Abstract
Objective
To determine the procedural feasibility of a pharmacist-led interdisciplinary service for providing genotype-guided warfarin dosing for hospitalized patients newly starting warfarin.
Design
Prospective observational study
Setting
483-bed hospital affiliated with a large academic institution
Participants
Eighty patients started on warfarin and managed by a newly implemented pharmacogenetics service.
Intervention
Routine warfarin genotyping and clinical pharmacogenetics consultation
Measurements and Main Results
The primary outcomes were percent of genotype-guided dose recommendations available prior to the second warfarin dose and adherence of the medical staff to doses recommended by the pharmacogenetics service. Of 436 genotype orders during the first 6 months of the service, 190 were deemed appropriate. For 80 patients on the service who consented to data collection, 77% of genotypes were available prior to the second warfarin dose. The median (range) time from the genotype order to the genotype result was 26 (7 to 80) hours, and the time to genotype-guided dosing recommendation was 30 (7 to 80) hours. Seventy-three percent of warfarin doses ordered by the medical staff were within 0.5 mg of the dose recommended by the pharmacogenetics consult service.
Conclusions
Providing routine genotype-guided warfarin dosing supported by a pharmacogenetics consult service is feasible from a procedural standpoint, with the majority of genotypes available prior to the second warfarin dose and good adherence to genotype-guided dose recommendations by the medical staff.
Keywords: warfarin, pharmacogenetics, CYP2C9, VKORC1, genotype feasibility, implementation, pharmacogenetics service
Warfarin is a difficult drug to dose, and inappropriate dosing can have significant adverse consequences. In fact, warfarin ranks as a leading drug-related cause of serious adverse events and hospitalizations in older United States adults.1 Warfarin dosing is complicated by the significant inter-patient variability in the dose necessary to achieve therapeutic anticoagulation, generally defined as an international normalized ratio (INR) of 2 to 3 for most indications. Clinical factors, such as age, body size, liver function, and concomitant medications, only partially explain the inter-patient variability in dose requirements.2
Substantial data demonstrate that genetic variation in cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase complex 1 (VKORC1) contributes to the inter-patient variability in warfarin dose requirements.3 CYP2C9 polymorphisms affect S-warfarin clearance, whereas VKORC1 genotype impacts sensitivity to warfarin. While other genes, most notable CYP4F2, have also been associated with warfarin dose requirements, CYP2C9 and VKORC1 are the strongest genetic predictors of warfarin maintenance dose.4-6
The recently revised Food and Drug Administration (FDA)-approved warfarin labeling provides dosing recommendations based on CYP2C9 and VKORC1 genotypes.7 In addition, guidelines by the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommend genotype-guided warfarin dosing when genetic information is available for clinical use.8 The CPIC guidelines further recommend using a pharmacogenetic-based dosing algorithm to assist with warfarin dosing. Two algorithms shown to be the most accurate for dose prediction are freely available through the http://www.warfarindosing.org website.2, 9, 10 Despite FDA labeling revisions, consensus genotype-based dosing guidelines, and readily available warfarin pharmacogenetic dosing algorithms, few medical centers have incorporated genotype-guided warfarin dosing into clinical practice. Thus, there is a paucity of data on the feasibility of genotype-guided warfarin dosing outside of the controlled research environment.
Starting in August 2012, all patients newly starting warfarin during their hospitalization at the University of Illinois Hospital & Health Sciences Center (UI-Health) are genotyped, with the goal of improving warfarin dosing and preventing adverse consequences associated with inappropriate dosing. In conjunction with routine genotyping, a clinical pharmacogenetics consult service was implemented to assist with genotype interpretation, provide genotype-guided dosing recommendations, and assist with anticoagulation management. Herein, we describe the process and procedural feasibility of implementing warfarin pharmacogenetics into clinical practice.
Methods
Description of the warfarin pharmacogenetics service
UI-Health consists of a 483-bed tertiary hospital affiliated with the University of Illinois and its seven health sciences colleges. Prior to implementation, the structure and procedures for providing genotype-guided warfarin therapy, including the pharmacogenetics consult service, were approved by the Anticoagulation Task Force and Pharmacy and Therapeutics Committee at UI-Health. Informatics and clinical decision support (CDS) tools were built into the electronic health record (EHR) to trigger an automatic order for genotyping and a pharmacogenetics consultation in response to an initial warfarin order for a hospitalized patient newly initiated on warfarin (i.e. with no history of warfarin use within our medical center or recorded as historical use during medication reconciliation within the previous 6 months). An automated alert appears in the EHR to inform the physician placing the warfarin order that genotyping and a consultation with the pharmacogenetics service has been ordered (Figure 1). The computer also provides an initial dose recommendation based on conventional clinical factors including patient age, height, weight, self-reported race, smoking status, amiodarone use, and the presence of a recent normal INR when these data are available in the EHR. This initial dose is calculated based on a published algorithm with the exception of indication of venous thromboembolism (VTE) since this diagnosis is not always readily retrievable from the EHR early in an admission.2 When any of these clinical data are missing, the computer alert appears without an initial dose recommendation. In this instance, the ordering physician is referred to the institutional guidelines for warfarin use to assist with selection of the first dose, which are directly accessible through an evidence link at the bottom of the electronic alert.
Figure 1.

Alert to the clinician in response to an initial warfarin order for a patient with no history of warfarin use within 6 months and no genetic information recorded in the electronic medical record. The dose provided is for a 50 year old Caucasian male with a height of 5’3” and weight of 130 pounds who smokes and is taking amiodarone.
The pharmacogenetics consult service operates 7 days a week and is jointly staffed by faculty from the Colleges of Pharmacy, Medicine, and Pathology (Figure 2). Physician coverage is provided by the UI-Health cardiology and hematology services, which alternate coverage on a 4-week basis. The clinical pharmacy team providing coverage remains the same to provide continuity in care. Table 1 describes the functions and responsibilities of the pharmacogenetics consult service.
Figure 2.
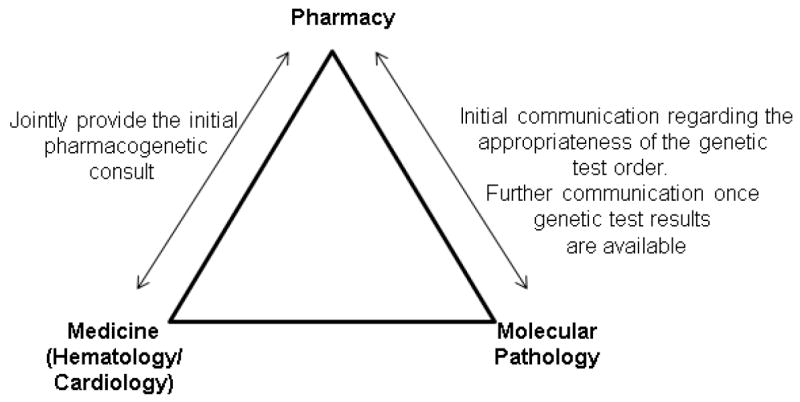
Warfarin pharmacogenetics consult team
Table 1.
Functions and responsibilities of the pharmacogenomic consult service
|
Figure 3 shows the workflow and procedures of the service. A clinical pharmacist on the service is alerted to each new warfarin genotyping order and pharmacogenetics consult via an alert to a service inbox set up in the EHR. At the same time, the clinical laboratory receives an alert for each genotyping order. The pharmacist manually screens the EHR to determine the appropriateness of genetic testing and cancels orders deemed unnecessary (e.g. warfarin use from an outside facility not recorded on admission) or inappropriate (e.g. warfarin order for suspected heparin induced thrombocytopenia in which the serotonin release assay was negative) and communicates this information to the laboratory. For orders deemed appropriate, blood is drawn by the phlebotomy service at the next scheduled collection time and sent to the Molecular Pathology Laboratory for genotyping.
Figure 3.
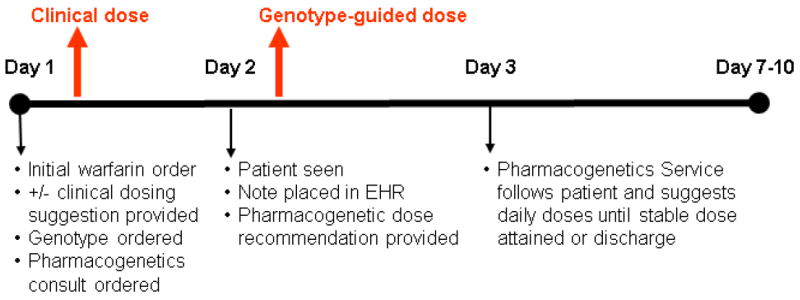
Warfarin pharmacogenetic consult timeline
The pharmacogenetics consult team provides an initial patient assessment (consisting of a review of the medical history, medications, laboratory results, patient demographics, physical assessment) and clinical dose recommendation for the first dose (when not provided automatically via the pharmacogenetics alert). The team targets completion of the initial assessment within 24 hours of the consult order. Genotyping is performed by the Molecular Pathology Laboratory, with results being available within 6-7 hours after receiving a sample. Genotype results are expected to be available in time to inform the second dose of warfarin. Generally, genotyping is started at 10 AM each morning for any sample available, with results available by 4 pm, well before the scheduled administration of warfarin at 9 pm. Genotyping is done using an eSensor® XT-8 system (GenMark DX, Carlsbad, CA), which captures the following genotypes: VKORC1 c.-1639G>A; CYP2C9 *2, *3, *5, *6, *11, *14, *15, and *16; and CYP4F2 p.V433M. Once results are available, the laboratory alerts the consult service pharmacist and enters the results into the EHR, and the pharmacist enters the genotype information into the consult note along with a genotype-guided dose recommendation. In cases where the genotype reveals extreme sensitivity to warfarin (e.g. VKORC1 -1639AA genotype) or reduction in warfarin clearance (e.g. CYP2C9 *2/*2, *1/*3, or *3/*3), the pharmacist also contacts the primary team directly to alert them.
The clinical pharmacist on the team continues to provide daily patient assessment and dose recommendations, which are refined based on INR response to previous doses and documented in the patient’s EHR for the initial 7 days of warfarin therapy or until the patient is discharged. Previously published algorithms, freely available through www.warfarindosing.org89 along with clinical factors not included in the algorithms (e.g. renal function, additional interacting medications) are used to assist with refined dose recommendations. In the event that genotype results become available after the patient is discharged, results are mailed to the patient with instructions to share them with their health care provider managing warfarin, and this information is stored in the EHR. For all other patients, the genotype results are included with the discharge instructions. In addition, patients are provided with written educational materials at discharge, customized to include the patient’s genotype. An example of information included in the patient educational material is shown in Figure 4.
Figure 4.

Information included in the patient educational information on warfarin pharmacogenetics. The information is customized to include the patient’s unique genetic information.
Patient recruitment and data collection
Patients receiving genotype-guided warfarin therapy were approached about providing written, informed consent for the use of their data and leftover genetic sample for research purposes. For consenting patients, time of the initial genotype and consult order, time to the genotype results appearing in the EHR, time to the initial consult and genotype-guided dose recommendation, and warfarin doses prescribed by the primary team were recorded. This research was approved by the University of Illinois at Chicago Institutional Review Board.
Genotyping
We did not have IRB approval to report genotypes from the EHR that were determined as part of clinical care; however all patients enrolled provided consent for the use of their leftover DNA sample for research purposes. Thus, patients were genotyped for research purposes in the College of Pharmacy Pharmacogenomics Laboratory, which previously served as the validation site for genotypes on the clinical platform. Patients were genotyped for the same variants on the clinical platform (VKORC1 -1639G>A; CYP2C9*2, *3, *5, *6, and *11; and CYP4F2 V433M), using previously described methods.11-13
Data analysis
Genotype frequencies were calculated and compared between race groups by Chi square analysis. The primary outcome was the percent of time that the genotype-guided dose recommendations were available prior to the second warfarin dose. Procedural feasibility was prospectively defined as at least 70% of recommendations provided before the second dose. The secondary outcome was the acceptance of genotype-guided dose recommendations by the primary team, defined as the percent of time the dose ordered was within 0.5 mg of the dose recommended by the team. Again, the goal acceptance rate was prospectively determined to be at least 70%. Another outcome of interest was recommendations provided by the pharmacogenetics team beyond warfarin dosing guidance.
Results
Patient population
In the initial 6 months of the warfarin pharmacogenetics service, a total of 436 warfarin genotype and consult orders were placed. Of these, 190 (44%) were deemed appropriate. Of the 246 (56%) orders deemed inappropriate, the primary reason for an inappropriate order was use of warfarin on admission from an outside medical center (n=223). Other reasons were genotype already available from a prior admission (n=6), patient discharged prior to blood for genotyping being drawn (n=3), substitution of an alternative anticoagulant (n=2), and genotyping not felt to be indicated in a patient with significant multiple comorbidities affecting warfarin disposition (n=1). In the remaining 11cases, the pharmacogenetics team recommended against warfarin use, such as in the case of active cancer, where low molecular weight heparin would be more appropriate (n=5) and a high suspicion for active bleeding (e.g. recent large hemoglobin drop), for which the team recommended withholding warfarin while the potential source for bleeding was interrogated (n=5);and a warfarin order for suspected heparin induced thrombocytopenia in which the serotonin release assay was negative (n=1).
Of the 190 patients with appropriate genotyping and consult orders, 65 (34%) were receiving warfarin for prophylaxis of venous thromboembolism after major orthopedic surgery and were not approached for study participation because of differing INR goals. Twenty (11%) were discharged prior to being approached about study participation; this included patients who were admitted and discharged over a weekend or holiday when research staff were unavailable to obtain consent. A total of 105 (55%) patients were approached about study participation, and 80 patients provided written, informed consent for collection and use of their clinical data and left over genetic sample for research purposes. Characteristics of the patients enrolled are shown in Table 2; most were of African ancestry. Allele frequencies in the population overall were 0.23 for VKORC1 -1639A, 0.08 for CYP2C9*2, 0.01 for CYP2C9*3, and 0.17 for CYP4F2 433M. None of the patients had a CYP2C9*5, *6, or *11 allele. African Americans had lower frequencies of the VKORC1 -1639A (0.07 versus 0.44, p<0.001), CYP2C9*2 or *3 (0.01 versus 0.17, p<0.001) and CYP4F2 433M (0.04 versus 0.33, p<0.001) alleles than patients of other ancestries.
Table 2.
Characteristics of patients enrolled
| Characteristic | Total n=80 |
|---|---|
| Age (years) | 50 ± 18 |
| Female sex | 43 (54) |
| Self-reported race/ethnicity | |
| African American | 45 (56) |
| European | 13 (16.5) |
| Hispanic | 13 (16.5) |
| Other | 9 (11) |
| Warfarin indication | |
| Deep vein thrombosis | 25 (31) |
| Pulmonary embolism | 20 (25) |
| Atrial fibrillation | 12 (15) |
| VTE prophylaxis after major orthopedic surgery | 11 (14) |
| Other | 12 (15) |
| Genotype | |
| VKORC1 -1639G>A | |
| AA | 8 (10) |
| AG | 21 (26) |
| GG | 51 (64) |
| CYP2C9 | |
| *1/*1 | 68 (85) |
| *1/*2 | 10 (13) |
| *1/*3 | 1 (1) |
| *2/*2 | 1 (1) |
| CYP4F2 V433M | |
| VV | 56 (70) |
| VM | 21 (26) |
| MM | 3 (4) |
Procedural feasibility data
Among the 80 patients enrolled, the median time from the pharmacogenomic consult order to the initial consultation, in which a clinical dose recommendation was provided, was 14 hours (range of 18 minutes to 28 hours). Eighty-nine percent of consultations were provided within our goal time of 24 hours from when the initial consult was placed. The time from the genotype order to the genotype result appearing in the EHR was 26 hours (range of 7 to 80 hours); 77% of genotypes were available prior to the second dose of warfarin. The primary reason for delay was blood not reaching the laboratory in a timely manner. Of 62 (78%) patients still hospitalized at the time the genotype results were returned, the time from the genotype order to the genotype-guided dose recommendation provide by the pharmacogenetics consult team was 30 (7 to 80) hours.
The median length of stay for the 80 patients enrolled was 5.5 days (range of <24 hours to 97 days). A total of 353 dose recommendations were provided for these patients. This includes the recommendation provided in the initial clinical consult by the pharmacogenetics team and subsequent daily dose refinement recommendations provided by the team pharmacist via a follow-up note in the EHR. For patients hospitalized for >7 days, dosing recommendations were provided for the initial 7 days or until a therapeutic INR was achieved for 2 consecutive days. Figure 5 shows the doses recommendation by the pharmacogenetics team versus the doses ordered by the medical staff. Overall, during the initial 6 months of the service, 73% of warfarin doses ordered by the primary team were within 0.5 mg of the dose recommended by the pharmacogenetics service. In 9 (3%) cases, the pharmacogenetics team recommended a warfarin dose, but the primary team subsequently decided to hold warfarin for a procedure, patient refusal to take warfarin, or other reason. Adherence with dose recommendations increased over time, with 66% adherence in months 1 to 2, 76% in months 3 to 4, and 80% in months 5 to 6.
Figure 5.
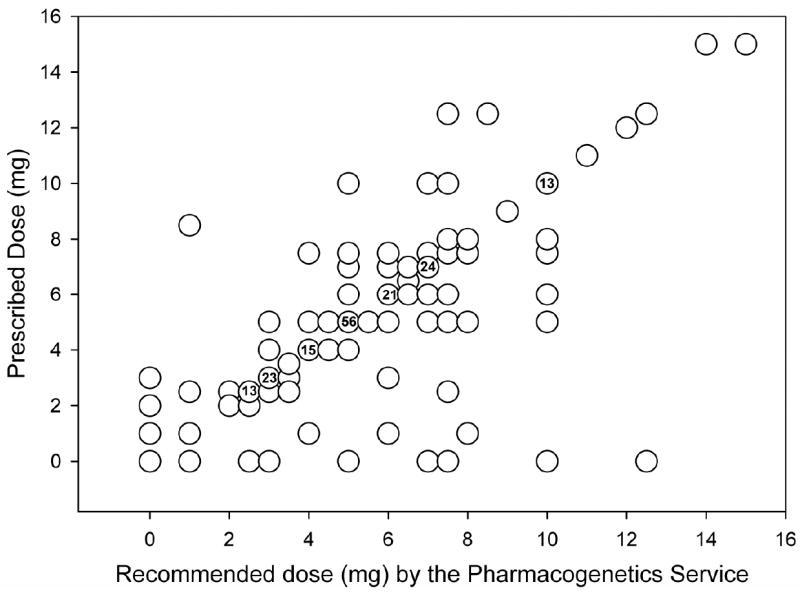
Scatter plot of dose recommended by the pharmacogenetics service versus the dose ordered by the primary team. The numbers within the circles indicate the number of times that dose combination occurred. For circles without numbers, the combination only occurred once.
Discussion
To our knowledge, we are among the first academic medical center to incorporate an interdisciplinary genotype-guided consult service as part of standard care for patients newly starting warfarin. As evidenced at our center, a routine clinical warfarin pharmacogenetics service is feasible from a procedural standpoint, with the majority of genotype-guided recommendation provided prior to the second warfarin dose. In addition, the medical staff were, in most cases, adherent to dose recommendations by the pharmacogenetics service, demonstrating acceptance of the pharmacogenetics service and dose recommendations. Importantly, adherence to dose recommendations increased over time indicating that lower adherence in the early weeks of the service may have been due to unfamiliarity with the new procedure for warfarin dosing. Our approach to providing a formal clinical consultation for each patient initiating warfarin is also unique and allows for interventions beyond providing pharmacogenetic dose recommendations. This is best illustrated in cases where warfarin is deemed inappropriate or unnecessary by the pharmacogenetics service who communicates this with the primary team, such as the case with active cancer or a recent drop in hemoglobin.
A major practical challenge with clinical implementation of warfarin pharmacogenetics is obtaining genotype results in a timely manner. However, previous data indicate that even when genotype results are not available until 4 to 7 days after warfarin initiation, genotype-guided dosing still allows for better prediction of the warfarin maintenance dose than dosing based on clinical factors alone.14, 15 Nonetheless, it is desirable to obtain genotype results sooner in order to better inform initial warfarin doses and avoid supra-therapeutic dosing in patients with the extremely sensitive VKORC1 -1639AA genotype or delays in reaching therapeutic anticoagulation in patients with insensitive genotypes. Thus, our goal is to obtain genotype results in time to inform the second warfarin dose, and our data show this was achievable nearly 80% of the time. Since the primary reason for a delay in obtaining genotype is a delay in sample collection, mechanisms to reduce the time for blood collection in response to a genotype order are being investigated.
Currently, one patient on average is genotyped each day as part of the warfarin pharmacogenetic service. This may be perceived as an inefficient use of hospital resources. However, our existing platform also allows for CYP2C19 testing for antiplatelet therapy, which we have recently implemented. Thus, on some days, multiple genotypes for multiple patients requiring warfarin or antiplatelet therapy may be determined simultaneously, improving the efficiency of the genotyping process. In addition, we are exploring approaches for preemptive genotyping, as is being done at other medical centers, in which multiple variants are determined ahead of time to inform future prescribing of warfarin and other drugs.16, 17 This would minimize the need for rapid genotyping for one patient at a time.
While we focused on procedural feasibility in this paper, the financial feasibility also deserves mention. Payment for consultations and genetic testing at our institution varies by third party payer. As such, this could affect the ability to implement a similar approach in other health systems. However, reimbursement by their party payers may change pending the outcome of ongoing clinical trials of genotype-guided warfarin therapy.
Another challenge is establishing the clinical decision support for genotype-based warfarin dosing. Currently, pharmacists are extracting clinical data from the EHR and entering these data in addition to genotype into pharmacogenetic dosing algorithms freely available through the world wide web. Such an approach is time consuming and subject to error in transferring data from one source to another. In addition, this approach is not ideal for situations when a pharmacist is not available. Thus, we are exploring means of integrating a pharmacogenetic dosing algorithm directly into the EHR, as we have done for the initial clinically-based warfarin dose. In addition, we are exploring a means of electronically and securely transferring data from the EHR to an external site (e.g. warfarindosing.org) where a dose may be calculated and returned to the EHR.
Changing the perception and behavior of the ordering physician to increase adherence to dose recommendations by the pharmacogenomics service can also be difficult. Strategies implemented by UI-Health to address this challenge include interdisciplinary continuing education programs focused on warfarin pharmacogenomics before and shortly after service inception. Additionally, several in-services and grand rounds were delivered during the initial weeks of the service to various medical services and units that care for patients requiring warfarin, including cardiology, neurology, medicine, and orthopedics. An internal website was also developed prior to service inception to provide information about warfarin pharmacogenetics and access to relevant guidelines, including institutional clinical warfarin use guidelines. These latter guidelines were updated to incorporate dosing and monitoring procedures based on warfarin pharmacogenetics. The website is directly and conveniently accessible through the evidence link on the automated alert to the physician, triggered by the initial warfarin order for a patient (Figure 1). The pharmacogenetics service is staffed by residency-trained pharmacists from 8 am to 5 pm 7 days a week. Clinical pharmacists are oncall and available by phone at all other times to consult with the medical staff and assist with any questions regarding warfarin pharmacogenetics; our resident pharmacist-on-call program with faculty service as back-up serves a vital role in this regard. Finally, in the initial weeks of the service, a screen saver in the EHR was designed for all hospital computers to alert the house staff of the new approach to warfarin dosing. The high adherence rate to dose recommendations suggests that this combination of strategies was successful. However, further or alternative education or communication models are likely still warranted to near our goal of 100% adherence.
Conclusions and Future Directions
In summary, we have found that implementation of warfarin pharmacogenetics into routine clinical practice is feasible from a procedural standpoint and well accepted by our medical staff. The components of the service described herein, including the genotyping procedure, CDS tools, and patient educational materials, may be exported to other institutions interested in implementing a warfarin pharmacogenetic programs. In addition, the interdisciplinary approach to the UI-Health pharmacogenetics service represents a model for team-based care including pharmacists, physicians, and laboratory personnel. Through the provision of personalized warfarin dosing, the goal is to improve anticoagulation management and reduce the risks for adverse events, namely bleeding and thrombosis, as a result of inappropriate warfarin dosing. A future aim, therefore, is to examine the effects of genotypeguided warfarin dosing in our diverse patient population on clinical outcomes, such as time to achieve stable dosing and risk for adverse warfarin-related sequelae.
Acknowledgments
funding sources:
The warfarin pharmacogenetics service is funded by the University of Illinois Hospital & Health Sciences System office of the Vice President for Health Affairs.
EN is supported by an NIH NHLBI award (K23HL112908). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Budnitz DS, Lovegrove MC, Shehab N, Richards CL. Emergency hospitalizations for adverse drug events in older Americans. N Engl J Med. 2011;365:2002–2012. doi: 10.1056/NEJMsa1103053. [DOI] [PubMed] [Google Scholar]
- 2.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorgensen AL, FitzGerald RJ, Oyee J, et al. Influence of CYP2C9 and VKORC1 on patient response to warfarin: a systematic review and meta-analysis. PLoS One. 2012;7:e44064. doi: 10.1371/journal.pone.0044064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper GM, Johnson JA, Langaee TY, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coumadin (warfarin sodium) package insert. Princeton, NJ: Bristol-Myers Squibb; 2010. Jan, [Google Scholar]
- 8.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkelman BS, Gage BF, Johnson JA, et al. Genetic warfarin dosing: tables versus algorithms. J Am Coll Cardiol. 2011;57:612–618. doi: 10.1016/j.jacc.2010.08.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hruska MW, Frye RF, Langaee TY. Pyrosequencing method for genotyping cytochrome P450 CYP2C8 and CYP2C9 enzymes. Clin Chem. 2004;50:2392–2395. doi: 10.1373/clinchem.2004.040071. [DOI] [PubMed] [Google Scholar]
- 12.Aquilante CL, Langaee TY, Lopez LM, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Cavallari LH, Butler C, Langaee TY, et al. Association of apolipoprotein E genotype with duration of time to achieve a stable warfarin dose in African-American patients. Pharmacotherapy. 2011;31:785–792. doi: 10.1592/phco.31.8.785. [DOI] [PubMed] [Google Scholar]
- 14.Horne BD, Lenzini PA, Wadelius M, et al. Pharmacogenetic warfarin dose refinements remain significantly influenced by genetic factors after one week of therapy. Thromb Haemost. 2012;107:232–240. doi: 10.1160/TH11-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenzini P, Wadelius M, Kimmel S, et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin Pharmacol Ther. 2010;87:572–578. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JA, Burkley BM, Langaee TY, et al. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther. 2012;92:437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieb W, Volzke H, Pulley JM, et al. Strategies for personalized medicine-based research and implementation in the clinical workflow. Clin Pharmacol Ther. 2012;92:443–445. doi: 10.1038/clpt.2012.119. [DOI] [PubMed] [Google Scholar]


