cell suppressive functions via expression of inducible forms of nitric oxide and arginase, and in some cases TGF-β (1, 2, 41). MDSCs have been identified in many tumor types (41), including experimental EL4 and LLC1 tumors. In the current study, T-cell immunity is not identified a major contributor to tumor growth modulation, and the tumor-promoting CD11b+Ly6C+Ly6G− cells do not express high-level Nos2 and Arg1 typical of MDSCs. Rater, the cells resemble phenotypically tumor-derived M2-type macrophages (2), which express the Notch ligand Dll4, similar to monocytes exposed to pro-inflammatory signals (42).
Recent reports and the current work show that Notch activity is a contributor to progression of some cancers (43), albeit not all cancers (44–48). It is currently unclear whether Notch-activating signals from the tumor microenvironment contribute to tumor progression. This was suggested in myeloma through tumor-stromal cells interactions (49). Using genetic, biochemical and functional approaches our results show that Dll4-expressing myeloid cells induce Notch stimulation in LLC1 tumor cells, which allows paracrine TGF-β signaling and growth in the tumor cells. This provides evidence for yet another mechanism of tumor progression by tumor-infiltrating myeloid cells. Our results argue that this function of tumor-infiltrating myeloid cells is most critical at the locally invasive edge of the tumor where single cells migration is a principal mode of invasion (28), and there is opportunity for myeloid-tumor cells interaction (27, 50).
In conclusion, our results provide mechanistic insights into TGF-β tumor-promotion by linking TGF-β and Notch signaling, and disclose an activating Dll4/Notch signaling axis linking tumor-promoting myeloid cells and tumor cells. This raises the possibility of dual targeting of Notch and TGF-β signaling in cancers where TGF-β plays Notch-dependent pro-tumorigenic functions, as in head and neck squamous cell carcinoma, which expresses components of the TGF-β and Notch signaling pathways at abnormally high levels. It is fortunate that inhibitors of TGF-β and Notch signaling (43) have been developed and hold promise in clinical development.
Supplementary Material
Acknowledgements
We thank the animal care personnel; Dr. J. Zhou, I-I Chen, M. Di Prima, G. Ghilardi and S. Park for their help; Dr. D. Lowy for research support; Dr. M. Elkin and members of the LCO for sharing reagents and insightful discussions.
Grant support
The work was supported by the Intramural Research Program of CCR/NCI/NIH and NINDS/NIH; H.O. was supported in part by Grant-in-Aid for Scientific Research (S2207), JSPS.
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose
References
- 1.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012 Mar 20;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008 Jul 24;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012 Jul 6;150(1):165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011 Jun;1(1):54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature. 2005 Dec 15;438(7070):967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 6.Chen Q, Zhang XH, Massague J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 2011 Oct 18;20(4):538–549. doi: 10.1016/j.ccr.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shojaei F, Wu X, Zhong C, Yu L, Liang XH, Yao J, et al. Nature. 2007 Dec 6;450(7171):825–831. doi: 10.1038/nature06348. [DOI] [PubMed] [Google Scholar]
- 8.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006 Oct;6(10):764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 9.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012 Oct;11(10):790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz LH, Li Y, Chen JS, Munoz NM, Majumdar A, Chen J, et al. Targeting TGF-beta signaling in cancer. Expert Opin Ther Targets. 2013 Jul;17(7):743–760. doi: 10.1517/14728222.2013.782287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006 Mar;6(3):227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 12.Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003 Jan;18(1):109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 13.Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW, et al. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet. 2002 Mar;30(3):295–300. doi: 10.1038/ng831. [DOI] [PubMed] [Google Scholar]
- 14.Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, Lendahl U, et al. Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol. 2003 Nov 24;163(4):723–728. doi: 10.1083/jcb.200305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh F, Itoh S, Goumans MJ, Valdimarsdottir G, Iso T, Dotto GP, et al. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. Embo J. 2004 Feb 11;23(3):541–551. doi: 10.1038/sj.emboj.7600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moya IM, Umans L, Maas E, Pereira PN, Beets K, Francis A, et al. Stalk cell phenotype depends on integration of Notch and Smad1/5 signaling cascades. Dev Cell. 2012 Mar 13;22(3):501–514. doi: 10.1016/j.devcel.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De La Luz Sierra M, Gasperini P, McCormick PJ, Zhu J, Tosato G. Transcription factor Gfi-1 induced by G-CSF is a negative regulator of CXCR4 in myeloid cells. Blood. 2007 Oct 1;110(7):2276–2285. doi: 10.1182/blood-2007-03-081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Jankovic D, Grinberg A, Guo L, Paul WE. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18214–18219. doi: 10.1073/pnas.0608981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segarra M, Ohnuki H, Maric D, Salvucci O, Hou X, Kumar A, et al. Semaphorin 6A regulates angiogenesis by modulating VEGF signaling. Blood. 2012 Nov 8;120(19):4104–4115. doi: 10.1182/blood-2012-02-410076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stathopoulos GT, Psallidas I, Moustaki A, Moschos C, Kollintza A, Karabela S, et al. A central role for tumor-derived monocyte chemoattractant protein-1 in malignant pleural effusion. J Natl Cancer Inst. 2008 Oct 15;100(20):1464–1476. doi: 10.1093/jnci/djn325. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Cambronero J, Horn J, Paul CC, Baumann MA. Granulocyte-macrophage colony-stimulating factor is a chemoattractant cytokine for human neutrophils: involvement of the ribosomal p70 S6 kinase signaling pathway. J Immunol. 2003 Dec 15;171(12):6846–6855. doi: 10.4049/jimmunol.171.12.6846. [DOI] [PubMed] [Google Scholar]
- 22.Satoh Y, Matsumura I, Tanaka H, Ezoe S, Sugahara H, Mizuki M, et al. Roles for c-Myc in self-renewal of hematopoietic stem cells. J Biol Chem. 2004 Jun 11;279(24):24986–24993. doi: 10.1074/jbc.M400407200. [DOI] [PubMed] [Google Scholar]
- 23.Alexandrow MG, Moses HL. Transforming growth factor beta 1 inhibits mouse keratinocytes late in G1 independent of effects on gene transcription. Cancer Res. 1995 Sep 1;55(17):3928–3932. [PubMed] [Google Scholar]
- 24.Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002 Jul 12;110(1):19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 25.Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004 Jan 1;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang CV. MYC on the path to cancer. Cell. 2012 Mar 30;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wels J, Kaplan RN, Rafii S, Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes Dev. 2008 Mar 1;22(5):559–574. doi: 10.1101/gad.1636908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010 Jan 11;188(1):11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, et al. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene. 2000 Jun 29;19(28):3220–3224. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]
- 30.Marcus B, Arenberg D, Lee J, Kleer C, Chepeha DB, Schmalbach CE, et al. Prognostic factors in oral cavity and oropharyngeal squamous cell carcinoma. Cancer. 2004 Dec 15;101(12):2779–2787. doi: 10.1002/cncr.20701. [DOI] [PubMed] [Google Scholar]
- 31.Rothenberg SM, Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. J Clin Invest. 2012 Jun 1;122(6):1951–1957. doi: 10.1172/JCI59889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie W, Aisner S, Baredes S, Sreepada G, Shah R, Reiss M. Alterations of Smad expression and activation in defining 2 subtypes of human head and neck squamous cell carcinoma. Head Neck. 2013 Jan;35(1):76–85. doi: 10.1002/hed.22924. [DOI] [PubMed] [Google Scholar]
- 33.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012 Oct;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L, Pang Y, Moses HL. TGF-beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010 Jun;31(6):220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008 Jun;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin Cell Dev Biol. 2005 Dec;16(6):663–672. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Sjolund J, Bostrom AK, Lindgren D, Manna S, Moustakas A, Ljungberg B, et al. The notch and TGF-beta signaling pathways contribute to the aggressiveness of clear cell renal cell carcinoma. PLoS One. 2011;6(8):e23057. doi: 10.1371/journal.pone.0023057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. Embo J. 2004 Mar 10;23(5):1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K, Ibanez CF, et al. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development. 2003 Dec;130(24):6089–6099. doi: 10.1242/dev.00834. [DOI] [PubMed] [Google Scholar]
- 40.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010 Apr 2;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012 Apr;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fung E, Tang SM, Canner JP, Morishige K, Arboleda-Velasquez JF, Cardoso AA, et al. Delta-like 4 induces notch signaling in macrophages: implications for inflammation. Circulation. 2007 Jun 12;115(23):2948–2956. doi: 10.1161/CIRCULATIONAHA.106.675462. [DOI] [PubMed] [Google Scholar]
- 43.Espinoza I, Miele L. Notch inhibitors for cancer treatment. Pharmacol Ther. 2013 Aug;139(2):95–110. doi: 10.1016/j.pharmthera.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Lin L, Shanker A, Malhotra A, Yang L, Dikov MM, et al. Resuscitating cancer immunosurveillance: selective stimulation of DLL1-Notch signaling in T cells rescues T-cell function and inhibits tumor growth. Cancer Res. 2011 Oct 1;71(19):6122–6131. doi: 10.1158/0008-5472.CAN-10-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sriuranpong V, Borges MW, Ravi RK, Arnold DR, Nelkin BD, Baylin SB, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001 Apr 1;61(7):3200–3205. [PubMed] [Google Scholar]
- 46.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004 Oct 8;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 47.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996 Jul;122(7):2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 48.Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, et al. Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basal-like breast cancer. Cancer Cell. 2012 May 15;21(5):626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jundt F, Probsting KS, Anagnostopoulos I, Muehlinghaus G, Chatterjee M, Mathas S, et al. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004 May 1;103(9):3511–3515. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- 50.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012 Mar;33(3):119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.