Abstract
We show that a higher incidence of campylobacteriosis is found in young children (age, <5 years) living in rural, compared with urban, areas. Association of this difference with particular animal sources was evaluated using multilocus sequence typing. This evaluation was achieved by comparing Campylobacter isolates originating from these children, retail poultry, and a range of animal sources by use of source attribution and phylogenetic analysis methods. The results indicate that chicken is a major source of infection in young urban children, although not in their rural counterparts, for which ruminant and other avian sources are more important.
Campylobacter infection in humans is the leading cause of recognized bacterial gastroenteritis in the developed world, and, in 2006, it accounted for 12.7 and 95.3 cases per 100,000 individuals in the United States and Scotland, respectively [1, 2]. Infection is commonly thought to result from foodborne pathways, particularly chicken [3], with some cases associated with environmental exposure (e.g., farm animals [4] and water [5]). Campylobacter organisms are widely found in both wild and domesticated farm animals, and, with multiple pathways for infection in humans, the source of most individual infections is unknown [3]. The relative importance of each source therefore remains unclear.
Spatial epidemiology is the study of the spatial distribution of disease and associated risk factors. The spatial epidemiologic profile of human campylobacteriosis suggests that its incidence is higher in young children in rural areas of Denmark than in young children in more highly populated areas of Denmark [6]. The reasons for this observation are unknown. However, it is likely that environmental sources of Campylobacter organisms in rural areas play a role in human infection; however, associations between spatial epidemiology and molecular typing data have not yet been robustly applied.
Human infections can be attributed to specific sources by microbial subtyping source attribution. In this approach, isolates are obtained from infected humans and from potential sources of infection and are then categorized as various subtypes by use of typing information. The relative contributions of different sources to human disease are inferred from the quantification of subtype sharing between the isolates obtained from humans and those obtained from the sources of infection. Reliable bacterial subtyping methods, based on serotype and phage-type determination, allow Salmonella infections in humans to be attributed to particular sources [7]. For Campylobacter infections, however, source attribution has been difficult to achieve because of the unavailability of an agreed-upon typing methodology, the lack of clonality, and the extent of recombination within the genus.
Multilocus sequence typing (MLST) [8] is a potentially useful subtyping method for use in source-attribution studies of Campylobacter infection [9]. MLST is a DNA sequence–based subtyping method that yields reliable, transferable data describing bacterial pathogens. Indeed, the source specificity of Campylobacter strains subtyped using MLST is evident in isolates recovered from cattle, sheep, and poultry [9] and from other avian sources, including gulls and pigeons [10].
Northeast Scotland (i.e., Grampian) is a suitable area in which to study campylobacteriosis in humans (including young children <5 years of age), because of the demographic characteristics and the range of risk factors to which the population is exposed. The urban area of the city of Aberdeen has a population of ~212,500 (including 10,500 children <5 years of age) and adjoins a predominantly rural area (size, 8700 km2) with a comparable population of ~328,000 (including 18,300 children <5 years of age). The environmental risk factors for Campylobacter infection facing the rural population of Grampian (e.g., direct or indirect contact with farm animals) are representative of rural areas with extensive mixed farming. In addition, ~10% of households in rural areas have private water supplies (mainly springs or wells) that are not part of public water treatment schemes and are located adjacent to fields with farm animals.
In the present study, we investigated the differences in cases of human campylobacteriosis in rural and urban populations in Grampian, as stratified by age. We then focused on young children (previously reported to have a high incidence of Escherichia coli O157 in rural areas [11]) by applying MLST to characterize their Campylobacter isolates. We visualized the differences in the genetic relatedness between human isolates obtained from urban and rural cohorts and a database of isolates from known animal sources. Finally, we used a quantitative source-attribution approach to determine whether the sources of infection for these urban and rural young children are different.
Methods
Anonymous data on human campylobacteriosis for the years 2000–2006 (5019 cases comprising 112 urban and 316 rural children <5 years of age), which included patient age group data, were collected from National Health Service Grampian, Aberdeen, United Kingdom. Cases were categorized as occurring in urban (the city of Aberdeen ) or rural (population density, <200 km−2) areas and were segregated by age group. Annual incidence rates were calculated using population data from the 2001 Scottish Census [12]. One-sample t tests were performed to determine whether the ratio of rural/urban case incidences was significantly different from 1. All Campylobacter isolates that were available and viable from cases involving 225 rural children and 85 urban children <5 years of age during 2000–2006 were obtained from the National Health Service Grampian Microbiology Laboratories and were typed by 7-locus MLST performed using standard methods and published primers, as described elsewhere [13].
MLST profiles of Campylobacter isolates (table 1) previously obtained from retail poultry (n = 277) [13], cattle (n = 104), sheep (n = 97), pigs (n = 27), unidentified birds (n = 27), duck (n = 12), geese (n = 17), gulls (n = 48), and feral pigeons (n = 71) [10] were used as the reference database for sources of infection. These isolates were collected predominantly from Grampian from June 2005 through September 2006.
The clonal genealogy of sequence types (STs) was estimated using a model-based approach for determining bacterial microevolution: ClonalFrame software (version 1.0; http://www.xavierdidelot.xtreemhost.com/clonalframe.htm [14]. This approach calculates clonal relationships with improved accuracy, compared with standard phylogenetic techniques for Campylobacter organisms, because it differentiates between point mutations and recombination events. Recombination in bacteria affects a contiguous sequence of DNA but leaves the remainder of the circular bacterial genome unaffected. The model determines the genealogy that has not undergone recombination and hence, in effect, estimates the extent of the clonal frame. This estimation is achieved by statistical inference using Bayesian methods. Analysis was performed for concatenated sequences representing the STs from the human and animal typed isolates listed above. The program was run with 50,000 burn-in iterations followed by 50,000 data collection iterations. The consensus trees represent combined data from 3 independent runs, with 75% consensus required for inference of relatedness.
Structure software (version 2.2; http://pritchardlab.stanford.edu/structure.html) [15] was used to assign human MLST isolate data to infection sources. This was achieved by initial determination of the genotype frequencies among each of the animal sources, and the uncertainty regarding this determination of genotype frequency results from the number of samples. This information is then used to estimate the source of each human isolate. The output gives the probability of the human isolate coming from each of the animal sources. The methodology used is a modification of that described elsewhere [9]. In brief, a no-admixture model assuming uncorrelated gene frequencies between populations was used. The probability of a human isolate originating from each particular animal source was estimated using Structure software, and estimation of this probability was repeated for all human isolates. The proportion of the source of human isolates from rural and urban areas was then calculated. This calculation was repeated using 10,000 Monte Carlo steps, to determine the average proportion attributed by source and the corresponding 95% confidence intervals (CIs).
Results
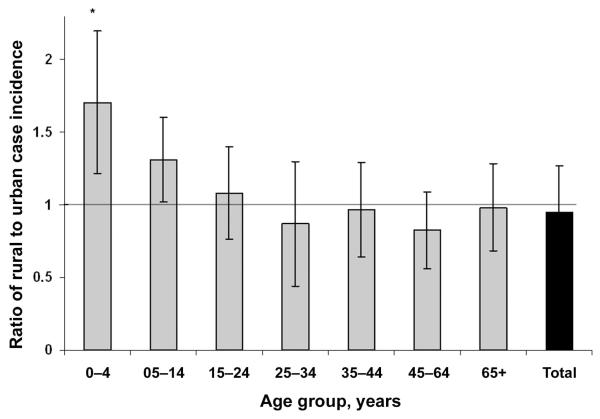
Children <5 years of age had the highest excess incidence of campylobacteriosis in rural (234 cases/100,000 population) versus urban (151 cases/100,000 population) areas (figure 1) (as determined by t test, t6 = 2.78; P = .03). The 6 older age groups all had ratios closer to 1, denoting no clear trend (P > .05).
Figure 1.
The ratio of rural/urban incidences of campylobacteriosis in Northeast Scotland in 2000–2006, as stratified by age. Error bars denote the SEM, and the asterisk denotes the statistical difference from 1, as determined by t test after sequential Bonferroni correction).
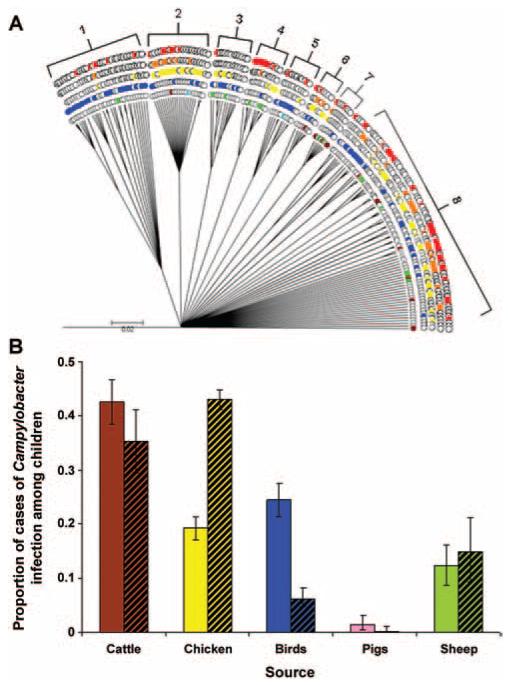
ClonalFrame successfully differentiated Campylobacter jejuni and Campylobacter coli clades, and the largest C. jejuni clade (figure 2A) showed some evidence of heterogeneity in source assignment. For example, subclade 1 is dominated by wild bird STs, and STs from rural children are more common than STs from urban children (5 vs. 1). Subclade 2 is dominated by chicken, with both urban and rural STs represented in similar numbers, despite the presence of fewer urban STs overall. Subclade 8 comprises a genetically diverse range of STs, which are represented across a variety of sources. The largest C. coli clade (data not presented) shows that, although chicken and pig isolates are widely distributed, human isolates are represented by only 3 STs.
Figure 2.
A, ClonalFrame tree of the main Campylobacter jejuni clade showing distribution of rural and urban young children with animal source sequence types (STs) (rural children are shown in red; urban children, orange; chicken, yellow; avian sources excluding chicken, blue; cattle, brown; pigs, pink; sheep, green; cattle and avian sources, gray; and pets, light blue). B, Attribution of the source of infection in children 0–4 years of age in rural areas and urban areas (bars with black diagonal lines), by probabilistic assignment of multilocus sequence typing allele frequency by use of Structure software.
According to the Structure analyses performed at the allele level (figure 2B), the 4 highest proportions of isolates recovered from young children (<5 years of age) in rural areas were attributed to cattle (42%), nonchicken avian sources (24%), chicken (19%), and sheep (12%). The same analysis revealed a different order for the attribution of isolates recovered from young children (<5 years of age) in urban areas: chicken (43%), cattle (35%), sheep (15%), and nonchicken avian sources (6%). Few isolates recovered from young children (<1.4%) were attributed to pigs in both rural and urban areas.
Discussion
The greater incidence of Campylobacter infection in young children from rural versus urban areas is consistent with the previous finding of a higher incidence of infection among young rural children in Denmark [6]. This increased incidence could not be explained by consumption of chicken or by foreign travel. Consequently, other environmental sources of infection (e.g., direct contact with farm animals or their feces or consumption of contaminated water) are likely to be more important in rural areas. Rural areas of Grampian have a high density of private water supplies (10% of the population), a large proportion of which are untreated. Furthermore, Grampian has intensive cattle (n = 364,500) and sheep (n = 632,500) farming. The high incidence of Campylobacter infection reported in young rural children in the present study is also consistent with the findings of epidemiologic studies of E. coli O157 [11]. However, the higher incidence of E. coli O157 in adults in rural, compared with urban, areas in Grampian is not observed in Campylobacter infection. This finding suggests that, in the adult population, Campylobacter infection occurring through the foodborne route is more important than infection occurring through contact with animals and water; however, further research is required to validate this finding.
In the present study, attribution of the source of infection in children shows that avian (nonpoultry) sources are more important in rural areas (24.4%) than in urban areas (6.4%). Interestingly, the importance of ruminants is high for both rural and urban cohorts. Surveys of contamination of red meat by Campylobacter organisms have found low values of prevalence and bacterial concentrations, and red meats have not been widely reported as a risk factor in case-control studies. These findings suggest that infection with ruminant strains is likely to occur via an indirect route (e.g., waterborne transmission or direct contact with animals, produce, or raw milk [although the latter cannot legally be sold in Scotland]) that cannot be quantified using the source attribution approach applied in the present study. The Structure attribution analysis suggests that the incidence of porcine sources of infection was low across Grampian, despite the relatively high numbers of pigs (n = 278,000) reared in rural areas. Because Campylobacter isolates recovered from pigs are dominated by C. coli, the low numbers of C. coli infections in humans in our data set results in a low proportion of cases attributable to infection acquired from pigs. Furthermore, all but one of the pig C. coli STs was absent from human isolates (supplementary data).
Genotyping performed with the use of ClonalFrame enabled the data to be visualized phylogenetically rather than by frequency-based Structure analysis. It suggested that there were some differences in the sources of infection between rural and urban children in subclade 1 but that this did not appear to be evident in other subclades. The use of concatenated sequence data, rather than the ST identifier, enables the genetic relatedness of strains to be determined at the highest resolution possible by use of MLST data. Source attribution by use of Structure was performed at the allele level, because Campylobacter alleles are evolutionarily more dynamic than STs as recombination and because mutations can yield source-specific regions of DNA that do not always cause a change of ST or clonal complex. As a result, the resolution of attribution using alleles is more robust. It should be noted that source misclassification can occur using Structure, when the frequencies of particular alleles between sources are similar and when there is not sufficient genetic information to discriminate between sources. Indeed, some Campylobacter strains are found across several sources. The current method of using 7 loci mitigates against this [9], but using additional loci is likely to improve robustness.
These findings demonstrate that the main sources of Campylobacter infections in young children in Grampian depend on residence location. Furthermore, the data indicate that animal sources excreting Campylobacter organisms into the environment can explain the increased incidence in rural areas and that food (i.e., retail chicken) is a larger source of infection in urban dwellers. Interventions that reduce Campylobacter organisms in highly contaminated foods, such as retail poultry [13], will help to reduce morbidity; however, in rural areas, educational campaigns that raise the awareness of environmental sources of Campylobacter organisms and the resulting risk of infection among young children may be useful. Hence, public health authorities should target their advice accordingly.
Supplementary Material
Acknowledgments
This publication made use of the Campylobacter jejuni and Campylocbacter coli MLST Home Page [16] developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford.
Financial support: The Food Standards Agency, Scotland.
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Centers for Disease Control and Prevention Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2006. MMWR Morb Mortal Wkly Rep. 2007;56:336–9. [PubMed] [Google Scholar]
- 2.Locking M, Browning L, Smith-Palmer A, Brownlie S. Gastro-intestinal and foodborne infections. HPS Weekly Report. 2007;41:3–4. [Google Scholar]
- 3.Rodrigues LC, Cowden JM, Wheeler JG, et al. The study of infectious intestinal disease in England: risk factors for cases of infectious intestinal disease with Campylobacter jejuni infection. Epidemiol Infect. 2001;127:185–93. doi: 10.1017/s0950268801006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley K, Jones K. Cattle and sheep farms as reservoirs of Campylobacter. J Appl Microbiol. 2003;94:104S–13S. doi: 10.1046/j.1365-2672.94.s1.12.x. [DOI] [PubMed] [Google Scholar]
- 5.Furtado C, Adak GK, Stuart JM, Wall PG, Evans HS, Casemore DP. Outbreaks of waterborne infectious intestinal disease in England and Wales, 1992–5. Epidemiol Infect. 1998;121:109–19. doi: 10.1017/s0950268898001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ethelberg S, Simonsen J, Gerner-Smidt P, Olsen KE, Molbak K. Spatial distribution and registry-based case-control analysis of Campylobacter infections in Denmark, 1991–2001. Am J Epidemiol. 2005;162:1008–15. doi: 10.1093/aje/kwi316. [DOI] [PubMed] [Google Scholar]
- 7.Hald T, Vose D, Wegener HC, Koupeev T. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 2004;24:255–69. doi: 10.1111/j.0272-4332.2004.00427.x. [DOI] [PubMed] [Google Scholar]
- 8.Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–5. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy ND, Colles FM, Dingle KE, et al. Host-associated genetic import in Campylobacter jejuni. Emerg Infect Dis. 2007;13:267–72. doi: 10.3201/eid1302.060620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogden ID, Forbes KJ, Gormley FJ, et al. Prevalence, enumeration and molecular subtyping of Campylobacter from environmental and retail food sources in Scotland—a CaMPS study. Zoonoses Pub Health. 2007;54:43. [Google Scholar]
- 11.Strachan NJ, Dunn GM, Locking ME, Reid TM, Ogden ID. Escherichia coli O157: burger bug or environmental pathogen? Int J Food Microbiol. 2006;112:129–37. doi: 10.1016/j.ijfoodmicro.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 12.General Register Office for Scotland [Accessed 30 January 2009];Census. Available at: http://www.gro-scotland.gov.uk/census/index.html.
- 13.Gormley FJ, Macrae M, Forbes KJ, Ogden ID, Dallas JF, Strachan NJ. Has retail chicken played a role in the decline of human campylobacteriosis? Appl Environ Microbiol. 2008;74:383–90. doi: 10.1128/AEM.01455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–66. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. [Accessed January 30 2009];Campylobacter jejuni and Campylocbacter coli MLST home page. Available at: http://pubmlst.org/campylobacter/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.