Abstract
Personalized cancer medicine requires the development of tumor-specific biomarkers to optimize selection of targeted therapies and to better assess response to therapy. Current efforts in several tumor types have shown that patients in whom circulating tumor cells (CTCs) are detected have an inferior prognosis relative to those in whom CTCs are not detected and that the elimination or decrease of CTCs following treatment is associated with improved clinical outcomes. Technological advances in the detection, isolation, capture, and characterization of CTCs from phlebotomy samples obtained in a routine clinical practice setting have enabled the evaluation of different CTC biomarkers. Unmet needs in cancer diagnosis and treatment where CTC biomarkers have been studied include determining prognosis, assessing the effects of treatment, and as a source of tumor for the biologic identification and characterization of determinants to predict sensitivity to one form of treatment versus another and to understand mechanisms of treatment resistance.
At present, there is no single definition of a CTC and no single CTC “biomarker.” Rather, multiple assays (tests) are in development for CTC biomarkers. However, before the role of any biomarker in medical decision making can be determined, it is essential that the assays used to measure the biomarker are analytically validated in a sequence of trials to generate the evidence to support the biomarker’s use in the given context of use. It is against this background that this review focuses on the process of developing CTC biomarker assays, with the objective of outlining the necessary steps to qualify specific CTC tests for medical decision making in clinical practice or drug development. The potential for point-of-care tests is clear.
Keywords: Circulating tumor cells, biomarker, regulatory qualification, personalized medicine
At some point in the development and progression of a cancer, cells from the primary tumor reach the circulation, a proportion of which have the ability to seed and proliferate in distant sites. Interest in circulating tumor cells (CTCs) is longstanding1 but has increased dramatically because of recent technological advances that have further enabled their detection, isolation, capture, and characterization from phlebotomy samples obtained in a routine clinical practice setting. Circulating tumor cells are estimated to account for at most one cell in a hundred million to a billion of the cells that are circulating in the blood2 and encompass the spectrum of the malignant phenotype including tumor-initiating cells with stem or stem cell–like properties, the full range of undifferentiated to differentiated phenotypes that lack tumor-initiating capabilities, and cells that have undergone an epithelial-to-mesenchymal transition (EMT).3,4 But, despite their rarity, phenotypic diversity, and heterogeneity, data are evolving rapidly that CTCs can serve as biomarkers for a range of clinical contexts with the potential to address unmet needs in cancer diagnosis and treatment. These contexts include establishing a diagnosis, determining prognosis, assessing the effects of treatment, and serving as a source of tumor for the biologic identification and characterization of determinants to predict sensitivity to one form of treatment versus another and understanding the mechanisms of treatment resistance.5 Importantly, there is no single definition of a “CTC” and no single CTC “biomarker.” Rather, there are assays (tests) for CTC biomarkers that may or may not prove to have clinical utility.
Establishing the role of a diagnostic test in medical decision making is not straightforward. For CTCs, the situation is complicated by the broad range of assays and devices currently in use and under development. Underappreciated is that different assays, be they for enumeration or biologic profiling, may not be evaluating the same cells or the same determinant in cells and as such are reporting different biomarkers. Underestimated is the complexity of establishing the analytical validity of an assay so that it can be used in rigorous series of trials needed to establish the role of the test in the clinic. Problematic as well is the paucity of dedicated trials designed specifically to address CTC biomarker questions. The result is a literature that is replete with reports describing “significant results,” whose clinical significance has not yet been established. It is in this background that this review is focused on the process of developing CTC biomarker assays, with the objective to outline the necessary steps to qualify specific CTC tests for medical decision making in clinical practice or drug development.
CTC BIOLOGY
CTCs in the Metastatic Process
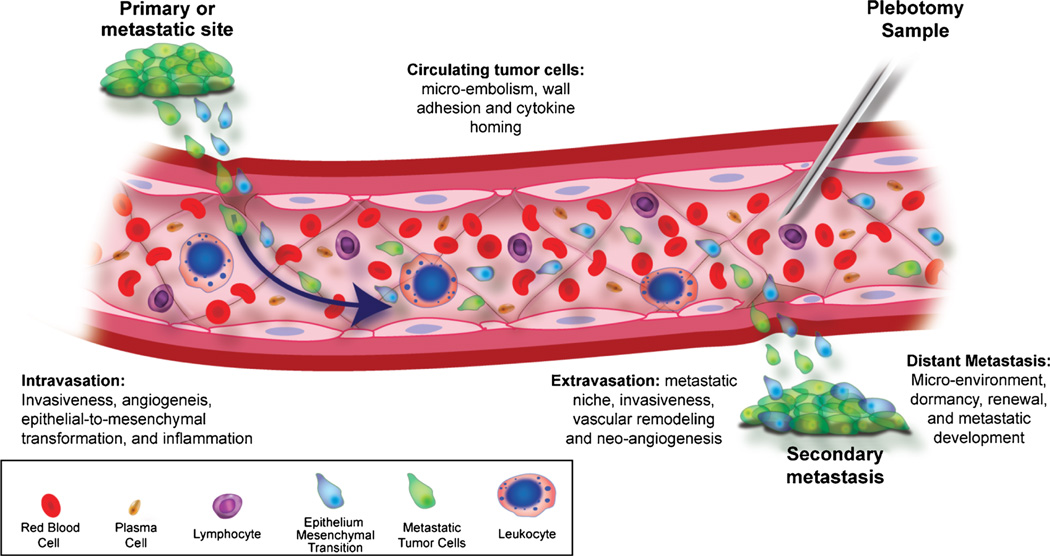
Circulating tumor cells originate from the primary tumor or metastatic deposits after invading and intravasating through the tumor vasculature by mechanisms that are not completely understood (Fig. 1). The hematogenous spread of a tumor results from CTC,6 which can also reseed the organ of origin to form new tumors in this location.7 Most CTCs die in the circulation, but a proportion are preprogrammed with homing receptors (e.g., integrins and chemokines) that enable them to attach to the vasculature in specific organs, following which they adhere and extravasate, co-opt the capillary microenvironment, and proliferate to form metastases.7–9 As an example, the CXCR4 chemokine receptor has been involved in the development of metastatic lesions from breast and prostate cancer CTCs.10 That these cells are viable has been shown by short-term proliferation experiments6,11 and by the ability of the cells in culture to secrete both tumor-specific markers, such as prostate-specific antigen (PSA) and stem-cell–like factors.12,13 More recently, CTC clusters have been described, but at this point, it is unclear whether they are an artifact of the method used to isolate them, or whether cluster formation leads to entrapment in the microcirculation or enables tumor cell survival resulting in an increased metastatic propensity.14,15
FIGURE 1.
Circulating tumor cells in the process of metastatic progression. Tumor cells transition from the primary or metastatic site into circulation to establish secondary sites. Sampled by phlebotomy at the time when treatment is being considered, CTCs have the potential to provide tumor material for molecular profiling for biomarkers informative of tumor sensitivity to the targeted therapy being considered.
Bone marrow (BM) is a major site of spread of both breast and prostate cancer, and recent studies have emphasized that the detection of tumor cells in this location, termed disseminated tumor cells (DTCs), at the time of diagnosis is associated with an increased risk of tumor recurrence.16–18 Indeed, multiple studies have shown that DTCs are detected more frequently in BM than CTCs are found in concurrently drawn blood samples.19,20 Although this may simply be a function of the sensitivity of the assays used to detect tumor cells in the BM versus the blood, it does suggest that the BM can serve as a reservoir organ for further dissemination to other sites in the BM as well as other organs.6,21,22 Detection of DTCs in the BM in experimental models and in the clinic is also a significant risk factor for relapse in the breast, further evidence in support of CTC reseeding.7,10,22 After seeding into a metastatic location,23 tumor cells have to adapt to survive in inhospitable conditions where there is low blood perfusion or pH,24 and to remain in a viable yet dormant state for extended periods.20,25 In both breast and prostate cancer, DTCs have been detected in the BM years after the primary diagnosis in patients who are clinically relapse-free.26,27 At this time however, assays to detect tumor cells in the BM are used only in the routine management of patients with leukemias and lymphomas. They are not a part of the routine management of solid tumors limiting its evaluation in large-scale trials.28
CTCs: One Biomarker or Many?
The range of CTC phenotypes that includes cells with stem or stem cell–like properties and those with an EMT phenotype has important implications. Epithelial-to-mesenchymal transition describes the phenotypic change of epithelial tumor cells to a fibroblastic cell morphology that is accompanied by a decrease in expression of epithelial adhesion molecules (e.g., E cadherin) and increased expression of mesenchymal molecules (e.g., vimentin), leading to increased mobility and invasiveness.3,4 It has been also proposed as an important cancer stem cell property.29,30 Epithelial-to-mesenchymal transition is important not only mechanistically, but also from the point of view of CTC detection because many of the isolation and capture technologies utilize antibodies to epithelial cell adhesion molecule (EpCAM), which, although commonly expressed on the majority of primary and metastatic tumor cells, may be down-regulated or absent on the subsets of CTCs that have undergone EMT.31,32 A recent study using the CellSearch system (Veridex LLC, Raritan, NJ) has shown that a basal-like breast cancer cell line with features of EMT expresses EpCAM levels that are too low to allow capture using such antibodies.33 Similarly, cells with stem or stem cell–like properties may also lack EpCAM or cytokeratin expression.34 To capture these cells requires non–EpCAM-based methods such as negative depletion strategies that remove leukocytes and staining the remaining mononuclear cell fraction with antibodies to markers specific to this specific CTC phenotype.12,13 Although expression of vimentin in cancer cells is an indicator of EMT, vimentin is also expressed by surrounding leukocytes and is therefore not a suitable marker for CTCs.
Thus, new markers for CTCs that are specific for tumor cells but not repressed during EMT are urgently needed. Moreover, at present it is not clear whether cancer cells with down-regulated EpCAM expression have stemness properties, as EpCAM has been included in a breast cancer stem cell signature.35 Although EpCAM might be down-regulated during EMT, the current view is that tumor cells with a partial EMT (or “intermediate phenotype” between epithelial and mesenchymal) are the most aggressive subclones. In addition, DTC homed in secondary organs such as the BM must undergo the reversal of EMT (called MET) to form a solid metastasis. It is therefore possible that cancer cells with a high plasticity to undergo both EMT and MET are the “real” metastasis-initiator cells.
It follows that there can be no one definition of a CTC or CTC biomarker that encompasses the range of clinical phenotypes and genotypes. Different assays detect different subsets of cells and different biologic determinants in respective subsets and cannot be assumed to provide equivalent information.11,15,32,36 As such, individual reports must be evaluated critically from the point of view of which subsets of cells are being captured and/or analyzed, the performance characteristics of the assay that is used for what type of measurement, how it is reported, and what clinical question or context of use the results of the assay are purporting to inform.
CONTEXTS OF USE FOR BIOMARKERS
Contexts of Use for Biomarkers
A biomarker is a characteristic that is objectively measured and evaluated as an indicator of normal biological or pathogenic processes or pharmacologic responses to therapeutic intervention.37 As drugs are approved for indications, biomarkers are qualified for a specific context of use as detailed in the Food and Drug Administration (FDA) Critical Path.38 Contexts of use represent specific clinical settings for which the results of the test will be used to inform a medical decision and can be divided into 5 categories.
Biomarkers: The Vocabulary of an Evolving Language
Diagnostic: demonstrates the presence of a malignancy and potentially establishes the tissue of origin.
Prognostic: informs the natural history of disease in the absence of therapy, including the risk or probability of disease occurrence, relapse, progression, or survival. It may also inform where the disease is most likely to occur.
Prediction: the likelihood of response to a specific therapy to enable the practice of more “personalized medicine.” Optimal utilization of a targeted approach requires a demonstration that the target is present in a patient’s tumor at the time treatment is considered. Both the natural evolution of a cancer and the specific therapies to which it has been exposed contribute to disease heterogeneity. Tracking and directing therapies to this changing biology is problematic without serial sampling of multiple metastatic sites over time and potentially better addressed using blood-based diagnostics that can be performed serially with minimal patient discomfort.
Pharmacodynamic: a posttreatment assessment that shows that a biologic response or activity has occurred, with the caveat that the “response” or “activity” observed does not necessarily imply patient benefit. Demonstrating a change in phosphorylation status following treatment with a tyrosine kinase inhibitor is one example, a change in proliferation status or apoptotic rate another.
Surrogate: a biomarker measured after treatment that is intended to substitute for a clinical endpoint at some later time point. A clinical benefit surrogate, one that represents how a patient feels, functions, or how long he/she survives, could enable more rapid drugs approvals.
ROADMAP TO BIOMARKER QUALIFICATION
A qualified biomarker is one whose measurement can be relied on to have a specific interpretation for a specific context of use (Fig. 2). To achieve qualification requires measurement of the biomarker using an analytically valid assay and a sequence of clinical studies to generate the evidence to support its use in the given context (Table 1). The regulatory implications are significant, because once a biomarker is qualified, the results of the evaluation with the biomarker can be used in regulatory filings without a rereview of the data supporting its use in the specified context. As an example, a qualified surrogate biomarker for survival is one measured earlier in the course of treatment, enabling an earlier analysis that, if successful, can be used in Investigational New Drug and New Drug Application/Biologic License Application submissions to accelerate drug approvals.38–41
FIGURE 2.

Specific context of use for validation and utilization of biomarkers. A qualified biomarker is one whose measurement can be relied on to have a specific interpretation for a potential clinical application.
TABLE 1.
Level-of-Evidence Scale and Group the Current CTC Tests
| Phase I: New CTC test is developed (and refined) in a laboratory, specificity and sensitivity shown, first clinical data on cancer patients, first publication(s). |
| Phase II: New CTC test compared with existing CTC test (criterion standard, e.g., CellSearch) and “superior” findings demonstrated with regard to clinical relevance in a limited study. |
| Phase III: New CTC test is disseminated to other CTC-specialized laboratories, and results of the initial studies are reproduced (e.g., specificity, sensitivity, reproducibility in blinded ring experiments). |
| Phase IV: New CTC test is implemented in large-scale clinical trial(s) with defined endpoints (e.g., progression-free or overall survival) side-by-side with most established CTC test for the particular cohort of patients and stage and treatment. Circulating tumor cell testing in specialized laboratories. |
ANALYTICAL VALIDATION
Analytical validation starts with the discovery of the biomarker to be measured and the development of a robust assay that provides consistent and reproducible results across multiple systems and laboratories42,43 The process is outlined in the Oncology Biomarker Qualification Initiative of the U.S. FDA, Centers for Medicare & Medicaid Services, and National Cancer Institute and detailed in the FDA Critical Path Initiative.38 For qualification, the assays are performed in Clinical Laboratory Improvement Amendments (CLIA)–certified laboratories. A draft guidance issued by the FDA40,44,45 outlined 3 steps:
Preanalytical assessment of specimen selection, handling, processing, and storage parameters, for the measurement to be assessed in the same context each time. For example, a blood specimen must be appropriately anticoagulated, stored at appropriate temperature, and processed in accordance to a strict standard operating procedures (SOP). The collection tube must be standardized. The effects of storage condition (time and temperature) must be determined.
- Validation of the analytical characteristics to meet CLIA regulatory requirements, establishing the performance characteristics of the assay to be comparable each time it is performed in the same laboratory, as well as in independent laboratories. Standardization requires the development of a comprehensive quality control program to
- assess reproducibility: desirable intra-assay and interassay variability
- quality controls should resemble patient sample
- internal quality control concentrations: negative and low positive controls reflecting marker concentration
- assess potential sources of assay interference: for example, for an antibody-based assay, test heterophilic, human anti–mouse antibody or other antibody
Establishment of standards for data management and storage: data reduction, interpretation, and reference interval.
Of note is that most biomarkers are measured using a device of some type to perform the actual measuring procedure, such as numerical counts of tumor cells of some specific phenotype in a blood sample. Review of the device and authorization for marketing is regulated by the Office of In Vitro Diagnostics and is entirely separated from the qualification process. A recent change in regulatory requirements resulted from the broadened definition of a significant risk In Vitro Diagnostic (one that is, “for a use of substantial importance in diagnosing, curing, mitigating, or treating disease or otherwise preventing impairment of human health and presents a potential for serious risk to the health, safety, or welfare of a subject.”) As an example, if the test result will be used for the context of prediction, to determine whether to offer one treatment versus another, it is subject to regulatory review.40
CLINICAL QUALIFICATION
Once the analytical performance of an assay has been established, clinical testing can begin. Whether the assay in question needs full CLIA certification before clinical testing is controversial but is a regulatory requirement for qualification. The first consideration is the context of use for which the test is intended (Fig. 2). Once established, components of the clinical validation process include sensitivity, specificity, false-positive and false-negative results, and positive and negative predictive values in relation to established standards if any. For the analysis of a predictive factor, this might include a comparison of the results of an analysis of CTCs relative to a biopsy of a metastatic lesion at the same time.
Our initial studies of CTC in prostate cancer focused on the detection of tumor cells using a reverse transcription–polymerase chain reaction (RT-PCR)–based assay for the messenger RNA (mRNA) for PSA.46 The preliminary results showed a higher frequency of detection in more advanced stages of disease as expected, but more interesting was the finding that mRNA for PSA could be detected in patients who had no detectable PSA following hormone therapy. This suggested to us that the process of tumor shedding might provide unique clinical information relative to the measurement of PSA or imaging, but the assay itself did not perform to a level that enabled widespread testing. Other assay methods were similarly inadequate, and it was not until the CellSearch assay was FDA cleared47 that more extensive clinical testing became feasible.
Clinical qualification follows a strict evidentiary process linking the biomarker with biological processes and clinical endpoints specific for the context of use that the test result will be used.18 Analogous to our approach with the evaluation of drugs, our biomarker studies are conducted in defined patient populations using strict eligibility criteria under institutional review board–approved protocols with patient informed consent. For our initial exploratory studies, eligibility includes patients with newly diagnosed, recurrent, or progressing disease. Starting with patients with established metastatic disease that were progressing with castrate levels of testosterone, we showed first that the cells captured using CellSearch expressed PSA and α-methylacyl-CoA-racemase that are unique to prostate cells, as well as prostate-specific genomic abnormalities such as androgen receptor (AR) copy number amplifications, phosphatase and tensin homolog (PTEN) deletions, and TMPRSS:ETV fusion products.48,49
Next, using duplicate samples obtained at the same blood draw, we showed that the results obtained in the Clinical Laboratory at Memorial Sloan-Kettering Cancer Center (MSKCC) were similar to the Reference Laboratory and that consistent results were obtained between samples processed on the day of and up to 72 hours after the blood was obtained from the patient. This enabled samples to be shipped from remote sites and eliminated the need for placing the technology in multiple locations. Samples that do not provide similar results on the day of draw and subsequent days are still useful but must be processed on the same day, limiting broader testing. Samples that can be stored and analyzed at a future date can be used for retrospective analyses, assuming the clinical specimens are annotated in what are termed retrospective prospective trials. Additional questions of importance that are often not considered are reproducibility and consistency of results on the same day in the same laboratory, over a 24-hour period, and the potential effect of other confounding factors that may alter the test results independent of disease biology. The latter might include the timing and relationship of the biomarker next to a biopsy or surgical procedure, radiation, or recent drug administration.
Next, we asked how often the test could detect CTCs at different points in the disease continuum. Figure 2 outlines discrete clinical states of a cancer including localized disease, biochemical recurrence in which there is an abnormal blood test (e.g., elevated PSA, carcinoembryonic antigen (CEA) or carbohydrate antigen 19-9 but no evidence of disease on imaging, established metastatic disease about to receive a first-line therapy, and established metastatic disease about to receive a second-or third-line therapy. It follows that CTC assays that do not detect cells at particular points in an illness would not be useful whether the context of use relates to drug development or clinical practice. In clinical trials, higher detection rates also reduce the number of patients needed to demonstrate the role of a test in medical decision making. As most CTC assays do not detect cells at a high frequency in localized disease states, one area of focus is to identify analytically valid assays that are more sensitive and demonstrate that detection rates are in fact increased.
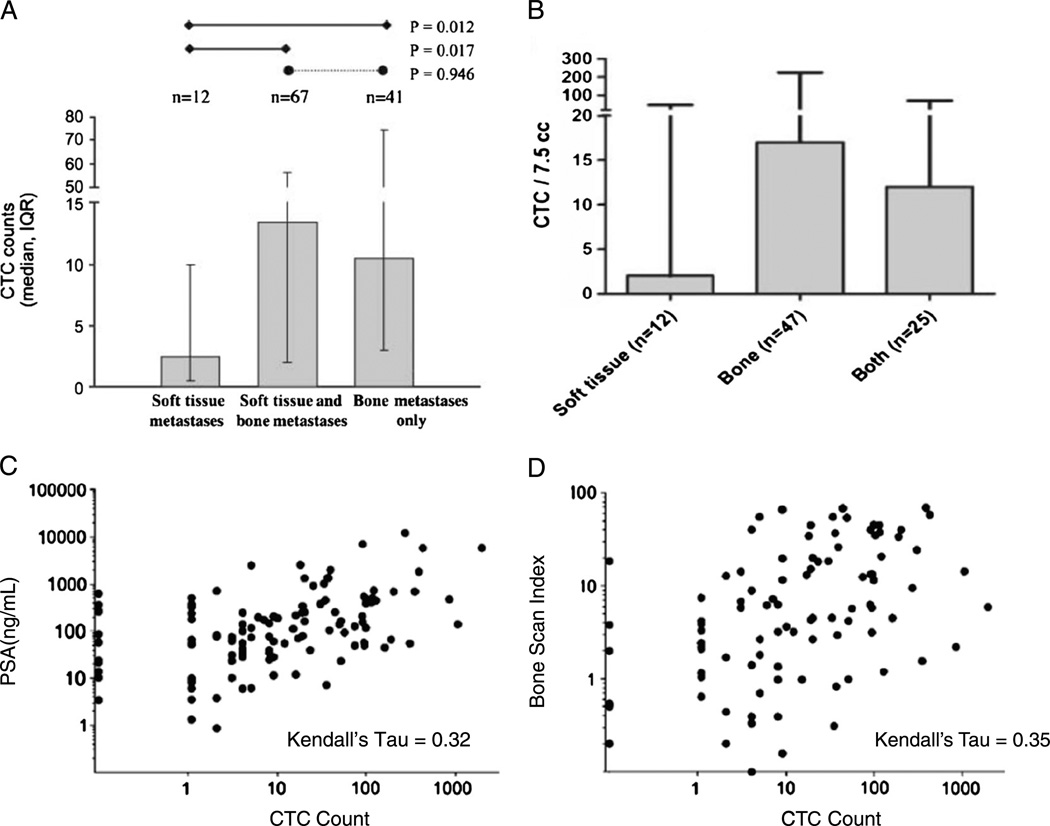
What constitutes a “positive” test differs for different assays. Some assays report a “number” of cells per given volume of blood, others an “all or none” or “positive or negative,” and others a gradation based on expression of a specific gene. Again using CellSearch, we showed that CTC number and level of PSA were modestly associated, as were CTC cell number and extent of BM involvement by the tumor on bone scan.50 This latter finding was independently confirmed.51,52 Overall, more cells are isolated from more patients with bone and visceral metastases, compared with patients with lymph node disease alone, consistent with the known routes of seeding prostate cancer cells by hematogenous versus lymphatic spread, respectively (Fig. 3).50,51,53 Next, the prognostic significance of CTC number was explored both before and after treatment in a series of trials of similar design in cohorts of patients with breast, colorectal, and subsequently prostate cancer with progressive disease about to start a new chemotherapy. The results showed the prognostic significance of the test, both pretreatment and posttreatment, and ultimately led to the FDA clearance of the assay as an aid to monitoring.53
FIGURE 3.
Circulating tumor cells as an intrinsic property of an individual patient’s tumor. Independent trials show that the CTC number varies with pattern of spread (A and B) and is modestly associated with tumor burden, as measured by PSA (C) or bone scan (D) (from Danila et al50 [A, C, and D]; Part B is Adapted and reprinted by permission from the American Association for Cancer Research: Goodman OB Jr, Fink LM, Symanowski JT, et al. Circulating tumor cells in patients with castration-resistant prostate cancer baseline values and correlation with prognostic factors. Cancer Epidemiol Biomarkers Prev. 2009;18:1904–1913.
Importantly, clearance is not a qualification, and the advice from FDA was to come back with a drug. A qualified biomarker is one that is measured in an analytical test system with well-established performance characteristics and for which there is an established scientific framework or body of evidence that elucidates the physiologic, toxicologic, pharmacologic, or clinical significance of the test result. The biomarker must provide consistent, plausible results in multiple prospective trials and must be accepted by scientific community at large to predict clinical or preclinical outcomes.54 The regulatory requirements for the qualification of a biomarker, like those for approval of a device, vary as a function of risk.42
For predictive biomarkers of sensitivity, an additional consideration is how often the marker is present at particular points in the illness and whether it changes as the disease progresses. For those determinants present at the same frequency in the primary tumor and in a recurrent metastatic state, profiles of the latter are not necessary. For those that change over time, due either to the intrinsic biology of the tumor itself or as an adaptive or selective change because of the specific therapy a patient has received, molecular profiling of the tumor at the time treatment is considered is essential. It is for this context that blood-based diagnostics are most critical. De novo and second site mutations in receptor tyrosine kinases shown to predict sensitivity to specific tyrosine kinase inhibitors in lung and colorectal cancer (CRC) are key examples.55–57 It is for the latter that the analysis of CTC biomarkers is likely to play their most critical role.
Criteria are under development to rate the data available for a specific assay applied to a specific context. These include the performance characteristics of the assay itself, as well as statistical methods used to evaluate results in different patient populations with or without treatment. For those that are particularly robust, and recognizing that the field in general is evolving very rapidly, the FDA initiated a Voluntary Exploratory Data Submission program to share pharmacogenomics data with the agency “to ensure that regulatory scientists are familiar with and prepared to appropriately evaluate future genomic submissions.”58 These submissions are evaluated on an individual basis. For those biomarkers with broader applications, the FDA Center for Drug Evaluation and Research has also developed a separate Biomarker Qualification program.40
TECHNOLOGIES TO ISOLATE, CAPTURE, AND CHARACTERIZE CTCS
There are many approaches to detect, isolate, enumerate, and characterize CTCs from the peripheral blood, but the development of robust assays for broad clinical testing has proven difficult. Because of their rarity,2 enrichment is generally needed to increase sensitivity to an acceptable level. Approaches that have been used for enrichment and capture of CTC include immune affinity using magnetic beads or fluorescent labeling, size filtration or adhesiveness, or density gradients or viscosity. One type of enrichment relies on antibody capture of cell surface markers (immunoseparation). Other methods for enrichment are based solely on morphologic/physical criteria, such as cell size or density, whereas others use acoustic and dielectrophoresis. Negative selection methods in which mononuclear cells are depleted have also been used.
After enrichment, CTCs are identified through putative tumor-specific or tissue-specific markers by nucleic acid–based techniques, such as RT-PCR methods, acoustic and dielectrophoresis methods, or cytometric methods, such as flow cytometry, laser-scanning cytometry, or semiautomated immunocytochemistry microscopy. Unfortunately, no one feature universally distinguishes CTCs from the normal, nonmalignant blood elements, and different histological and molecular types of tumors express different arrays of markers. At present, cytokeratins are the most commonly used epithelial markers for CTC detection in cancer patients.59 Cytokeratins, however, belong to a large family that consists of at least 20 different proteins, and individual cytokeratins (e.g., CK8 or CK18) can be down-regulated in carcinoma cells,60 which may lead to false-negative findings.
Current technical limitations in analysis of CTC and contaminating leukocytes present in the sample emphasize the need for improved isolation and characterization methods to enable clinical applications. It has already been demonstrated that the analysis of cancer-related alterations of DNA and protein from CTCs is feasible in a hospital-based clinical laboratory. Nevertheless, these assays are exploratory; none has been analytically validated. Sensitivity and specificity are also an issue with all of the techniques because of heterogeneity of tumors in cell size, density, and marker expression. Consequently, some tumor cell loss is likely to occur irrespective of the enrichment technique used.61
We briefly describe some of the CTC methods that are at relatively advanced stages of development, and in particular CellSearch, which presently is the only assay that is cleared for enumeration by the U.S. FDA. The other assays described here are in different phases of development and validation, along the pathway of reducing the CTC biomarker technology to clinical practice (Fig. 4).
FIGURE 4.
Technology maturation. Circulating tumor cell methods are in different phases of development and validation, along the pathway of reducing the CTC biomarker technology to clinical practice.
Immunoselection-Based Technologies
CellSearch
The CellSearch (Veridex LLC, Warren, NJ) immunomagnetic isolation technique, equipment, and software for identifying CTCs were developed by the Immunicon Corporation (Huntingdon Valley, PA) and is currently marketed by Veridex LLC. The assays process uses antibodies to epithelial cell adhesion molecule (EpCAM) conjugated to magnetic beads for the immunomagnetic capture of EpCAM-expressing epithelial cells. The EpCAM-expressing cells are automatically displayed visually and manually scored by a trained technologist as a CTC based on strict morphologic and immunohistochemical staining characteristics. A captured cell is classified as a CTC if it expresses cytokeratin, displays a nucleus when stained with 4′,6-diamidino-2-phenylindole (DAPI), and is not a white blood cell as determined by a lack of expression of CD45.62 The results are reported as the number of cells meeting the definition per 7.5 mL of blood.
Studying patients with a diagnosis of prostate cancer who were about to start a new line of therapy, the frequency of detection of CTC was limited to more advanced, castration-resistant metastatic disease state (Table 2). Importantly, the number of cells meeting the criteria for a CTC is only a small proportion of the EpCAM positive events displayed.63 The clinical development of the assay is discussed further in the case study section. As the assay uses a positive selection based on immunomagnetic enrichment for tumor cells expressing EpCAM, for patients with tumors lacking expression of EpCAM, such as malignant melanoma, a different “hook” is necessary to capture tumor cells from circulation.
TABLE 2.
CTC Enrichment by CellSearch: Frequency of CTC Detection in Different Clinical States Using CellSearch in Patients With a Diagnosis of Prostate Cancer (MSKCC Data)
| CellSearch CTC* |
Localized (n = 37) |
Rising PSA (n = 75) |
Non-Castrate Mets (n = 127) |
mCRPC Chemotherapy Exposure | ||
|---|---|---|---|---|---|---|
| Pre (n = 127) | 1st Line (n = 144) | 2nd Line (n = 250) | ||||
| <5 | 36 (97) | 73 (97) | 109 (86) | 94 (75) | 71 (49) | 132 (53) |
| ≥5–9 | 0 (0) | 2 (3) | 11 (9) | 15 (11) | 21 (14) | 21 (8) |
| 10–49 | 1 (3) | 0 (0) | 3 (2) | 13 (10) | 30 (20) | 56 (22) |
| ≥50 | 0 (0) | 0 (0) | 4 (3) | 5 (4) | 22 (15) | 41 (16) |
Number of events /7.5 mL of blood that meet the Veridex definition of a CTC.
Values are n (%).
mCRPC indicates metastatic CRPC.
Micro-Posts and Herringbone CTC Chips
Micro-posts and herringbone CTC chips allow sensitive and selective detection of CTC for enumeration and further molecular profiling at genomic, transcriptional, and translational levels in CTC isolated from patients with lung, breast, and prostate cancer.15,64,65 Reports using this technology have shown CTC isolation and capture rates at points in an illness where CellSearch does not detect cells meeting the FDA-cleared definition of a CTC, such as localized prostate cancer.11 A more recent development was the CTC chip that uses a herringbone configuration designed to create turbulence to maximize gentle cell contact with the surface of the chip while minimizing cell trauma. Used for blood samples from patients with metastatic non-small cell lung cancer (median number, 74 cells/mL) allowed viable cells to be captured that were in turn analyzed for the epidermal growth factor receptor (EGFR) T790M activating mutation66 that predicts for sensitivity to gefitinib and erlotinib.57 Additional designs stagger obstacles to optimize cell size–dependent flow to increase contact with capture antibody-laden surfaces, where prostate-specific membrane antigen–based immunoselection allows enrichment of EpCAM-negative tumor cells.67
Immunomagnetic Enrichment of Melanoma Cells
Immunomagnetic enrichment of melanoma cells that do not express EpCAM was reported in an exploratory study that analyzed blood samples from patients with cutaneous and uveal melanoma based on expression of a melanoma-associated chondroitin sulfate proteoglycan. Light microscopy was used to recognize circulating melanoma cells (CMCs) based on morphology.68 This method allowed the capture and characterization of tumor cells in more advanced stages of the disease. Furthermore, the tumor cells isolated in this study showed chromosomal aberrations typical for melanoma by comparative genomic hybridization, which may confer a predictive potential.
Flow Cytometric Methods
Flow cytometric methods are used to enrich for more homogeneous and highly purified tumor cell populations separated based on multiple cell surface and intracellular markers. The technique is useful for the evaluation expression profiles in CTC.69–71 Whole-genome amplification from the cellular events isolated by sorting also enables mutational analyses and studies of copy number alterations at the level of single cells, as prognostic and predictive biomarkers.72
Functional Assays
EPithelial ImmunoSPOT
EPithelial ImmunoSPOT (EPISPOT) assays detect CTC based on the release of full-length CK19 by carcinoma cells of various origins.13 Studies of BM samples from patients with breast cancer detected CK19-releasing cells in 44% to 70% of cases, which was associated with a higher rate of development of overt metastases and a reduced survival relative to patients in whom CK19-releasing cells were not detected (P = 0.025 by log-rank test; P = 0.0019; hazard ratio [HR], 4.7). In blood samples, CTCs were detected based on expression of tumor antigen mucin 1 (MUC1) or PSA in the majority of patients with metastatic breast and prostate (83.3%) cancer, respectively, whereas the test results were negative in healthy controls or in patients with benign prostatic hyperplasia.73
Collagen Adhesion Matrix
Collagen adhesion matrix assays allow enriching CTC based on their ability to ingest and invade into the connective tissue–like material. Although in initial development phase, this method could allow for a function separation of more aggressive, invasive type of CTC from patients with prostate or breast cancer.74
Size- and Biophysical-Based Assay
Dielectrophoretic Array
Dielectrophoretic array technologies displace blood cells when they pass over microelectrode array configurations designed to separate cells as a function of cell size and electrostatic charges. Cells isolated this way can be characterized by microscopy or could be analyzed for predictive molecular biomarkers based on relative fluorescence parameters combined with morphology.75–77
Microfiltration Devices
Microfiltration devices capture CTC for enumeration and on-filter characterization based on their larger size and inflexible biophysical proprieties relative to hematopoietic cells when passed through a porous membrane. Several different pore sizes and shapes have been tested. Advantages of these systems are the ability to perform CTC analysis, including immunohistochemistry, directly on membrane, as well as the agnostic enrichment of CTC, indifferent of EpCAM expression that allows collection of EMT and CTC with stem cell–like features.78–81
Microfluidics Separation
Microfluidics separation exploits the unique differences in size and deformability of CTC compared with peripheral blood cells in microchannels optimized by computational analysis to enhance the isolation efficiency. The isolated cells are amenable to direct visualization for immunofluorescence microscopy or can be retrieved and further cultured or profiled for specific tumor predictive biomarkers.82,83
Genomic, Transcriptional, or Translational-Based Assay
Reverse Transcriptase–Polymerase Chain Reaction
Reverse transcriptase–polymerase chain reaction assays have been long used to detect low levels of cancer-specific mRNA in the mononuclear cell fraction of the blood that are presumed to be from CTC. In some cases, the tumor-specific mRNA detected in the mononuclear cell fraction represents phagocytosed tumor cells in macrophages and does not represent intact tumor cells. Messenger RNA for PSA has been tested in peripheral blood of patients with prostatic cancer,46,84 as well as detecting CEA, CK, and CD133 in patients with CRC.85 The most frequently studied marker of CMCs is the melanocyte-specific tyrosinase, followed by melanoma antigen gene family members, glycoprotein gp100, melanotransferrin p97, and tyrosinase-related proteins.86 Additional markers are under investigation for detection of CTC from patients with breast, prostate, lung, and head and neck cancers.87–94
Sarcoma-Specific Gene Fusions
Sarcoma-specific gene fusions allow identification of CTC or DTC in the peripheral blood or BM, respectively. An RT-PCR method targeting EWS-FLI-1 or EWS-ERG transcripts was studied in patients with Ewing tumors to detect evidence of occult tumor cells.95 Flow cytometry or fluorescence in situ hybridization–based methods to identify sarcoma-specific fusion genes are currently being tested.96–98
Imaging-Based Profiles of CTCs at the Protein and Chromosomal Level
The CellSearch Digital image analysis identifies the cells likely to be tumor cells using 3 channels for CK, CD45, and DAPI staining, which also enables additional molecular analysis in the fourth channel.66,99 In this channel, immunofluorescence microscopy can be used for protein staining for human EGFR 2 (HER2), insulin-like growth factor 1 receptor, AR, or markers of cell cycling, apoptosis, and DNA damage such as phosphorylated γ-H2AX in patients treated with chemotherapeutic agents.100–102
Fiberoptic array scanning technology of immunofluorescent staining of peripheral mononucleated blood cells has been used to identify CTC based on particular cytomorphologic features present in the patient’s primary or metastatic tumor tissue.103 Fluorescence in situ hybridization analysis of CTCs from patients with prostate, breast, kidney, sarcoma, and colon cancer is being used to identify gene copy number alterations or chromosomal abnormalities.49,56,104–111
Contexts of Use
As noted, a key consideration for the clinical evaluation of any biomarker is the context of use for which it is being studied (Fig. 2). No single assay can fulfill the requirements for clinical validation or qualification for all contexts of use. The trial designs needed to establish clinical validity and qualification would also differ as a function of what if any assay is available that addresses the context of use. To be broadly applicable, however, the specific biomarker question generally requires to be embedded in large-scale randomized trials powered on a clinical endpoint to which the biomarker result will be associated. Also required are that consistent results are obtained across multiple trials, with as broad a range of agents and diseases as possible.
Prognosis and Response-Indicator Biomarkers
In a series of clinical trials of similar design, patients with metastatic breast,112–115 prostate,53,116 and colorectal117,118 cancer about to start a new line of therapy had CTC measured at baseline and with subsequent follow up visits. Disease-specific cutoffs were used to define patient groups with a favorable (≤4 CTCs per 7.5 mL of blood for breast and prostate cancers, and ≤2 for CRC) and unfavorable (≥5 for breast and prostate and ≥3 for CRC) prognosis both before and after therapy. In this context, CTC enumeration was prognostic of median progression-free survival and overall survival (Table 3).113,119 Posttreatment CTC enumeration remains prognostic at 4, 8, or 12 weeks after initiating treatments in this context.53,116–118,120 These studies resulted in the FDA clearance of the CellSearch system to measure CTC as a marker of prognosis in the monitoring of patients with metastatic breast cancer, prostate cancer, and CRC, and as a response-indicator biomarker by serial testing for CTC in conjunction with other clinical methods, as described in the 510(k) document.47
TABLE 3.
Context of Use for Baseline CTC Counts: a Prognosis Biomarker for Overall Survival for Patients With Metastatic Disease
| Baseline CTC Counts in Patients With Metastatic Cancer |
Threshold for Unfavorable CTC |
No. Patients (%) With Unfavorable Baseline Counts |
Overall Survival Based on CTC Counts (95%CI), mo |
||
|---|---|---|---|---|---|
| Favorable | Unfavorable | Cox HR; Log-Rank P | |||
| Prostate cancer53,116 | ≥5 | 219 (43) | 21.7 (21.3 to >26) | 11.5 (9.3–13.7) | 3.3; P < 0.0001 |
| Breast cancer115 | ≥5 | 177 (49) | 21.9 (20.1 to >25) | 10.9 (6.4–15.1) | 2.45 (1.64–3.65); P < 0.0001 |
| Colorectal cancer117,118 | ≥3 | 413 (26) | 18.5 (15.5–21.2) | 9.4 (7.5–11.6) | 2.45 (1.77–3.39); P < 0.0001 |
Separately, PCR-based assays have been tested as biomarkers of prognosis and response indicators. The frequency of detection of PSA mRNA in peripheral blood of patients with prostatic cancer correlated with tumor stage.46,84 Detecting CEA, CK 19, CK20, and/or CD133 mRNA by RT-PCR demonstrated a significant prognostic factor for overall survival (HR, 3.84; 95% confidence interval [CI], 2.41–6.22; P < 0.001) and disease-free survival (HR, 3.02; 95% CI, 1.83 to 5.00; P < 0.001) in particular in patients with Dukes stage B and C CRC.85 Serial monitoring by RT-PCR of a multimarker panel for CMCs during biochemotherapy may be useful for predicting therapeutic efficacy and disease outcome in patients with metastatic melanoma, although additional studies are necessary.121
Predictive Biomarkers
To predict the likelihood of a patient to benefit from a targeted therapy requires a demonstration that the “target” is present in the tumor when treatment is considered. Biological determinants currently under analysis in CTCs include HER2 protein expression for patients with breast cancer.56,66,99,122 In prostate cancer, recent reports evaluated alterations in the AR at the protein, RNA, and DNA levels,49,104 IGF1R expression,100 and TMPRSS2-ETV fusions,49,123 which have been postulated to increase the likelihood of response to a therapy targeting the specific signaling pathway. Detection of kinase mutation predictive of tumor sensitivity in CTCs has potential for clinical application in selecting patients with metastatic colorectal or lung cancer most likely to benefit from EGFR-targeted therapies.55–57 Each of these questions has important clinical implications, and each represents a specific clinical context where the results of a test would be useful for medical decision making. There are many more potential contexts, and the analytical validation of these assays is ongoing.
CASE STUDY: THE EFFORT TO QUALIFY CELLSEARCH CTC ENUMERATION AS A BIOMARKER IN PROSTATE CANCER
Unmet Need for Surrogate Cancer-Related Endpoints
A critical unmet need in prostate cancer drug development and treatment is for outcome measures that reflect clinical benefit. As a bone-dominant disease, prostate cancer metastases are difficult to assess using conventional imaging modalities.124,125 Determination of the level of PSA, despite extensive investigation, has not been established as a surrogate endpoint for regulatory approval, as only part of the treatment effect is reflected in PSA changes.126
In the phase II trial (IMMC-38, trial registration ID: NCT00133900) of prostate cancer that led to the FDA clearance in this disease, the results of the posttherapy CTC number analyzed using CellSearch at all time points were more prognostic than a 50% or greater decline in PSA.127 A subsequent analysis, limited to patients receiving their first cytotoxic drug, confirmed the association of higher counts with a worsening prognosis in a continuous manner, but at the same time showed that favorable counts did not guarantee a long survival,116 arguing against the use of discrete cutoff values. When CTC count was tested along with various established predictors in a multivariate analysis, the only factors independently prognostic of survival were CTC count and lactate dehydrogenase, whereas PSA was not an independent predictor.
Initial Studies in the Efficacy-Response Context
With the recent approval of 4 treatments for patients with metastatic prostate cancer, it is critical to develop clinical endpoints that can be used for drug approvals, short of overall survival.128–130 For an efficacy-response biomarker to be qualified as surrogate, it requires an additional level of evidence before it can be used in a regulatory filing.
Based on the initial IMMC-38 trial, CTC enumeration was prospectively embedded in clinical trials to be studied as efficacy-response biomarkers. Two phase II postchemotherapy trials of patients with prostate cancer treated with abiraterone acetate, an oral inhibitor of 17α hydroxylase and C17,20-lyase, showed similar frequency of baseline unfavorable CTC counts (Table 4): 79% in the Royal Marsden Hospital–chaired trial (trial registration ID: NCT00474383) and 69% in the MSKCC-chaired trial (trial registration ID: NCT00485303). The treatment-induced CTC conversion rates in the 2 studies from unfavorable to favorable were also similar at 41% and 34%, respectively.131,132 Separately, phase I/II data of treatment with MDV3100, another AR-directed therapy specifically developed by investigators at MSKCC for activity in prostate cancer cells with overexpressed AR, showed similar posttherapy CTC conversion rates in 25 (49%) of 51 patients with castration-resistant prostate cancer (CRPC) with baseline unfavorable counts (trial registration ID: NCT00510718).120 With these results, the decision was made to incorporate CTC enumeration as an efficacy-response biomarker in the phase III registration trial of abiraterone acetate (Cougar AA-301, trial registration ID: NCT00638690).
TABLE 4.
Abiraterone Acetate Shows a High CTC Conversion Rate in Post–Chemotherapy-Treated Patients With Metastatic CRPC
| Trial | No. Patients | No. Patients (%) With ≥50% PSA Decline From Baseline |
Percentage of Patients With Unfavorable Baseline Counts (No. Patients With CTC ≥5/No. Patients With CTC Enumerated) |
CTC Conversion From ≥5 to <5 With Treatment (No. Patients) |
|---|---|---|---|---|
| Reid et al131 | 47 | 24 (51) | 79% (27/37) | 41% (11) |
| Danila et al132 | 58 | 25 (43) | 69% (29/42) | 34% (10) |
CTCs were studied as an endpoint because of uncertainty of PSA-based endpoint for an AR signaling–directed therapy.
Current and Future Studies
Qualifying CTC enumeration as an efficacy-response surrogate marker for survival requires consistent results in multiple phase III clinical trials where the biomarker is embedded. A unique opportunity arose from the availability of 2 phase III survival trials of similar design in the same clinical context of patients with postchemotherapy metastatic CRPC: Cougar AA-301 and the registration trial of MDV3100 (trial registration ID: NCT00974311). The data analysis of the first phase III trial powered on survival and incorporating the CTC biomarker question, where patients with post–chemotherapy-treated CRPC were randomized to receive abiraterone acetate or placebo, was discussed in a face-to-face meeting at the Center for Drug Evaluation and Research. The trial showed prolonged survival (HR, 0.65; 95% CI, 0.54–0.77), time to PSA progression (HR, 0.58; 95% CI, 0.46–0.73), and radiographic progression-free survival (HR, 0.67; 95% CI, 0.58–0.78).133 For the biomarker portion of the study, the first question will be which specific posttherapy CTC biomarker criterion best associates with overall survival.134 Next will be an analysis of whether the prognostic value of the criterion is increased if it is combined with other biomarkers (e.g., lactate dehydrogenase, hemoglobin, PSA). If it is, then the best combination of markers will be used as a CTC-based biomarker panel in subsequent trials.135,136
CONCLUSIONS
Recent work in several tumor types has shown that patients in whom CTCs are detected with a variety of different assays have an inferior prognosis relative to those in whom they are not detected and that the “elimination or decrease” of CTCs following treatment is associated with improved clinical outcomes.137 The observation in a prostate cancer population that CTCs could be detected in patients with no measurable PSA on hormonal therapy suggests that seeding could continue, despite seemingly successful therapy, and that the continued detection of CTCs could provide unique information. Equally intriguing is the potential for the biologic characterization of CTCs to provide a profile of an individual patient’s tumor that can be used to guide treatment selection: a liquid biopsy to enable personalized medicine where the potential for point-of-care tests is clear.
Acknowledgments
Source of Funding: Department of Defense Prostate Cancer Research Program Physician Research Award W81XWH-09-1-0307 (Dr Danila), Memorial Sloan-Kettering Cancer Center Special Program of Research Excellence in Prostate Cancer (Grant No. P50 CA92629) (Drs Fleisher and Scher), Department of Defense Prostate Cancer Research Program (Grant No. PC051382) (Dr Scher), Prostate Cancer Foundation (Drs Danila, Fleisher, and Scher) Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research (Drs Danila and Scher). The Experimental Therapeutics Center, and the Geoffrey Beene Cancer Research Center of Memorial Sloan-Kettering Cancer Center (Drs Danila, Fleisher, and Scher). Research support: Veridex, LLC, Raritan, NJ (Drs Pantel, Fleisher, and Scher). Honoraria: Veridex, LLC, Raritan, NJ (Drs Pantel and Fleisher). Consultancies: Ortho Biotech Oncology Research and Development, New Brunswick, NJ (Compensated), and Medivation, San Francisco, CA (Dr Scher, Uncompensated).
Footnotes
Conflicts of Interest: No other conflicts of interest were reported.
REFERENCES
- 1.Ashworth T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J. 1869;14:146–149. [Google Scholar]
- 2.Ross AA, Cooper BW, Lazarus HM, et al. Detection and viability of tumor cells in peripheral blood stem cell collections from breast cancer patients using immunocytochemical and clonogenic assay techniques. Blood. 1993;82:2605–2610. [PubMed] [Google Scholar]
- 3.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 4.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keiser J. Cancer’s circulation problem. Science. 2010;327:1072–1074. doi: 10.1126/science.327.5969.1072. [DOI] [PubMed] [Google Scholar]
- 6.Muller V, Stahmann N, Riethdorf S, et al. Circulating tumor cells in breast cancer: correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin Cancer Res. 2005;11:3678–3685. doi: 10.1158/1078-0432.CCR-04-2469. [DOI] [PubMed] [Google Scholar]
- 7.Kim MY, Oskarsson T, Acharyya S, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 9.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Comen E, Norton L, Massague J. Clinical implications of cancer self-seeding. Nat Rev Clin Oncol. 2011;8:369–377. doi: 10.1038/nrclinonc.2011.64. [DOI] [PubMed] [Google Scholar]
- 11.Stott SL, Lee RJ, Nagrath S, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra3. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alix-Panabieres C, Vendrell JP, Pelle O, et al. Detection and characterization of putative metastatic precursor cells in cancer patients. Clin Chem. 2007;53:537–539. doi: 10.1373/clinchem.2006.079509. [DOI] [PubMed] [Google Scholar]
- 13.Alix-Panabieres C, Vendrell JP, Slijper M, et al. Full-length cytokeratin-19 is released by human tumor cells: a potential role in metastatic progression of breast cancer. Breast Cancer Res. 2009;11:R39. doi: 10.1186/bcr2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 15.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun S, Vogl FD, Naume B, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 17.Weckermann D, Polzer B, Ragg T, et al. Perioperative activation of disseminated tumor cells in bone marrow of patients with prostate cancer. J Clin Oncol. 2009;27:1549–1556. doi: 10.1200/JCO.2008.17.0563. [DOI] [PubMed] [Google Scholar]
- 18.Kollermann J, Weikert S, Schostak M, et al. Prognostic significance of disseminated tumor cells in the bone marrow of prostate cancer patients treated with neoadjuvant hormone treatment. J Clin Oncol. 2008;26:4928–4933. doi: 10.1200/JCO.2007.15.0441. [DOI] [PubMed] [Google Scholar]
- 19.Riethdorf S, Wikman H, Pantel K. Review: biological relevance of disseminated tumor cells in cancer patients. Int J Cancer. 2008;123:1991–2006. doi: 10.1002/ijc.23825. [DOI] [PubMed] [Google Scholar]
- 20.Pantel K, Alix-Panabieres C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6:339–351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 21.Pierga JY, Bonneton C, Vincent-Salomon A, et al. Clinical significance of immunocytochemical detection of tumor cells using digital microscopy in peripheral blood and bone marrow of breast cancer patients. Clin Cancer Res. 2004;10:1392–1400. doi: 10.1158/1078-0432.ccr-0102-03. [DOI] [PubMed] [Google Scholar]
- 22.Bidard FC, Vincent-Salomon A, Gomme S, et al. Disseminated tumor cells of breast cancer patients: a strong prognostic factor for distant and local relapse. Clin Cancer Res. 2008;14:3306–3311. doi: 10.1158/1078-0432.CCR-07-4749. [DOI] [PubMed] [Google Scholar]
- 23.Husemann Y, Geigl JB, Schubert F, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Bartkowiak K, Wieczorek M, Buck F, et al. Two-dimensional differential gel electrophoresis of a cell line derived from a breast cancer micrometastasis revealed a stem/progenitor cell protein profile. J Proteome Res. 2009;8:2004–2014. doi: 10.1021/pr8009758. [DOI] [PubMed] [Google Scholar]
- 25.Uhr JW, Pantel K. Controversies in clinical cancer dormancy. Proc Natl Acad Sci U S A. 2011;108:12396–12400. doi: 10.1073/pnas.1106613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan TM, Lange PH, Porter MP, et al. Disseminated tumor cells in prostate cancer patients after radical prostatectomy and without evidence of disease predicts biochemical recurrence. Clin Cancer Res. 2009;15:677–683. doi: 10.1158/1078-0432.CCR-08-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan TM, Lange PH, Vessella RL. Detection and characterization of circulating and disseminated prostate cancer cells. Front Biosci. 2007;12:3000–3009. doi: 10.2741/2290. [DOI] [PubMed] [Google Scholar]
- 28.Scher HI, Pantel K. Bone marrow aspiration for disseminated tumor cell detection: a must-have test or is the jury still out? J Clin Oncol. 2009;27:1531–1533. doi: 10.1200/JCO.2008.21.2092. [DOI] [PubMed] [Google Scholar]
- 29.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 31.Thurm H, Ebel S, Kentenich C, et al. Rare expression of epithelial cell adhesion molecule on residual micrometastatic breast cancer cells after adjuvant chemotherapy. Clin Cancer Res. 2003;9:2598–2604. [PubMed] [Google Scholar]
- 32.Deng G, Burgess D, Manna E, et al. A new system for enrichment and detection of circulating tumor cells in the peripheral blood of patients-with metastatic breast cancer. Breast Cancer Res Tr. 2007;106:S24. [Google Scholar]
- 33.Sieuwerts AM, Kraan J, Bolt J, et al. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikolajczyk SD, Millar LS, Tsinberg P, et al. Detection of EpCAM-negative and cytokeratin-negative circulating tumor cells in peripheral blood. J Oncol. 2011;2011:252361. doi: 10.1155/2011/252361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 36.Yu M, Stott S, Toner M, et al. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atkinson AJ, Colburn WA, DeGruttola VG, et al. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 38.FDA. Critical Path Initiative. [Accessed November 11, 2011];2011 Available at: http://www.fda.gov/ScienceResearch/SpecialTopics/CriticalPathInitiative/default.htm.
- 39.U.S. Department of Health and Human Services, Food and Drug Administration. Guidance for industry: patient-reported outcome measures: use in medical product development to support labeling claims: draft guidance released for comment. [Accessed November 11, 2011];2006 Feb; doi: 10.1186/1477-7525-4-79. Available at: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM193282.pdf. [DOI] [PMC free article] [PubMed]
- 40.FDA. Guidance for Industry. E16 biomarkers related to drug or biotechnology product development: context, structure, and format of qualification submissions. [Accessed November 11, 2011];2011 Aug; Available at http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM267449.pdf. [PubMed]
- 41.Fleming TR, DeMets DL. Surrogate end points in clinical trials: are we being misled? Ann Intern Med. 1996;125:605–613. doi: 10.7326/0003-4819-125-7-199610010-00011. [DOI] [PubMed] [Google Scholar]
- 42.Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genet Med. 2009;11:3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.FDA. Guidance for Industry Pharmacogenomic Data Submissions. [Accessed November 11, 2011];2005 Available at: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM079849.pdf.
- 44.CDER F. Guidance for Industry. Bioanalytical Method Validation. [Accessed November 11, 2011];2001 Available at: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm070107.pdf.
- 45.Food and Drud Administration. Guidance for Industry Qualification Process for Drug Development Tools. FDA; 2010. Oct, [Accessed November 11, 2011]. [Google Scholar]
- 46.Ghossein RA, Scher HI, Gerald WL, et al. Detection of circulating tumor cells in patients with localized and metastatic prostatic carcinoma: clinical implications. J Clin Oncol. 1995;13:1195–1200. doi: 10.1200/JCO.1995.13.5.1195. [DOI] [PubMed] [Google Scholar]
- 47.Veridex. 510k for CellSearch Veridex. [Accessed November 11, 2011];2008 Available at: http://www.accessdata.fda.gov/cdrh_docs/pdf7/K073338.pdf.
- 48.Shaffer DR, Leversha MA, Danila DC, et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:2023–2029. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 49.Attard G, Swennenhuis JF, Olmos D, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 50.Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 51.Goodman OB, Jr, Fink LM, Symanowski JT, et al. Circulating tumor cells in patients with castration-resistant prostate cancer baseline values and correlation with prognostic factors. Cancer Epidemiol Biomarkers Prev. 2009;18:1904–1913. doi: 10.1158/1055-9965.EPI-08-1173. [DOI] [PubMed] [Google Scholar]
- 52.Imbriaco M, Larson SM, Yeung HW, et al. A new parameter for measuring metastatic bone involvement by prostate cancer: the bone scan index. Clin Cancer Res. 1998;4:1765–1772. [PubMed] [Google Scholar]
- 53.de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 54.Fleming TR. Surrogate endpoints and FDA’s accelerated approval process. Health Aff (Millwood) 2005;24:67–78. doi: 10.1377/hlthaff.24.1.67. [DOI] [PubMed] [Google Scholar]
- 55.Yang MJ, Chiu HH, Wang HM, et al. Enhancing detection of circulating tumor cells with activating KRAS oncogene in patients with colorectal cancer by weighted chemiluminescent membrane array method. Ann Surg Oncol. 2010;17:624–633. doi: 10.1245/s10434-009-0831-8. [DOI] [PubMed] [Google Scholar]
- 56.Punnoose EA, Atwal SK, Spoerke JM, et al. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010;5:e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.FDA. Voluntary Exploratory Data Submission 2010. [Accessed November 11, 2011]; Available at: http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083673.htm.
- 59.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 60.Willipinski-Stapelfeldt B, Riethdorf S, Assmann V, et al. Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin Cancer Res. 2005;11:8006–8014. doi: 10.1158/1078-0432.CCR-05-0632. [DOI] [PubMed] [Google Scholar]
- 61.Naume B, Borgen E, Beiske K, et al. Immunomagnetic techniques for the enrichment and detection of isolated breast carcinoma cells in bone marrow and peripheral blood. J Hematother. 1997;6:103–104. doi: 10.1089/scd.1.1997.6.103. [DOI] [PubMed] [Google Scholar]
- 62.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 63.Coumans FA, Doggen CJ, Attard G, et al. All circulating EpCAM+ CK+CD45− objects predict overall survival in castration-resistant prostate cancer. Ann Oncol. 2010;21:1851–1857. doi: 10.1093/annonc/mdq030. [DOI] [PubMed] [Google Scholar]
- 64.Stott SL, Hsu CH, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Helzer KT, Barnes HE, Day L, et al. Circulating tumor cells are transcriptionally similar to the primary tumor in a murine prostate model. Cancer Res. 2009;69:7860–7866. doi: 10.1158/0008-5472.CAN-09-0801. [DOI] [PubMed] [Google Scholar]
- 66.Giordano A, Giuliano M, De Laurentiis M, et al. Circulating tumor cells in immunohistochemical subtypes of metastatic breast cancer: lack of prediction in HER2-positive disease treated with targeted therapy. Ann Oncol. 2011 doi: 10.1093/annonc/mdr434. [DOI] [PubMed] [Google Scholar]
- 67.Gleghorn JP, Pratt ED, Denning D, et al. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip. 2010;10:27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulmer A, Schmidt-Kittler O, Fischer J, et al. Immunomagnetic enrichment, genomic characterization, and prognostic impact of circulating melanoma cells. Clin Cancer Res. 2004;10:531–537. doi: 10.1158/1078-0432.ccr-0424-03. [DOI] [PubMed] [Google Scholar]
- 69.Racila E, Euhus D, Weiss A, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci U S A. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scholtens TM, Schreuder F, Ligthart ST, et al. CellTracks TDI: an image cytometer for cell characterization. Cytometry A. 2011 doi: 10.1002/cyto.a.21024. [DOI] [PubMed] [Google Scholar]
- 71.Bocsi J, Varga VS, Molnar B, et al. Scanning fluorescent microscopy analysis is applicable for absolute and relative cell frequency determinations. Cytometry A. 2004;61:1–8. doi: 10.1002/cyto.a.20061. [DOI] [PubMed] [Google Scholar]
- 72.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alix-Panabieres C, Rebillard X, Brouillet JP, et al. Detection of circulating prostate-specific antigen–secreting cells in prostate cancer patients. Clin Chem. 2005;51:1538–1541. doi: 10.1373/clinchem.2005.049445. [DOI] [PubMed] [Google Scholar]
- 74.Paris PL, Kobayashi Y, Zhao Q, et al. Functional phenotyping and genotyping of circulating tumor cells from patients with castration resistant prostate cancer. Cancer Lett. 2009;277:164–173. doi: 10.1016/j.canlet.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 75.Shim S, Gascoyne P, Noshari J, et al. Dynamic physical properties of dissociated tumor cells revealed by dielectrophoretic field-flow fractionation. Integr Biol (Camb) 2011;3:850–862. doi: 10.1039/c1ib00032b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gel M, Kimura Y, Kurosawa O, et al. Dielectrophoretic cell trapping and parallel one-to-one fusion based on field constriction created by a micro-orifice array. Biomicrofluidics. 2010;4 doi: 10.1063/1.3422544. pii: 022808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borgatti M, Altomare L, Abonnec M, et al. Dielectrophoresis-based ‘Lab-on-a-chip’ devices for programmable binding of microspheres to target cells. Int J Oncol. 2005;27:1559–1566. [PubMed] [Google Scholar]
- 78.Lecharpentier A, Vielh P, Perez-Moreno P, et al. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer. 2011;105:1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin HK, Zheng S, Williams AJ, et al. Portable filter-based micro-device for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng S, Lin HK, Lu B, et al. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 2011;13:203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan SJ, Lakshmi RL, Chen P, et al. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens Bioelectron. 2010;26:1701–1705. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 83.Tan SJ, Yobas L, Lee GY, et al. Microdevicez for the isolation and enumeration of cancer cells from blood. Biomed Microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 84.Helo P, Cronin AM, Danila DC, et al. Circulating prostate tumor cells detected by reverse transcription-PCR in men with localized or castration-refractory prostate cancer: concordance with CellSearch assay and association with bone metastases and with survival. Clin Chem. 2009;55:765–773. doi: 10.1373/clinchem.2008.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iinuma H, Watanabe T, Mimori K, et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J Clin Oncol. 2011;29:1547–1555. doi: 10.1200/JCO.2010.30.5151. [DOI] [PubMed] [Google Scholar]
- 86.Mocellin S, Hoon D, Ambrosi A, et al. The prognostic value of circulating tumor cells in patients with melanoma: a systematic review and meta-analysis. Clin Cancer Res. 2006;12:4605–4613. doi: 10.1158/1078-0432.CCR-06-0823. [DOI] [PubMed] [Google Scholar]
- 87.Xi L, Nicastri DG, El-Hefnawy T, et al. Optimal markers for real-time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers. Clin Chem. 2007;53:1206–1215. doi: 10.1373/clinchem.2006.081828. [DOI] [PubMed] [Google Scholar]
- 88.Reinholz MM, Kitzmann KA, Tenner KS, et al. Cytokeratin-19 and mammaglobin gene expression in circulating tumor cells from metastatic breast cancer patients enrolled in North Central Cancer Treatment Group (NCCTG) trials, N0234/336/436/437. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-11-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sieuwerts AM, Mostert B, Bolt-de Vries J, et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin Cancer Res. 2011;17:3600–3618. doi: 10.1158/1078-0432.CCR-11-0255. [DOI] [PubMed] [Google Scholar]
- 90.Zhao S, Liu Y, Zhang Q, et al. The prognostic role of circulating tumor cells (CTCs) detected by RT-PCR in breast cancer: a meta-analysis of published literature. Breast Cancer Res Treat. 2011;130:809–816. doi: 10.1007/s10549-011-1379-4. [DOI] [PubMed] [Google Scholar]
- 91.Obermayr E, Sanchez-Cabo F, Tea MK, et al. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer. 2010;10:666. doi: 10.1186/1471-2407-10-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoon SO, Kim YT, Jung KC, et al. TTF-1 mRNA-positive circulating tumor cells in the peripheral blood predict poor prognosis in surgically resected non-small cell lung cancer patients. Lung Cancer. 2011;71:209–216. doi: 10.1016/j.lungcan.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 93.Ignatiadis M, Kallergi G, Ntoulia M, et al. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, mammaglobin A, and HER2 in early breast cancer. Clin Cancer Res. 2008;14:2593–2600. doi: 10.1158/1078-0432.CCR-07-4758. [DOI] [PubMed] [Google Scholar]
- 94.Nakagawa T, Martinez SR, Goto Y, et al. Detection of circulating tumor cells in early-stage breast cancer metastasis to axillary lymph nodes. Clin Cancer Res. 2007;13:4105–4110. doi: 10.1158/1078-0432.CCR-07-0419. [DOI] [PubMed] [Google Scholar]
- 95.Schleiermacher G, Peter M, Oberlin O, et al. Increased risk of systemic relapses associated with bone marrow micrometastasis and circulating tumor cells in localized ewing tumor. J Clin Oncol. 2003;21:85–91. doi: 10.1200/JCO.2003.03.006. [DOI] [PubMed] [Google Scholar]
- 96.Yaniv I, Cohen IJ, Stein J, et al. Tumor cells are present in stem cell harvests of Ewings sarcoma patients and their persistence following transplantation is associated with relapse. Pediatr Blood Cancer. 2004;42:404–409. doi: 10.1002/pbc.20022. [DOI] [PubMed] [Google Scholar]
- 97.Wong IH, Chan AT, Johnson PJ. Quantitative analysis of circulating tumor cells in peripheral blood of osteosarcoma patients using osteoblast-specific messenger RNA markers: a pilot study. Clin Cancer Res. 2000;6:2183–2188. [PubMed] [Google Scholar]
- 98.Dubois SG, Epling CL, Teague J, et al. Flow cytometric detection of Ewing sarcoma cells in peripheral blood and bone marrow. Pediatr Blood Cancer. 2010;54:13–18. doi: 10.1002/pbc.22245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pierga JY, Hajage D, Bachelot T, et al. High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann Oncol. 2011 doi: 10.1093/annonc/mdr263. [DOI] [PubMed] [Google Scholar]
- 100.de Bono JS, Attard G, Adjei A, et al. Potential applications for circulating tumor cells expressing the insulin-like growth factor-I receptor. Clin Cancer Res. 2007;13:3611–3616. doi: 10.1158/1078-0432.CCR-07-0268. [DOI] [PubMed] [Google Scholar]
- 101.Fehm T, Becker S, Duerr-Stoerzer S, et al. Determination of HER2 status using both serum HER2 levels and circulating tumor cells in patients with recurrent breast cancer whose primary tumor was HER2 negative or of unknown HER2 status. Breast Cancer Res. 2007;9:R74. doi: 10.1186/bcr1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rossi E, Basso U, Celadin R, et al. M30 neoepitope expression in epithelial cancer: quantification of apoptosis in circulating tumor cells by CellSearch analysis. Clin Cancer Res. 2010;16:5233–5243. doi: 10.1158/1078-0432.CCR-10-1449. [DOI] [PubMed] [Google Scholar]
- 103.Marrinucci D, Bethel K, Lazar D, et al. Cytomorphology of circulating colorectal tumor cells: a small case series. J Oncol. 2010;2010:861341. doi: 10.1155/2010/861341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leversha MA, Han J, Asgari Z, et al. Fluorescence in situ hybridization analysis of circulating tumor cells in metastatic prostate cancer. Clin Cancer Res. 2009;15:2091–2097. doi: 10.1158/1078-0432.CCR-08-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Flores LM, Kindelberger DW, Ligon AH, et al. Improving the yield of circulating tumour cells facilitates molecular characterisation and recognition of discordant HER2 amplification in breast cancer. Br J Cancer. 2010;102:1495–1502. doi: 10.1038/sj.bjc.6605676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Desitter I, Guerrouahen BS, Benali-Furet N, et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011;31:427–441. [PubMed] [Google Scholar]
- 107.Swennenhuis JF, Tibbe AG, Levink R, et al. Characterization of circulating tumor cells by fluorescence in situ hybridization. Cytometry A. 2009;75:520–527. doi: 10.1002/cyto.a.20718. [DOI] [PubMed] [Google Scholar]
- 108.Fehm T, Solomayer EF, Meng S, et al. Methods for isolating circulating epithelial cells and criteria for their classification as carcinoma cells. Cytotherapy. 2005;7:171–185. doi: 10.1080/14653240510027082. [DOI] [PubMed] [Google Scholar]
- 109.Vona G, Beroud C, Benachi A, et al. Enrichment, immunomorphological, and genetic characterization of fetal cells circulating in maternal blood. Am J Pathol. 2002;160:51–58. doi: 10.1016/S0002-9440(10)64348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang ZP, Eisenberger MA, Carducci MA, et al. Identification and characterization of circulating prostate carcinoma cells. Cancer. 2000;88:2787–2795. doi: 10.1002/1097-0142(20000615)88:12<2787::aid-cncr18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 111.Vona G, Sabile A, Louha M, et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Budd GT, Cristofanilli M, Ellis MJ, et al. Circulating tumor cells versus imaging—predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12:6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 113.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 114.Cristofanilli M, Hayes DF, Budd GT, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 115.Hayes DF, Cristofanilli M, Budd GT, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 116.Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 118.Cohen SJ, Punt CJ, Iannotti N, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol. 2009;20:1223–1229. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- 119.Botteri E, Sandri MT, Bagnardi V, et al. Modeling the relationship between circulating tumour cells number and prognosis of metastatic breast cancer. Breast Cancer Res Treat. 2010;122:211–217. doi: 10.1007/s10549-009-0668-7. [DOI] [PubMed] [Google Scholar]
- 120.Scher HI, Beer TM, Higano CS, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet. 2010;375:1437–1446. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Koyanagi K, O’Day SJ, Boasberg P, et al. Serial monitoring of circulating tumor cells predicts outcome of induction biochemotherapy plus maintenance biotherapy for metastatic melanoma. Clin Cancer Res. 2010;16:2402–2408. doi: 10.1158/1078-0432.CCR-10-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Riethdorf S, Muller V, Zhang L, et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res. 2010;16:3634–2645. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 123.Danila DC, Anand A, Sung CC, et al. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. Eur Urol. 2011;60:897–904. doi: 10.1016/j.eururo.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 125.Scher HI, Sawyers C. Biology of progressive castration resistant prostate cancer: directed therapies targeting the androgen receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 126.Newling DWW. Issues with the use of prostate-specific antigen as a surrogate end point in hormone-resistant prostate cancer. Eur Urol Suppl. 2009;8:13–19. [Google Scholar]
- 127.de Bono JS, Scher HI, Montgomery B, et al. Circulating tumor cells predict overall survival following treatment in metastatic prostate cancer. Clinical Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 128.Halabi S, Vogelzang NJ, Ou SS, et al. Progression-free survival as a predictor of overall survival in men with castrate-resistant prostate cancer. J Clin Oncol. 2009;27:2766–2771. doi: 10.1200/JCO.2008.18.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sternberg CN, Petrylak DP, Sartor O, et al. Multinational, double-blind, phase III study of prednisone and either satraplatin or placebo in patients with castrate-refractory prostate cancer progressing after prior chemotherapy: the SPARC trial. J Clin Oncol. 2009;27:5431–5438. doi: 10.1200/JCO.2008.20.1228. [DOI] [PubMed] [Google Scholar]
- 130.Schellhammer PF. Words of wisdom. Re: pathologic characteristics of cancers detected in the Prostate Cancer Prevention Trial: implications for prostate cancer detection and chemoprevention. Eur Urol. 2009;55:522–523. doi: 10.1016/j.eururo.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 131.Reid AH, Attard G, Danila DC, et al. Significant and sustained antitumor activity in post-docetaxel, castration-resistant prostate cancer with the CYP17 inhibitor abiraterone acetate. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.24.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Danila DC, Morris MJ, de Bono JS, et al. Phase II multicenter study of abiraterone acetate plus prednisone therapy in patients with docetaxel-treated castration-resistant prostate cancer. J Clin Oncol. 2010;28:1496–1501. doi: 10.1200/JCO.2009.25.9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scher H, Heller G, Molina A, et al. Evaluation of circulating tumor cell (CTC) enumeration as an efficacy response biomarker of overall survival (OS) in metastatic castration-resistant prostate cancer (mCRPC): planned final analysis (FA) of COU-AA-301, a randomized doubleblind, placebo-controlled phase III study of abiraterone acetate (AA) plus low-dose prednisone (P) post docetaxel. In: ASCO, editor. J Clin Oncol. 2011;29(suppl) abstr LBA4517. [Google Scholar]
- 135.Buyse M, Molenberghs G. Criteria for the validation of surrogate endpoints in randomized experiments. Biometrics. 1998;54:1014–1029. [PubMed] [Google Scholar]
- 136.Burzykowski T, Molenberghs G, Buyse M, et al. Validation of surrogate endpoints in multiple randomized clinical trials with failure-time endpoints. Appl Stat. 2001;50:405–422. [Google Scholar]
- 137.Pantel K, Riethdorf S. Pathology: are circulating tumor cells predictive of overall survival? Nat Rev Clin Oncol. 2009;6:190–191. doi: 10.1038/nrclinonc.2009.23. [DOI] [PubMed] [Google Scholar]