Abstract
Background
Wnt/PCP signaling plays a critical role in multiple developmental processes, including limb development. Wnt5a, a ligand of the PCP pathway, signals through the Ror2/Vangl2 or the Vangl2/Ryk complex to regulate limb development along the proximal-distal axis in mice. Based on the interaction between Van Gogh and Prickle in Drosophila, we hypothesized the vertebrate Prickle1 have similar function as Vangl2 in limb development.
Results
We show Prickle1 is expressed in the skeletal condensates that will differentiate into chondrocytes and later form bones. Disrupted Prickle1 function in Prickle1C251X/C251X mouse mutants alters expression of genes such as Bmp4, Fgf8, Vangl2 and Wnt5a. These expression changes correlate with shorter and wider bones in the limbs and loss of one phalangeal segment in digits 2-5 of Prickle1C251X mutants. These growth defects along the proximal-distal axis are also associated with increased cell death in the growing digit tip, reduced cell death in the interdigital membrane and disrupted chondrocyte polarity.
Conclusions
We suggest Prickle1 is part of the Wnt5a/PCP signaling, regulating cell polarity and affecting expression of multiple factors to stunt limb growth through altered patterns of gene expression, including the PCP genes Wnt5a and Vangl2.
Introduction
Prickle like 1 (Prickle1) is a core component in Wnt non-canonical planar cell polarity (PCP) pathway (Gubb et al., 1999; Tao et al., 2009), which regulates cell polarity and related cell activity in Drosophila and vertebrates (Barrow, 2006; Kestler and Kühl, 2008; McNeill and Woodgett, 2010). In Drosophila, prickle (pk) and Van Gogh (Vang) form a protein complex in the cell membrane opposite to the frizzled/disheveled protein complex, thus establishing cell polarity along a given axis (Bastock et al., 2003; Barrow, 2006). In zebrafish, knockdown of prickle1 or van gogh like2 (vangl2) expression affects the PCP related convergent-extension (CE) movements during gastrulation and facial branchial motor neuron migration (Carreira-Barbosa et al., 2003; Takeuchi et al., 2003; Veeman et al., 2003; Glasco et al., 2012). Most of our current understandings of Wnt/PCP function in mammals stem from Wnt5a and Vangl2 mouse mutants (Torban et al., 2004; Montcouquiol et al., 2006; Gao et al., 2011; Glasco et al., 2012; Deans, 2013) with almost no data from Prickle1 mutants (Gubb et al., 1999; Tao et al., 2009).
In mammalian limbs, disruption of Wnt5a, Vangl2 or Ror2 (receptor tyrosine kinase-like orphan receptor 2) leads to shorter limbs and brachydactyly (Yamaguchi et al., 1999; Schwabe et al., 2004; Witte et al., 2010; Gao et al., 2011; Wang et al., 2011). Ror2 mediates Wnt5a signaling via a dose-dependent Vangl2 phosphorylation, establishing a gradient of Vangl2 activity along the proximal-distal axis, and thus directs limb elongation (Gao et al., 2011). Mutations in the ROR2 gene in human cause brachydactyly type B or Robinow syndrome which affects long bones in arms and legs among other defects (Afzal et al., 2000; Oldridge et al., 2000; Schwabe et al., 2000; van Bokhoven et al., 2000; Afzal and Jeffery, 2003). Ryk (receptor-like tyrosine kinase) also plays a role in regulating PCP signaling (Andre et al., 2012). Although simple Ryk−/− mice do not have obvious defects in limb development, double null Ryk−/− ;Vangl2−/− mutants have more severe limb defects than Vangl2−/− mice, and are approaching the Wnt5a−/− mice phenotype in severity, presumably because Wnt5a potentiates the binding between Vangl2 and Ryk, promoting stability of Vangl2 (Andre et al., 2012).
The defects in limb development appear to be the consequences of disrupted signaling network controlling cell death and proliferation, such as Fgf and Bmp signaling (Witte et al., 2010; Wang et al., 2011). Fgf and Bmp signaling balance proliferation/growth, cell death in the digit and interdigital space and cell fate commitment to the cartilage lineage. These factors are important in growth and patterning of the limb bud along the PD axis (Niswander and Martin, 1993; Yokouchi et al., 1996; Macias et al., 1997; Wang et al., 2004; Bandyopadhyay et al., 2006). Consistent with the proposed molecular interaction, Bmp4 haploinsufficiency partially rescues phalangeal growth in Vangl2/looptail mutants (Wang et al., 2011).
Another proposed explanation for the long bone growth defects is disrupted chondrocyte polarity in the growth plate (Li and Dudley, 2009; Randall et al., 2012). Proliferative chondrocytes in the developing bones divide orthogonal to the long bone axis and then intercalate to form a column to promote long bone growth (Li and Dudley, 2009). Disrupted PCP signaling in chicken alters the cell division plane in the proliferating chondrocytes (Li and Dudley, 2009). Activated Wnt5a signaling through Fzd7, Ror2 and Vangl2 induces the formation of chondrocytes in columns in cultured rat growth plate (Randall et al., 2012). These results show that Wnt/PCP signaling is critical for polarity of proliferative chondrocytes in the growing long bones, regulating oriented cell divisions and thus leads to long bone growth.
Prickle expression has been reported in the developing limbs of mice (Bekman and Henrique, 2002) and chicken (Cooper et al., 2008). Unfortunately, the function of Prickle1, the presumed intracellular partner of Vangl2 (Jessen et al., 2002; Bastock et al., 2003; Carreira-Barbosa et al., 2003; Jenny et al., 2003) remains poorly understood. Supporting a fly-like interaction, Prickle1 or Vangl2 mutation causes similar phenotypes during the primitive streak formation in mice and fish and neural tube defects (NTD) in humans (Tao et al., 2009; Lei et al., 2010; Bosoi et al., 2011; Kibar et al., 2011; Mapp et al., 2011). In addition, Prickle1 is regulated by Smurf and asymmetrically localized in the cochlear hair cells through ubiquitin-mediated degradation and Vangl2lp/lp mice have disrupted hair cell polarity (Narimatsu et al., 2009; Yin et al., 2012). Beyond this correlative evidences, there is limited data on Prickle1 participation in Wnt/PCP signaling in mammals.
Prickle1 has a PET (Prickle, Espinas and Testin) domain in its N-terminus and three LIM (Lin11, Isl-1 and Mec3) domains in the C-terminus. It belongs to the group of LIM proteins that are associated with actin and the transcriptional machinery (Kadrmas and Beckerle, 2004; Zheng and Zhao, 2007) thus playing a dual role in gene regulation and cargo transportation. For example, LMCD1/dyxin, a protein closely related to Prickle1 (Bekman and Henrique, 2002) is known to interact with other transcription factors such as GATA factors (Rath et al., 2005). Consistent with the possible nuclear signaling of Prickle1 are several nuclear localization signals, N-glycosylation sites, cAMP-dependent protein kinase A sites and prenylation motif at the C-terminus (Shimojo and Hersh, 2003; Liu et al., 2013). These structures are necessary for the proteins to localize to the nucleus, which is critical for the Prickle1 protein function as a transcription modulator (Bassuk et al., 2008; Mapp et al., 2011; Tao et al., 2012; Liu et al., 2013). In fact, recent data suggest that vertebrate Prickle1's function might in part be mediated by nuclear signaling (Mapp et al., 2011; Tao et al., 2012). We therefore hypothesized that Prickle1 plays a role in Wnt/PCP signaling partially through gene regulation, possibly a neo-functionalization of Prickle1.
We approached this hypothesis by analyzing the Prickle1 C251X mouse line, which has a premature stop-codon in the third LIM domain, eliminating the third LIM domain and the nuclear localization sequence (Shimojo and Hersh, 2003; Shimojo and Hersh, 2006; Zhong et al., 2009; Mapp et al., 2011; Tao et al., 2011). Our results are consistent with the hypothesis that Prickle1 is associated with Wnt5a-PCP signaling in limb development. In addtion, mutated Prickle1 rapidly affects gene expression that regulates limb growth by altered apoptosis in both digit and interdigital regions and altered chondrocyte polarity of the long bones, leading to shorter and wider long bones in the limb and loss of one phalangeal segment in digits 2-5, a phenotype similar to human Robinow syndrome and brachydactyly type B.
Results
Mutation in Prickle1 causes growth defects
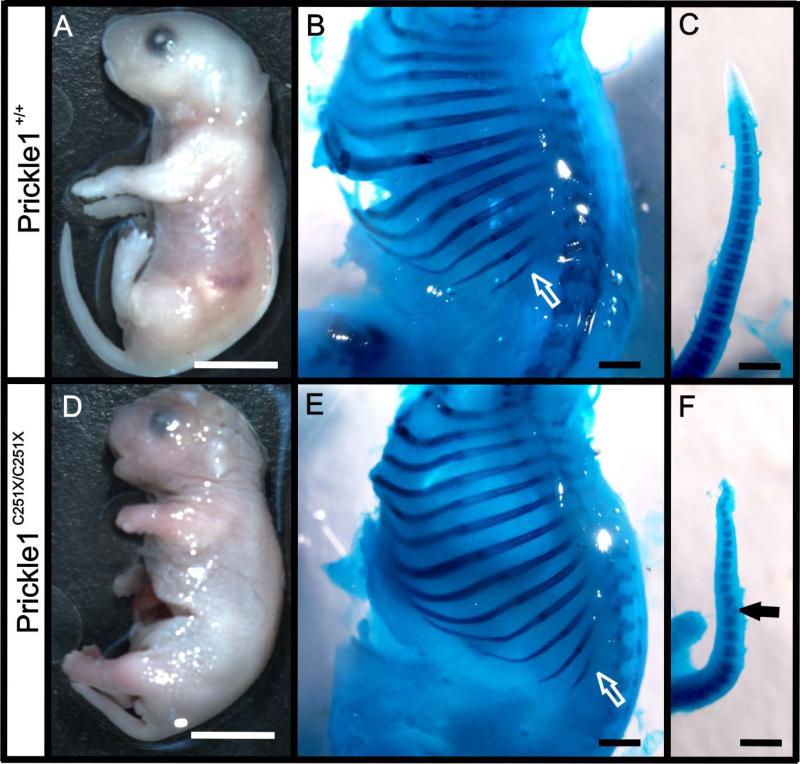
At embryonic day 18.5 (E18.5), Prickle1 C251X homozygous mutants (Prickle1C251X/C251X) had shorter snouts, limbs and tails (Fig. 1 A and D) compared with their wild-type littermates (Fig. 1 B and E). The shortening of the tail was due to smaller vertebrae (Fig. 1 C and F). In addition, some vertebrae in the mutant were deformed (Fig. 1F). In contrast to the vertebrae in the tail, vertebrae in the main body axis of the mutants showed no obvious difference from the wild-type (Fig. 1 B, E). Vertebrae number, size, ossification, and the distance between the neighboring vertebrae were all normal. This suggested that the growth defects in Prickle1C251X/C251X mice were not due to a general delayed development, but due to a specific role of Prickle1 in ossified structures extending away from the main body axis.
Figure 1.
Prickle1 C251X homozygotic mice have stunted growth of appendages extending from the axis. A and D: at E18.5, the Prickle1 C251X/C251X mice (D) have shorter snout, shorter limbs and shorter tail compared with wild-type (A). B, C, E and F: the skeleton from wild-type embryos (B-C) and mutant embryos (E-F) is double stained with Alcian blue and Alizarin red. While the total length of the rib cage does not show significant difference, ribs tend to be shorter in the mutant (compare while arrows in B and E). The mutant has shorter tail due to deformed (black arrow in F) and fewer segments (compare C and F). The scale bar is 5 mm in A and D and 1mm in B, C, E and F.
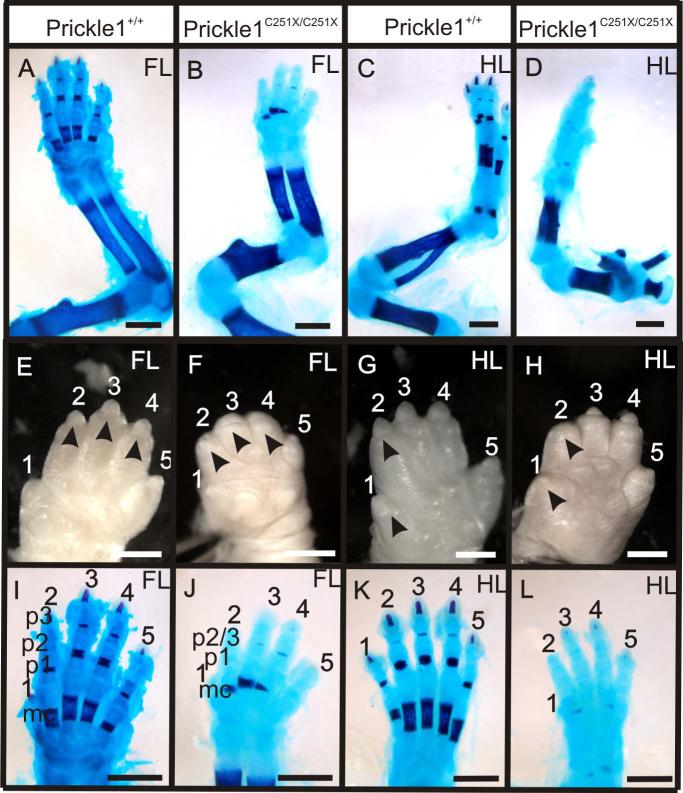
Since limb development is a molecularly well characterized process (Tabin and McMahon, 2008; Mao et al., 2009; Shubin et al., 2009), we examined the limb defects in detail and studied alterations of gene expressions in Prickle1 mutants. The limbs were stained with Alcian Blue and Alizarin Red double staining, which labels the cartilage and calcified bone matrix respectively. At E18.5, all the long bones in the limbs of Prickle1C251X/C251X mice were shorter (Fig. 2 A-D). The mutants also had abnormal primary claw fields at the digit tip (black arrowheads, Fig. 2 E-H), a structure homologous to finger nails in humans. In the mutant, the claw field was absent in digits 3 and 4 in the fore limb and digit 2 in the hind limb, but even though it was present in the rest of the digits, it appeared reduced in size. The defects are similar to the clinical symptom in some human Robinow syndrome and brachydactyly type B1 (BDB1) patients, as well as Vangl2 Lp/Lp and RorW745X/W745X mouse mutants (Afzal et al., 2000; Afzal and Jeffery, 2003; Schwabe et al., 2004; Witte et al., 2010; Wang et al., 2011).
Figure 2.
Prickle1C251X/C251X mice showed defective limb growth at E18.5. A-D: Alizarin red and Alcian blue staining showing the impaired limb growth in forelimb (A and B) and hindlimb (C and D). The long bones in the mutants (B and D) are shorter than those in the wild-type (A and C). E-H: the digits in the forelimb (E) and hindlimb (H) of the mutants are shorter than that of their wild-type littermates (E and G). The primary claw field is also impaired in the mutants (compare black arrows). I-L: Alizarin red and Alcian blue staining of the hand (I and J) and foot (K and L). There are metacarpal, p1, p2 and p3 in digits 2-5 in the wild-type (I and K), but in the mutant (J and L), there are only metacarpal, p1 and p2/3. Ossification is impaired in all the segments in all digits (red staining in mc, p1, p2 and p3). mc, metacarpal; p1-p3, phalange 1-3. The scale bar is 1mm.
We noticed in addition to an overall growth reduction, there was loss of one phalangeal bone in digits 2-5 in both forelimbs and hindlimbs of the mutants (Fig. 2 I-L). In wild-type or Prickle1C251X/+ mice, digits 2–5 consisted of the metacarpal (Fig. 2I, mc) and three phalangeal bones (Fig. 2I, p1, p2 and p3). In contrast, Prickle1 C251X/C251X limbs had only the metacarpal and two phalangeal bones (Fig. 2J, mc, p1 and p2/3). In addition, the mineralization of the digits in the mutant forelimbs was also impaired (Fig. 2, compare I and J, alizarin red staining). The shape of calcified bone in metacarpals was wedge shaped, and calcification was delayed in some phalanges, especially in the distal bones (Fig. 2, compare I and J). Mineralization in the autopod of the mutant hindlimbs was almost absent (Fig. 2, compare K and L).
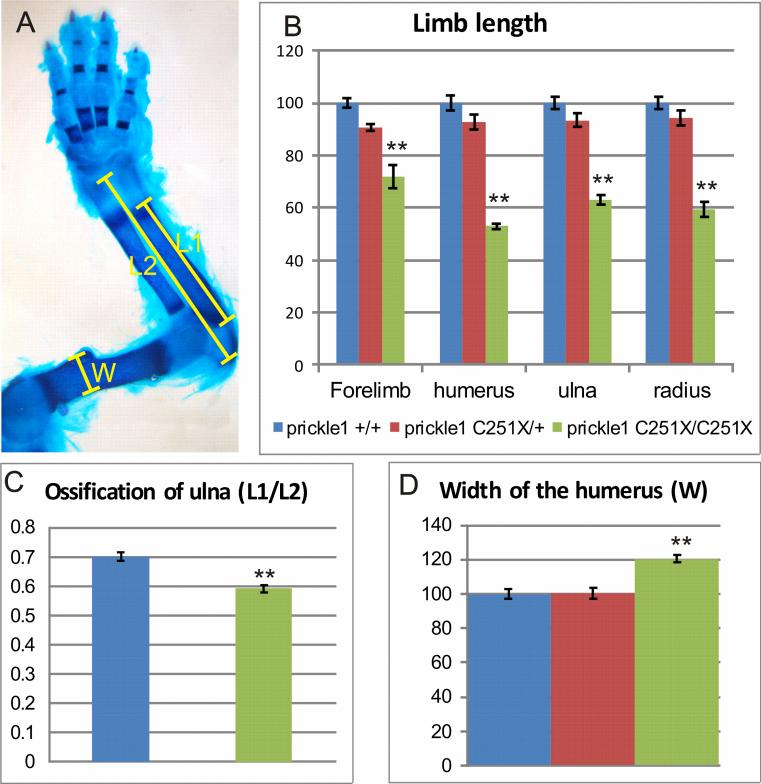
We quantified these defects in the forelimb (Fig. 3). The mutant forelimb was only about 70% of the wild-type's forelimb (Fig. 3B, n-8, p<0.01, ANOVA). We also quantified the length of long bones by measuring the mineralized part stained by alizarin red (Fig. 3A), including humerus, ulna and radius in the forelimb (Fig. 3 B). We only measured length of calcified bones because it was easy to define the borderline and it served to demonstrate the defects in mineralization. All long bones in the homozygous mutant limbs were about 40% shorter than the wild-types (Fig. 3B, n=8, p<0.01, ANOVA). This general reduction in the whole limb was similar to the Wnt5a−/− mutants (Yamaguchi et al., 1999; Gao et al., 2011; Wang et al., 2011), although the paw defects in Prickle 1 mutants were not as severe.
Figure 3.
Quantification of the limb growth defects in Prickle1 C251X mice at E18.5. A: Diagram showing the methods for quantification. L1 is the bone part that is stained red Alizarin red, which is mineralized bone. L2 is the total bone length, measured from joint to joint. W is the width of the widest part of the humerus. B: comparison of the normalized length of the forelimb, humerus, ulna and radius. Homozygotic mutant forelimb and long bones are significantly shorter than both wild-type and the heterozygous mice (n=8, p<0.01, ANOVA), but heterozygotic mutant are not significantly shorter than wild-type. C: The percentage of mineralized bone length in homozygotic mutants (60%) is smaller than the wild-type (70%) (n=8, p<0.01, t-test). D: The width of the humerus is compared, and the homozygotic mutants have significantly wider bone than the wild-type and the heterozygote (n=8, p<0.01, ANOVA). However, the heterozygotic mutant humeri are not significantly wider than the wild-type.
To show the defects in calcification, we also measured the ratio of mineralization of the ulna (Fig. 3 A and C). While calcified bone was about 70% of the whole bone in the wild-type, calcified bone was only about 60% of the whole bone (Fig. 3C, n=8, p<0.01, t-test). Since the bones in the mutants looked wider, we compared the bone width by measuring the widest point of the humerus (Fig. 3 A and D). The mutant humeri were significantly wider than the wild-type (Fig. 3 D, n=8, p<0.01, ANOVA).
Although some C251X heterozygous mutants had what appeared to be shorter limbs, neither the length nor the width was statistically significant different from the wild-type (Fig. 3 B and D, n=8, ANOVA). For this reason, we only focused on homozygous mutants in the following study.
Prickle1 mutation affects morphology of limb bud starting at E12.5
To understand how the Prickle1 C251X mutation alters limb development, we analyzed next the limb bud at early stages. Since Prickle1 is a member of PCP pathway, which regulates convergent-extension (CE) in various developmental processes, we asked whether the limb bud had CE defects as in Vangl2/looptail mutants. To see how early a morphological effect can be detected we quantified the size of the hand at various stages. We identified the area of autopod by defining a reference line. The widest line parallel to the reference line was measured and defined as the width of the autopod. From E11.5 to E13.5, the hand width was not statistically different between wild-type and Prickle1 C251X/C251X mutants (data not shown). We drew a line perpendicular to the reference line to define the longest point of the limb connecting to the reference line; this was defined as hand length. Although the mutant limbs were slightly shorter and narrower since E12.5, this difference was not statistically different until E14.5 (data not shown), which is different from Vangl2 lp/lp mutants that have CE defects starting at E11 (Wang et al., 2011). We also compared the thickness of limb bud along the dorsal-ventral (DV) axis at E12.5. The mutant limb buds were not obviously thicker than wild-type as in Vangl2 lp/lp mice (data not shown).
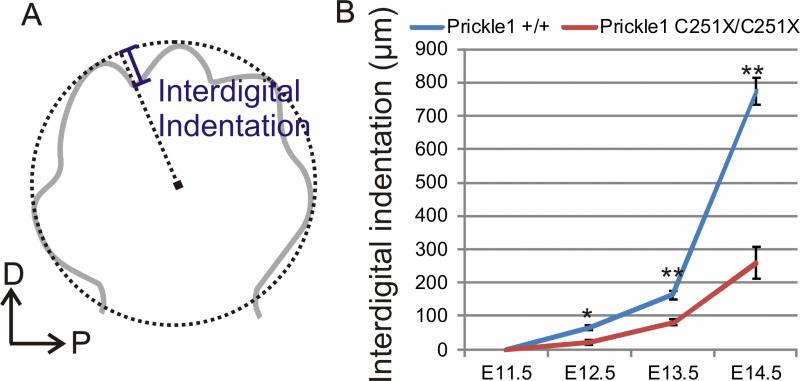
We quantified next the development of interdigital indentation in the mutants. We drew a circle that best fit the tip of digits 2-5, and measured the distance from the lowest point in the interdigital indentation between digits 2 and 3 along the radius from the center to the circle (Fig. 4A). At E11.5, either wild-type or mutant showed digit indentation. The interdigital indentation was significantly different beginning at E12.5 (Fig. 4B, n=4, p<0.05, t-test) and became even more profound at E13.5 (n=6, p<0.01, t-test) and E14.5 (n=4, P<0.01, t-test). Thus, changing the ratio of digit growth and interdigital tissue indentation is the first morphologically detectable effect of Prickle1C251X/C251X mice and suggested an even earlier onset of molecular defects prior to E12.5.
Figure 4.
Interdigital space indentation is impaired in Prickle1C251X/C251X mice. A: diagram showing how the quantification was performed. A circle that best fit all the digits is drawn. A radius across the lowest point between digits 2 and 3 is drawn. The length of interdigital indentation is defined by the distance from this lowest point to the circle. B: comparison of the interdigital space between wild-type and homozygotic mutants. From E12.5, the digit indentation is impaired in the mutants (n=4 and p<0.05 at E12.5, n=6 and p<0.01 at E13.5, n=4 and p<0.01at E14.5, t-test).
Wnt5a and Bmp4 expression change might be responsible for early limb development defects
Because Prickle1C251X mutation did not cause the obvious early limb bud dimensional changes observed in Vangl2/looptail mutants, we reasoned Prickle1 rather than acting as the intracellular partner of Vangl2, might regulate limb development using mechanisms different from that of Vangl2. Considering the loss of nuclear translocation signaling in our Prickle1C251X/C251X protein, we hypothesized Prickle1 might regulate gene expression at the transcription level. In addition, since molecular changes have to be earlier than morphological changes, we examined the expression of genes known to be important in the limb development at E12.5 and E11.5.
E12.5
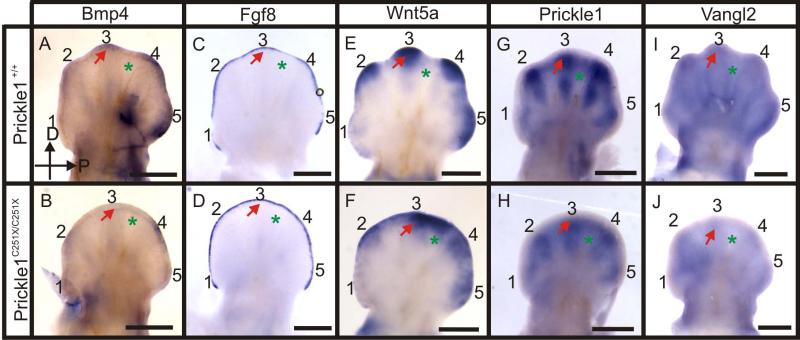
At E12.5 the expression of several genes was changed (Fig. 5). Bmp4 was expressed in the distal region of digits 2-5, and low expression was also visible in the developing digit rays (Fig. 5 A). In Prickle1C251X/C251X mice, the expression of Bmp4 in the digit rays was reduced, and the expression in the distal region was ablated in digit 3 and down-regulated in digits 2, 4 and 5 (Fig. 5 B). Fgf8 was expressed in the AER, but showed down-regulation in the interdigital space in the wild-type (Fig. 5C). However, the differential expression between digit and interdigital space was absent in the mutants (Fig. 5D). This failed differential expression might suggest delayed development in the mutants, but also might suggest patterning defects between digit and interdigital space. Wnt5a was expressed in the distal parts of the digits but not in the interdigital space in the wild-type (Fig. 5 E). Similarly to Fgf8, the differential expression of Wnt5a between digit and interdigital space was blurred in the mutant (Fig. 5F). In addition, the expression was reduced in the tip of digits 2-3 in the mutants (Fig. 5 E and F).
Figure 5.
Whole mount mRNA in situ hybridization shows the expression of several genes is affected by Prickle1C251X mutation at E12.5. A-B: Bmp4 is weekly expressed by developing digit condensation, but the expression in the distal tip of the mutant digits is greatly reduced (arrow). C-D: Fgf8 is down-regulated in interdigital space in the wild-type, but is still expressed in the mutant (asterisks). E-F: Wnt5a expression is down-regulated in the distal tip of digits 2-4 in the mutant, but the interdigital membrane of the mutant maintains Wnt5a expression (asterisks). G-H: Prickle1 is highly expressed by developing digit rays (arrow) and weekly in the interdigital membrane (asterisk) in the wild-type. However, the general expression is less intensive in the mutant. In addition, Prickle1 expression in the wild-type segments into two in digits 2-5, but this segmentation of expression is not obvious in the mutant. I-J: Vangl2 is expressed by the whole limb bud, but strongly expressed by the digit tips (arrow) in wild-type. The expression is down-regulated in mutants. The hand is imaged from the dorsal side, with distal up and posterior right. Numbers (1-5) indicate the digit number. Arrow, digit; asterisk, interdigital membrane. The scale bar is 500 μm.
Combined, the Fgf8, Bmp4 and Wnt5a expression pattern suggests that the differential growth of digit and interdigital space is associated with the differential expression of Bmp-Fgf-Wnt5a signaling in the distal digit (Fig. 5 A, C and E). This distinct expression pattern is disturbed in the mutant (Fig. 5 D, F and H), and this coincides with defects in digit growth and interdigital space indentation. However, whether gene expression change was the cause of the patterning defects or the consequence of patterning defects was still unclear as earlier expression alterations could have already induced these changes.
We therefore next investigated the expression of Prickle1 reasoning that expression of Prickle1 must be present before the mutation could elicit an effect. At E12.5, Prickle1 was highly expressed in the digit condensate. The digit's pre-chondrogenic condensate is a template of the future bone. At this stage, the condensate is a single unsegmented column known as the digit ray (Fig. 5 G). Therefore, the differential Prickle1 expression is associated with digit segmentation, and down-regulation of Prickle1 may be necessary for joint formation. Prickle1 was also expressed weakly in the distal mesenchyme in the interdigital region, connecting the expression in the digits (Fig. 5G).
It is known that the majority of nonsense transcripts are recognized and efficiently degraded through the nonsense-mediated mRNA decay (NMD) (Frischmeyer and Dietz, 1999). Therefore, we expected a reduced mRNA level in the mutants. Consistent with our prediction, the general expression of Prickle1 was lower in the mutants. In addition, the digit segmentation was not obvious, and the Prickle1 expressing region was shorter and wider, reflecting an altered growth of the digit condensates (compare Fig. 5 G and H).
Since Wnt5a expression was changed by Prickle1 C251X mutation, we asked whether Vangl2 expression was also changed by this mutation. At E12.5, Vangl2 expression was mostly concentrated in the digit tips (Fig. 5 I red arrow). Vangl2 expression was reduced in the posterior limb region in Prickle1 mutants (Fig. 5 J). Vangl2 and Prickle1 expression showed limited overlap in digit tips (Fig. 5 G and I). Importantly, the profound expression of Prickle1 in proximal digit regions and of Vangl2 in regions defining the digit boundary hardly overlap with each other. This limited overlap of expression reduces the probability that an interaction between Prickle1 and Vangl2 protein may cause the defects in Prickle1 C251X mutants. We suggest that the reduction in expression of both Vangl2 and Wnt5a in the Prickle1 C251X mutants could result in the PCP related defects of Prickle1 mutants.
E11.5
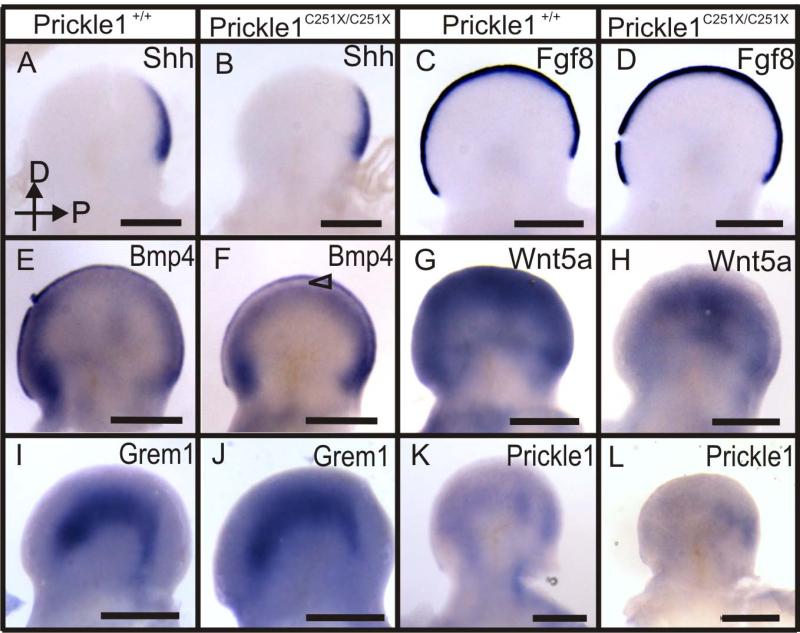
At E11.5, although Shh, Fgf8 and Grem1 expression were indistinguishable between control and Prickle 1 C251X mutants (Fig. 6 A-D, I-J), there was already a visible expression pattern change in Bmp4 and Wnt5a (Fig. 6 E-H). Bmp4 was expressed by the AER and subridge mesenchyme in the wild-type (Fig. 6E). In the mutant, there was an area in the subridge mesenchyme that was not expressing Bmp4 (Fig. 6F, triangle). In the wild-type, Wnt5a was highly expressed by the outer half of the autopod (Fig. 6G), but the expression was reduced in the mutants (Fig. 6H). Prickle1 expression was relatively low at this stage compared with E12.5. It was expressed in the mesenchyme, possibly the future digits (Fig. 6 K). However in the Prickle1C251X/C251X mutants, the mRNA expression was barely detectable (Fig. 6L).
Figure 6.
Whole mount mRNA in situ hybridization shows Bmp4 and Wnt5a mRNA expression is changed in C251X homozygotic mutants at E11.5 (A-L). A-D: The expression of Shh, and Fgf8 in the wild-type and the mutant do not show obvious differences. E-F: Bmp4 is expressed by the AER and subridge mesenchyme in the wild-type. However, its expression is reduced in the subridge mesenchyme in the mutant (F, triangle). G-H: Wnt5a is profoundly expressed by the outer half of the limb bud, but its expression is greatly reduced in the mutants. I-J: Gremlin1 is expressed in the mesenchyme and there is no obvious expression pattern difference between wild-type and mutant. K-L: Prickle1 is variably expressed in the limb bud. The expression pattern might be the developing skeleton condensates (K). Prickle1 mRNA expression is reduced in the mutants, especially the expression in the anterior limb bud (L). The hand is imaged from the dorsal side, with distal up and posterior to the right. Scale bar is 500 μm.
We did not detect any patterned expression of Prickle1 at E10.5 and thus assume that E11.5 demarcates the earliest relevant level of Prickle1 expression in the limbs. Since the most profound and earliest change detected in any of the genes surveyed here is the dramatic down-regulation of Wnt5a in Prickle1C251X/C251X mice, we suggest that Wnt5a expression changes might be responsible for subsequent change of other genes and thus much of the phenotype. The apparently fast and profound down-regulation of Wnt5a in Prickle1C251X/C251X mice suggests a rapid feedback loop for which Prickle1 is necessary to stabilize Wnt5a expression through an unknown mechanism.
The reduction in digit growth in C251X mutants is associated with changed apoptosis pattern
We reasoned that shorter bone length along PD axis could be due to either increased cell death or reduced cell proliferation. Either change would, in turn, result in reduced cell number and lead to defective growth and loss of a phalanx. After immunocytochemistry staining of the limb buds, the whole limb bud was imaged at 5μm interval along the DV axis. Three middle images from a sample were used for quantification, and the average of the three sections was taken as the value for that sample. To test proliferation, we examined dividing cells by labeling proliferating cells with PHH3 antibody, a mitotic marker. At E12.5 (data not shown), in a circle of 200 μm diameter in the tip of digit 3, there was no obvious difference in proliferating cell rate, suggesting that the proliferating cell population was not massively affected in Prickle1 C251X/C251X mutants (n=3). However, it should be noted that since the digits in the mutants were shorter than the wild-type, the total number of proliferating cells should nevertheless be reduced although the proliferation rate per volume was not obviously affected.
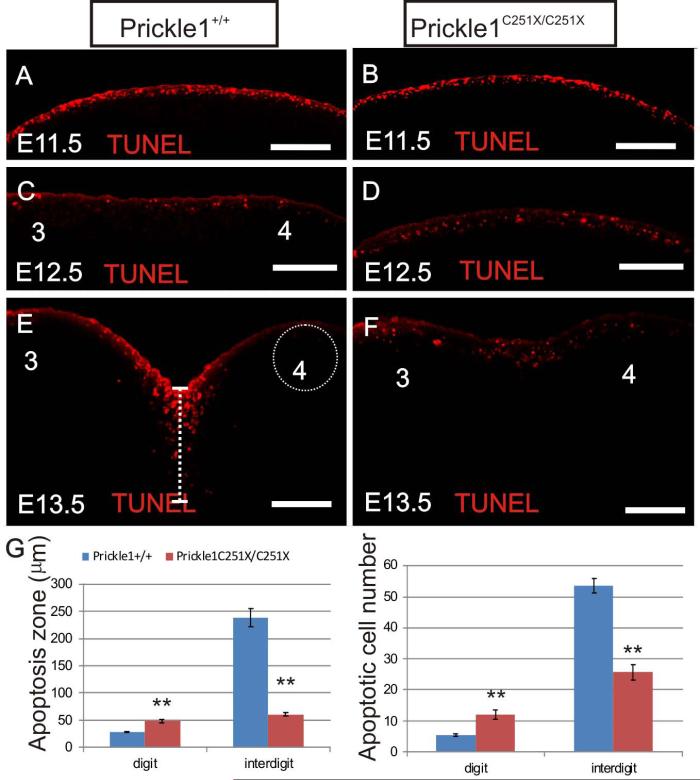
In our next attempt to account for the size reduction, we examined cell death in E11.5, E12.5 and E13.5 limbs with Click-iT TUNEL assay (Invitrogen). At E11.5, the cell death in the hand was restricted to the AER in both wild-type and mutant (Fig. 7 A and B). At E12.5, comparing with the uniformed cell death in AER, the cell death in the regions that were distal to the digit condensates was reduced in the wild-type, while the cell death in the forming interdigital space was increased (Fig. 7 C). Surprisingly, the cell death pattern in the mutant was still uniform in the subridge region resembling the cell death pattern of wild-type at E11.5 (Fig. 7D). In addition, the cells undergoing apoptosis expanded over a wider region relative to the epithelial cells (Fig. 7 D). At E13.5, apoptosis was restricted to the interdigital space in the wild-type (Fig. 7 E). In contrast, apoptosis was still detected in mutants in the digit region while the interdigital space had reduced level of apoptosis compared with wild-type (Fig. 7 F). We quantified the distance underneath the AER where apoptotic cells could be found (apoptosis zone) along PD axis as well as dying cell numbers within a 100 μm diameter circle underneath the AER along PD axis (Fig. 7 E, dashed line and dashed circle). There were more apoptotic cells in the digit tips but fewer apoptotic cells in the interdigital membrane in the mutant than the wild-type (Fig. 7 G, n=3, p<0.01, t-test). This patterning defects between digit and interdigital space may be due to interrupted Bmp4, Fgf8 and Wnt5a signaling described above. We suggest that the altered pattern of apoptosis might be responsible for delayed digit growth and progression of the indentation to carve out digits.
Figure 7.
TUNEL Click-iT test shows the abnormal apoptosis in Prickle1C251X/C251X limbs. A-B: at E11.5, apoptotic cells are uniform across the whole in the limb bud in both wild-type and mutants. C-D, at E12.5, apoptotic cells starts to be restricted to interdigital membrane in the wild-type (C), but this differential apoptosis pattern is absent in the mutant (D). E-F: at E13.5, there is profound cell death in the interdigital membrane between digits 3 and 4 but limited cell death in the digit tips in the wild-type (E). In the mutant (F), cell death is uniform in digit and interdigital membrane. G: The distance underneath AER where apoptotic cells are found is measured and defined as cell death zone (dashed line in E). Apoptotic cells within a circle of 100 μm diameter underneath AER are counted in both digit and interdigital space (dashed circle in E). At E13.5, the wild-type has significantly larger apoptosis zone and more apoptotic cells in the distal digit, but smaller apoptosis zone and fewer apoptotic cells in the interdigital space, leading to a more uniform cell death pattern (n=3, p<0.01, t-test). The scale bar is 100μm.
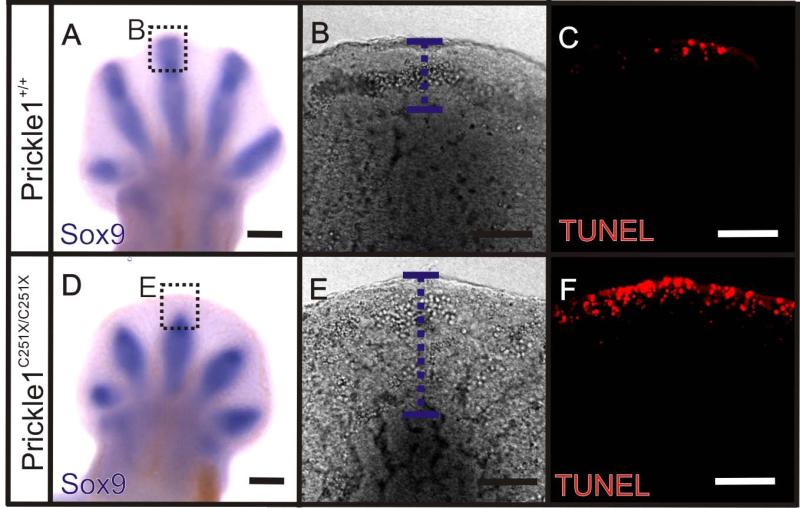
Sox9 is a transcription factor that activates chondrocyte-specific marker genes and thus is a marker for chondrogenesis (Wright et al., 1995; Akiyama et al., 2002). Post-mitotic cells that fail to up-regulate Sox9 are not committed to the chondrocyte lineage and thus undergo apoptosis. It also has been shown that Sox9 expression is down-regulated in the digit tip in Wnt5a mutants (Topol et al., 2003). We reasoned that a failure to up-regulate Sox9 might be responsible for the increased apoptotic cell numbers in the digit tip. Therefore, we examined the co-localization of Sox9 and apoptotic cells by running Sox9 whole amount in situ hybridization first and then TUNEL (Invitrogen) at E12.5. At this stage, Sox9 was expressed in the developing digit condensates in both wild-type and mutant (Fig. 8 A and D), but there was a wider area in the digit tip without Sox9 expression in the mutant (Fig. 8 A-B and D-E). Consistent with our hypothesis, Click-it TUNEL assay showed apoptotic cells in a wider space underneath AER in the mutant (Fig. 8 F). In the wild-type, the apoptotic cells were restricted to AER (Fig. 8 C).
Figure 8.
Distal mesenchyme cells fail to up-regulate Sox9 in digit tip at E12.5. A and D: Whole amount mRNA in situ hybridization shows that Sox9 is highly expressed by the developing digit rays in the wild-type (A) and the mutant (D), but there is a wider area not expressing Sox9 at the digit tip in the mutant (D). B-C and E-F: A single focal plane image of digit 3 tip shows the co-localization of Sox9 and apoptotic cells. B and E: There is a wider area not expressing Sox9 in the mutant (E, the purple dashed line) than the wild-type (B). C and F: Click-iT TUNEL shows that cell death is more restricted to the AER at this digit tip in the wild-type (C) than the mutant (F). Scale bar is 500 μm.
Prickle1 mutation affected directed long bone growth
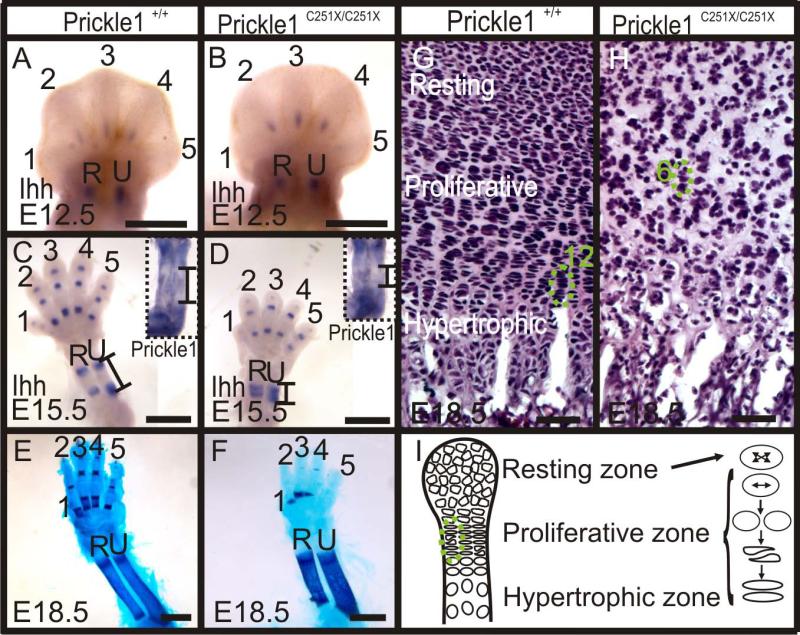
We have shown above that calcification was affected in the mutants (Fig. 2 and 3), which could be due to impaired deposit of calcium, impaired ossification or only secondary to development defects in the cartilage preformation. To search for the possible causes, we examined Ihh expression. Ihh is an important transcription factor required for the initiation of endochondral ossification (Vortkamp et al., 1996; St-Jacques et al., 1999). At E12.5, Ihh was expressed in ulna, radius, and one segment in digits 2-4 in both wild-type and mutant (Fig 9 A and B). However, at E15.5, compared with four segments of Ihh in the digits 2-5 in the wild-type (Fig. 9 C), there were only three segments of Ihh expression in the digits of homozygotic Prickle1C251X/C251X mice (Fig. 9 D). In ulna and radius, Ihh expression had split into two clearly separated ends of the growing bones (Fig. 9 C). However in the mutant, Ihh expression in the ulna and the radius was still closely spaced and near the middle of the forming bones (Fig. 9D). Although Ihh expression did not expand normally in the mutants as in the wild-types, the mineralization in ulna and radius at E18.5 mutants suggested that the mechanisms of endochondral ossification were not disrupted, but rather delayed, possibly secondary to defects in the development of cartilage formation (Fig. 2 and 9 E-F).
Figure 9.
Ossification is affected by Prickle1 C251X mutation. A-D: Whole mount mRNA in situ hybridization shows Ihh and Prickle1 expression at E12.5 and E15.5. A-B: At E12.5, there is no obvious difference between wild-type and mutants. C-D: At E15.5, Ihh expression has split into two ends in wild-type ulna and radius, but the spacing of the Ihh expression zones is almost absent in the mutant. Prickle1 is expressed at the two ends of these two long bones (inset). The black lines market the expression boundary for both Prickle1 and Ihh. It should be noted that the dark area in the center of the bone does not reflect staining but only the darker color of ossified bone. E-F: At E18.5, Alizarin red is detected in the ulna and radius in both the wild-type and the mutant. G: At this stage, the chondrocytes in the wild-type humerus could be easily separated into three zones according to cell morphology and distribution, and the proliferative and hypertrophic chondrocytes are organized into columns (12 cells in a column). H: In the mutant, the three zones could not be easily distinguished due to changed cell morphology and distribution. Notably, the proliferative chondrocytes are not organized into columns (6 cells in a column). I: modified after (Li and Dudley, 2009). The proliferative chondrocytes divide orthogonal to the long bone axis, and the daughter chondrocytes converge to form one column. Numbers 1-5 indicate digit numbers 1-5. R, radius; U, ulna. The scale bar is 500 μm in A-B, 1 mm in C-F and 100 μm in G-H.
The morphologic defects of shorter and wider bones (Fig. 3 and 9 E-F) have previously been associated with the Wnt/PCP signaling regulating the chondrocyte polarity (Li and Dudley, 2009; Randall et al., 2012). To further analyze this for limb growth, we examined Prickle1 expression in ulna and radius. Prickle1 expression was detected in the two ends of the bones, namely resting and proliferative chondrocytes and some hypertrophic chondrocytes (Fig. 9 C inset).This expression pattern suggests Prickle1 could affect long bone growth by acting on resting or proliferative chondrocytes. Therefore, we examined whether the polarity of chondrocyte division is affected by Prickle1 C251X mutation. We examined the cell morphology in the humeri of E18.5 mice. In the wild-type, the proliferating chondrocytes formed long columns parallel to the long bone axis (Fig. 9 G, green circle). Consistent with a function of Prickle1 as part of the PCP pathway, the organization of proliferative chondrocytes was disrupted in Prickle1 C251X mutants with shorter, disorganized chondrocyte columns (Fig. 9 H, green circle). This disorganization is possibly the outcome from dis-oriented cell division similar to those reported in other PCP mutants (Fig. 9 I) (Li and Dudley, 2009; Randall et al., 2012).
In summary, our data show that Prickle 1 mutation affects growth of limbs through alteration of digital apoptosis, Wnt5a and Bmp4 expression and chondrocyte organization. Early expression changes, altered pattern of cell death and disrupted cell polarity results in less directed bone growth and ultimately in shorter and wider bones in the limbs. The observed changes in Ihh expression only reflect the overall changes without causing them.
Discussion
Prickle1 is required for outgrowth of multiple structures
We show that Prickle1 is needed for normal snout, limb, rib, and tail growth. These structures are newly evolved extensions from the main body axis. The involvement of Prickle1 in these multiple outgrowth processes suggests Prickle1 has been recruited to assist this extended growth. Consistent with the shorter tail in Prickle1C251X/C251X mice, the secondary somitogenesis of the tail bud is defective. Each somite is smaller in size which leads to reduction in anterior-posterior length, but the rate of segmentation and thus the number of somites are not affected (Goldbeter and Pourquié, 2008; Maroto et al., 2012). The causality that leads to smaller somites only in the short tail in Prickle1 C251X homozygous mutants requires further work. Here we concentrate on the limb deformation.
The evolution of limbs from fins entails extended growth (Shubin et al., 2009; Sheth et al., 2012). Polarized cell activity combined with proliferation and differential cell death play an important role in limb growth and sculpting. The Wnt/PCP pathway, which regulates cell polarity as a prerequisite for coordinated migration, assists in limb growth. As in impaired tail vertebrae and impaired rib growth, our Prickle1 mutants suggests that this factor plays an essential role in this extension growth away from the body axis, possibly by regulating gene expression.
Functional consequences of Prickle 1 mutation
The Prickle1 C251X mouse line has a point mutation in exon 6, which changes the amino acid 251 cysteine to a premature stop signal. Prickle1 has three LIM domains, which are able to bind with other LIM-homeobox proteins; its C-terminal NLS and farnesylation site are also shown to be necessary for REST nuclear transportation (Shimojo and Hersh, 2003; Kadrmas and Beckerle, 2004; Shimojo and Hersh, 2006). . In another structurally similar protein TESTIN, LIM and PET domains have been shown to be necessary for nuclear localization (Zhong et al., 2009). Therefore, it is possible that Prickle1 could be translocalized to the nucleus and regulate gene expressions cooperating with other LIM-Homeobox domain genes. This strongly suggests some role of Prickle1 in gene expression regulation. Based on current understanding, nonsense mutation leads to unstable mRNA which is then degraded (Frischmeyer and Dietz, 1999; Chang et al., 2007). Therefore, Prickle1 mRNA level is expected to be lower in the mutant. Our mRNA in situ hybridization shows reduced Prickle1 expression in Prickle1C251X/C251X mutant, suggesting that even if the mutant form of Prickle1 protein could still perform some of its function, the amount of total protein is likely reduced.
If protein is made at all from the reduced amount of unstable mRNA, current literature of Prickle1 structure and function suggests that this mutant form of Prickle1 protein is possibly not able to translocate into the nucleus (Shimojo and Hersh, 2003; Shimojo and Hersh, 2006; Zhong et al., 2009; Mapp et al., 2011; Liu et al., 2013) and thus will not function as a transcription regulator. However, limited protein interaction in the cytoplasm would still be possible. Moreover, a recent paper (Liu et al., 2013) showed that the Prickle1 ΔLIM+CIIS mutant form, which has LIM and the C-terminal nuclear localization signals deleted, probably acts as a dominant-negative mutant and competes with normal Prickle1 protein function. Unfortunately, we could not substantiate our prediction due to a lack of specific antibodies for the mutant Prickle1 protein.
Prickle interacts with the Wnt5a signaling network in limb development
Prickle1 is strongly expressed in the digit condensates, with the highest expression in the distal tip. In the mutants, the expression of several genes in this region is severely affected, including Sox9, Bmp4, Wnt5a, Vangl2 and Fgf8. Limb development is regulated by Fgf-Shh-Bmp-Wnt signaling networks (Bénazet et al., 2009) in cooperation with Hox genes (Sheth et al., 2012). These signals interact to form feedback loops that can buffer against certain levels of perturbation but apparently not the perturbations generated by the Prickle1 C251X mutation. It should be noted that a point mutation usually causes more complicated phenotypes than simple deletion, for instance the Vangl2lp/lp mice have more severe phenotypes than Vangl2−/− (Wang et al., 2011; Andre et al., 2012; Yin et al., 2012). Therefore, it is possible that our Prickle1 C251X mutants have more complicated phenotypes than simple Prickle1 loss-of-function. This complicates our analysis of the signaling interactions. To test this, generating a complete deletion mutant mouse line is necessary.
Based on the in situ hybridization results, the signals that are affected first at E11.5 in Prickle1C251X/C251X mutants are Wnt5a and Bmp4. Currently, we do not know how directly or indirectly Prickle1 affects Wnt5a expression. Despite this unresolved issue, the obvious and rapid reduction in Wnt5a expression at the mRNA level in Prickle1C251X/C251X mice is compatible with a positive feedback loop whereby Prickle1 protein stabilizes, through unknown intermediates, Wnt5a expression. Supporting our data and conclusion, Wnt5a expression is also down-regulated in Vanlg2lp/lp mouse hindbrain (Glasco et al., 2012). Given that normal expression of Prickle1 is required to maintain Wnt5a expression levels, we propose that the limb phenotype in Prickle1C251X/C251X mutants is partially due to the reduced Wnt5a expression. Since Wnt5a dose-dependently increases the activity of Vangl2 and stabilizes Vangl2 protein through Ror2 and Ryk respectively (Gao et al., 2011; Andre et al., 2012), the down-regulation of Wnt5a in Prickle1 C251X mutants might leads to reduced Vangl2 protein amount and activity. To further complicate this issue, we find reduction of Vangl2 mRNA, which possibly leads to further down-regulation of Vangl2 protein. Combined with reduced level of dysfunctional Prickle1 protein and reduction of Wnt5a, any remaining interaction between Vangl2 and Prickle1 protein is likely compromised in our mutants. It is possible that more PCP pathway genes expression could be affected by Prickle1 C251X mutation, but with two expression changes already demonstrated it would not add further to our understanding of dysfunction of the PCP signaling.
Prickle1 C251X mutation affects limb development differently from Vangl2 mutation
Prickle1C251X mutation most closely resembles the combined effects of Ror2/Vangl2, or Ryk/Vangl2 but is more severe than Vangl2 looptail mutation (Yamaguchi et al., 1999; Gao et al., 2011; Andre et al., 2012), suggesting the function of Prickle1 is more complicated than simply mediating Vangl2 signaling. It could be another Vangl family member (Vangl1), a protein interacting with Vangl2 (Ror1/2), or another signal pathway independent of Vangl2 such as the proposed Prickle1 interaction with Disheveled (Narimatsu et al., 2009).
In addition, these mutants have subtle differences in cell death and proliferation pattern. Although there is increased cell death in all mutants, the pattern differs. In Prickle1C51X/C251X mutants, cell death is increased in the distal digit but decreased in the interdigital space, but there are no significant changes in cell proliferation rate. Similarly to Prickle1C251X/C251X mutants, in Ror2W749X/W749X mutants, the defects in digit elongation is due to mesenchymal cells failing to commit to the chondrocyte lineage (Witte et al., 2010). In Wnt5+/−; Vangl2lp/lp or Vangl2lp/lp mutants, in addition to increased cell death, there is also reduced cell proliferation (Wang et al., 2011). There is no obvious limb development defect in Ryk mutants, and cell proliferation or cell death has not been studied in Ryk−/−; Vangl2−/− mice (Andre et al., 2012).
Interesting, Vangl2 protein is uniformly expressed at E11.5 and the asymmetric cellular localization is established at E12.5 (Gao et al., 2011). If Vangl2 protein asymmetric distribution is necessary to establish cell polarity, their results (Gao et al., 2011) then suggest cell polarity does not play a role in limb development until after E11.5. However, at this stage, we already find Wnt5a and Bmp4 expression changes in Prickle1 C251X mutants. This suggests that Prickle1 mutation may predominantly affect limb development through a mechanism unrelated to asymmetric localization of the PCP protein Vangl2 but instead relates to down-regulation of PCP related genes. However, it is also possible that Vangl2 has other roles than establishing cell polarity before E12.5 since Vangl2 protein is expressed at E11.5. Taken together, although Prickle1 and Vangl2 mutants have somewhat similar limb development defects, subtle differences suggest a more complicated Prickle1-Vangl2 interaction rather than the protein binding model proposed for flies. We suggest that mammalian Prickle1 may have evolved novel functions that regulate expression levels of PCP pathway partners in addition to its protein-protein interactions.
Roles of Prickle1 in long bone development
In the long bones, the proliferative chondrocytes divide orthogonal to the long bone axis, which then converge to form columns parallel to the long bone axis (Fig. 9 I) (Li and Dudley, 2009). This process drives long bone growth. It has been shown that Wnt/PCP signaling, such as Wnt5a, Vangl2 and Ror2, plays a critical role in regulating the direction of chondrocyte divisions (Li and Dudley, 2009; Randall et al., 2012). This disrupted cell division leads to failure to form organized chondrocyte columns, resulting in stunted bone growth (Li and Dudley, 2009). We have shown here that chondrocytes in Prickle1C251X/C251X bones fail to form organized columns, possibly due to disrupted cell polarity. Because of the disorganized chondrocyte morphology and distribution, it is difficult to define exactly the length of proliferative and hypertrophic zones in the mutants (Fig. 9 G and H). With this caveat in mind, the mutant hypertrophic zone appears shorter than that in the wild-type, which suggests the mutant long bones have shorter cartilage template to start ossification. Therefore, the smaller portion of calcification defects shown by Alizarin Red is probably only secondary to the shortened cartilage template. In summary, undirected proliferation is at least partially responsible for the shorter and wider bones in the Prickle1 mutants, in agreement with the involvement of Wnt/PCP signaling in long bone growth.
In conclusion, our results support the hypothesis that Prickle1 modulates digit and interdigital membrane identity by regulating expression of other genes. Among the genes that we studied, the most profound and earliest affected gene is Wnt5a, which is generally believed to be the ligand of the PCP pathway. The down-regulation of Wnt5a suggests Prickle1 is required to maintain Wnt5a expression through unknown mechanisms. In fact, the down-regulation of Wnt5a and other genes (Fgf8, BMP4, Vanlg2) might be responsible for the growth defects in Prickle1 C251X/C251X mutants. Further, disrupted Prickle1 signaling leads to disorganized proliferative chondrocytes in the long bones, which is possibly responsible for the shorter and wider bones as well as later calcification.
Experimental Procedures
Mice
All animal treatment was approved by University of Iowa IACUC (ACURF 0804066) and (ACURF1109204). The generation of Prickle Cys251X mutant mice has been previously described (Tao et al., 2011). Briefly, a point mutation from C to A was generated by ENU-mutagenesis, which changes the reading frame from TGC to TGA, introducing a premature stop codon at the 251 amino acid from N-terminal. The point mutation line identified from the mutagenesis screen was backcrossed to C57/BL6 using a rapid congenic protocol [genome wide microsatellite markers used for genotyping available upon request (Tao et al., 2011)]. Homozygous mutants die around birth.
Embryos from timed breeding were fixed in 4% paraformaldehyde (PFA). Noon on the day of vaginal plug visualization was deemed as E0.5. Tails were collected for PCR and sequencing for genotyping. The PCR primers used were: P1 5’-TTTGTGCTCAGAGCCAGTG-3’, P2 5’-CAAGCGTTAAAGAAGCAAGG-3’. The PCR product was 378 bp, which was then sent to sequencing to verify the mutation.
Alizarin red and Alcian blue staining
Skeletal staining using Alcian blue and Alizarin red staining was performed as previously described (Ovchinnikov, 2009). Briefly, fixed samples were washed in tap water for 1 h and then scaled in hot tap water (65 °C – 70 °C) for 30 sec. Skin, muscles and all other soft tissues were removed as much as possible using forceps. The samples were then fixed in 95% ethanol overnight and then acetone overnight at room temperature. The samples were stained for cartilage in alcian blue stain solution for 24 h and then alizarin red stain solution overnight. Samples were cleared by 1% potassium hydroxide/20% glycerol for 2 days or longer and then imaged in glycerol: ethanol (1:1).
In situ hybridization
The probes for in situ hybridization were generated by in vitro transcription from the plasmid and then labeled with digoxigenin. Bmp4, Fgf8, Shh, Ihh, Tbx20, Sox9, Runx2, Wnt5a, Vangl2, and Prickle1 probes were previously described (Jones et al., 1991; Heikinheimo et al., 1994; Bitgood and McMahon, 1995; Marigo et al., 1995; Kraus et al., 2001; Coppola et al., 2005; Song et al., 2006; Tissir and Goffinet, 2006; Okuda et al., 2007).
Embryos were hemisected in 0.4% PFA. Opposite halves from mutant and wild-type littermate embryos were reacted in the same tube for the same probe to minimize the reaction variability. Samples were digested by 20 mg/ml of Proteinase K (Ambion, Austin, TX, USA) for 1 hour, and then hybridized overnight at 60 °C to the riboprobe in hybridization solution containing 50% (v/v) formamide, 50% (v/v) saline sodium citrate and 6% (w/v) dextran sulfate. After washing off the unbound probe, the samples were incubated overnight with an anti-digoxigenin antibody conjugated with alkaline phosphatase (Roche Diagnostics GmbH, Mannheim, Germany). The samples were reacted with nitroblue phosphate/5-bromo, 4-chloro, 3-indolil phosphate (BM purple substrate, Roche Diagnostics GmbH, Mannheim, Germany), which changed the color to purple by alkaline phosphatase. The reaction was stopped by 4% PFA. Samples were then mounted in glycerol and viewed in a Leica M205 FA microscope. Images were captured with Leica application suite V3. All the limb buds were imaged from the dorsal side, and then images taken from left limb buds were flipped horizontally to keep the same orientation using CorelDRAW14. At least three half embryos per genotype were used to verify the consistency of the pattern.
Immunohistochemistry
Limbs were bleached (10 mL 3% H2O2, 10 mL 10% KOH and 40 mL H2O) overnight at 4°C to eliminate the autofluorescence from red blood cells. Apoptotic cells were labeled with Click-it® TUNEL assay (Invitrogen) following the manufacturer's protocol. Proliferating cells were labeled with anti-phospho-histone H3 (pHH3) (1:400, Upstate Biotechnologies) with secondary antibody conjugated to Alexa 532 (Sigma, 1:500). Nuclei were counter stained with Hoechst (Polysciences, Inc.; 1:1000 in PBS for 10 min) and then rinsed briefly in PBS. Imaging was performed using Leica TCS SP5 confocal systems using appropriate filter settings and Leica AF software. Images were analyzed using ImageJ and CorelDRAW14.
For the co-localization of Sox9 and apoptotic cells, Sox9 in situ hybridization was performed and recorded with Leica M205 FA microscope. Then Click-iT TUNEL assay was performed and the samples were imaged using Leica TCS SP5 confocal microscope.
Histology
Humeri from E18.5 embryos were dissected and cryo-sectioned at 20μm thickness. The sections were then stained with Haematoxylin and Eosin staining. Samples were mounted in Permount and imaged using Nikon E800 compound microscope using Metamorph software.
Bullet points.
Prickle1 regulates limb outgrowth
Prickle1 is required for Wnt5a and Vangl2 expression
Prickle1 regulates chondrocyte polarity
Acknowledgements
We would like to express our gratitude to all those who have helped us. We are grateful for Shu Wu who helped a lot in mouse breeding. We thank Dr. Miyata, Dr. AP McMahon, Dr. A. Nifuji, Dr. P. Koopman, Dr. D. Epstein and Dr. A. Chandrasekhar for providing plasmids for in situ probe. We also would like to thank Dr. Sarit Smolikove for sharing the PHH3 antibody with us and Dr. Sigmar Stricker for comment on an earlier version of the manuscript. This work was supported by NIH R01 grants (R01 DC005590; P30 DC 010362) to BF and (NIH 1R01 NS064159-01A1) to AGB.
Reference
- Afzal AR, Jeffery S. One gene, two phenotypes: ROR2 mutations in autosomal recessive Robinow syndrome and autosomal dominant brachydactyly type B. Human Mutation. 2003;22:1–11. doi: 10.1002/humu.10233. [DOI] [PubMed] [Google Scholar]
- Afzal AR, Rajab A, Fenske CD, Oldridge M, Elanko N, Ternes-Pereira E, Tuysuz B, Murday VA, Patton MA, Wilkie AOM, Jeffery S. Recessive Robinow syndrome, allelic to dominant brachydactyly type B, is caused by mutation of ROR2. Nat Genet. 2000;25:419–422. doi: 10.1038/78107. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier M-C, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes & Development. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre P, Wang Q, Wang N, Gao B, Schilit A, Halford MM, Stacker SA, Zhang X, Yang Y. The Wnt coreceptor Ryk regulates Wnt/planar cell polarity by modulating the degradation of the core planar cell polarity component Vangl2. Journal of Biological Chemistry. 2012 doi: 10.1074/jbc.M112.414441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic Analysis of the Roles of BMP2, BMP4, and BMP7 in Limb Patterning and Skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow JR. Wnt/PCP signaling: A veritable polar star in establishing patterns of polarity in embryonic tissues. Seminars in Cell and Developmental Biology. 2006;17:185–193. doi: 10.1016/j.semcdb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Bassuk AG, Wallace RH, Buhr A, Buller AR, Afawi Z, Shimojo M, Miyata S, Chen S, Gonzalez-Alegre P, Griesbach HL, Wu S, Nashelsky M, Vladar EK, Antic D, Ferguson PJ, Cirak S, Voit T, Scott MP, Axelrod JD, Gurnett C, Daoud AS, Kivity S, Neufeld MY, Mazarib A, Straussberg R, Walid S, Korczyn AD, Slusarski DC, Berkovic SF, El-Shanti HI. A Homozygous Mutation in Human PRICKLE1 Causes an Autosomal-Recessive Progressive Myoclonus Epilepsy-Ataxia Syndrome. The American Journal of Human Genetics. 2008;83:572–581. doi: 10.1016/j.ajhg.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastock R, Strutt H, Strutt D. Strabismus is asymmetrically localised and binds to Prickle and Dishevelled during Drosophila planar polarity patterning. Development. 2003;130:3007–3014. doi: 10.1242/dev.00526. [DOI] [PubMed] [Google Scholar]
- Bekman E, Henrique D. Embryonic expression of three mouse genes with homology to the Drosophila melanogaster prickle gene. Mechanisms of Development. 2002;119(Supplement):S77–S81. doi: 10.1016/s0925-4773(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Bénazet J-D, Bischofberger M, Tiecke E, Gonçalves A, Martin JF, Zuniga A, Naef F, Zeller R. A Self-Regulatory System of Interlinked Signaling Feedback Loops Controls Mouse Limb Patterning. Science. 2009;323:1050–1053. doi: 10.1126/science.1168755. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. Hedgehog and Bmp Genes Are Coexpressed at Many Diverse Sites of Cell–Cell Interaction in the Mouse Embryo. Developmental Biology. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Bosoi CM, Capra V, Allache R, Trinh VQ-H, De Marco P, Merello E, Drapeau P, Bassuk AG, Kibar Z. Identification and characterization of novel rare mutations in the planar cell polarity gene PRICKLE1 in human neural tube defects. Human Mutation. 2011;32:1371–1375. doi: 10.1002/humu.21589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Barbosa F, Concha ML, Takeuchi M, Ueno N, Wilson SW, Tada M. Prickle 1 regulates cell movements during gastrulation and neuronal migration in zebrafish. Development. 2003;130:4037–4046. doi: 10.1242/dev.00567. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson ME. Annual Review of Biochemistry. Annual Reviews; Palo Alto: 2007. The nonsense-mediated decay RNA surveillance pathway. pp. 51–74. [DOI] [PubMed] [Google Scholar]
- Cooper O, Sweetman D, Wagstaff L, Münsterberg A. Expression of avian prickle genes during early development and organogenesis. Developmental Dynamics. 2008;237:1442–1448. doi: 10.1002/dvdy.21490. [DOI] [PubMed] [Google Scholar]
- Coppola E, Pattyn A, Guthrie SC, Goridis C, Studer M. Reciprocal gene replacements reveal unique functions for Phox2 genes during neural differentiation. EMBO J. 2005;24:4392–4403. doi: 10.1038/sj.emboj.7600897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans MR. A balance of form and function: Planar polarity and development of the vestibular maculae. Seminars in cell & developmental biology. 2013 doi: 10.1016/j.semcdb.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC. Nonsense-Mediated mRNA Decay in Health and Disease. Human Molecular Genetics. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, Andre P, Robinson J, Sood R, Minami Y, Economides AN, Yang Y. Wnt Signaling Gradients Establish Planar Cell Polarity by Inducing Vangl2 Phosphorylation through Ror2. Developmental Cell. 2011;20:163–176. doi: 10.1016/j.devcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasco DM, Sittaramane V, Bryant W, Fritzsch B, Sawant A, Paudyal A, Stewart M, Andre P, Cadete Vilhais-Neto G, Yang Y, Song M-R, Murdoch JN, Chandrasekhar A. The mouse Wnt/PCP protein Vangl2 is necessary for migration of facial branchiomotor neurons, and functions independently of Dishevelled. Developmental Biology. 2012;369:211–222. doi: 10.1016/j.ydbio.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Pourquié O. Modeling the segmentation clock as a network of coupled oscillations in the Notch, Wnt and FGF signaling pathways. Journal of Theoretical Biology. 2008;252:574–585. doi: 10.1016/j.jtbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Gubb D, Green C, Huen D, Coulson D, Johnson G, Tree D, Collier S, Roote J. The balance between isoforms of the Prickle LIM domain protein is critical for planar polarity in Drosophila imaginal discs. Genes & Development. 1999;13:2315–2327. doi: 10.1101/gad.13.17.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikinheimo M, Lawshé A, Shackleford GM, Wilson DB, MacArthur CA. Fgf-8 expression in the post-gastrulation mouse suggests roles in the development of the face, limbs and central nervous system. Mechanisms of Development. 1994;48:129–138. doi: 10.1016/0925-4773(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Jenny A, Darken RS, Wilson PA, Mlodzik M. Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 2003;22:4409–4420. doi: 10.1093/emboj/cdg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen JR, Topczewski J, Bingham S, Sepich DS, Marlow F, Chandrasekhar A, Solnica-Krezel L. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat Cell Biol. 2002;4:610–615. doi: 10.1038/ncb828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Lyons KM, Hogan BL. Involvement of Bone Morphogenetic Protein-4 (BMP-4) and Vgr-1 in morphogenesis and neurogenesis in the mouse. Development. 1991;111:531–542. doi: 10.1242/dev.111.2.531. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kestler HA, Kühl M. From individual Wnt pathways towards a Wnt signalling network. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:1333–1347. doi: 10.1098/rstb.2007.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibar Z, Salem S, Bosoi CM, Pauwels E, De Marco P, Merello E, Bassuk AG, Capra V, Gros P. Contribution of VANGL2 mutations to isolated neural tube defects. Clinical Genetics. 2011;80:76–82. doi: 10.1111/j.1399-0004.2010.01515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx20. Mechanisms of Development. 2001;100:87–91. doi: 10.1016/s0925-4773(00)00499-8. [DOI] [PubMed] [Google Scholar]
- Lei Y-P, Zhang T, Li H, Wu B-L, Jin L, Wang H-Y. VANGL2 Mutations in Human Cranial Neural-Tube Defects. New England Journal of Medicine. 2010;362:2232–2235. doi: 10.1056/NEJMc0910820. [DOI] [PubMed] [Google Scholar]
- Li Y, Dudley AT. Noncanonical frizzled signaling regulates cell polarity of growth plate chondrocytes. Development. 2009;136:1083–1092. doi: 10.1242/dev.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lin C, Whitaker DT, Bakeri H, Bulgakov OV, Liu P, Lei J, Dong L, Li T, Swaroop A. Prickle1 is expressed in distinct cell populations of the central nervous system and contributes to neuronal morphogenesis. Human Molecular Genetics. 2013 doi: 10.1093/hmg/ddt075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias D, Ganan Y, Sampath TK, Piedra ME, Ros MA, Hurle JM. Role of BMP-2 and OP-1 (BMP-7) in programmed cell death and skeletogenesis during chick limb development. Development. 1997;124:1109–1117. doi: 10.1242/dev.124.6.1109. [DOI] [PubMed] [Google Scholar]
- Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Developmental Cell. 2009;16:600–606. doi: 10.1016/j.devcel.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapp OM, Walsh GS, Moens CB, Tada M, Prince VE. Zebrafish Prickle1b mediates facial branchiomotor neuron migration via a farnesylation-dependent nuclear activity. Development. 2011;138:2121–2132. doi: 10.1242/dev.060442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V, Roberts DJ, Lee SMK, Tsukurov O, Levi T, Gastier JM, Epstein DJ, Gilbert DJ, Copeland NG, Seidman CE, Jenkins NA, Seidman JG, McMahon AP, Tabin C. Cloning, Expression, and Chromosomal Location of SHH and IHH: Two Human Homologues of the Drosophila Segment Polarity Gene Hedgehog. Genomics. 1995;28:44–51. doi: 10.1006/geno.1995.1104. [DOI] [PubMed] [Google Scholar]
- Maroto M, Bone RA, Dale JK. Somitogenesis. Development. 2012;139:2453–2456. doi: 10.1242/dev.069310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Woodgett JR. When pathways collide: collaboration and connivance among signalling proteins in development. Nat Rev Mol Cell Biol. 2010;11:404–413. doi: 10.1038/nrm2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Sans N, Huss D, Kach J, Dickman JD, Forge A, Rachel RA, Copeland NG, Jenkins NA, Bogani D, Murdoch J, Warchol ME, Wenthold RJ, Kelley MW. Asymmetric Localization of Vangl2 and Fz3 Indicate Novel Mechanisms for Planar Cell Polarity in Mammals. The Journal of Neuroscience. 2006;26:5265–5275. doi: 10.1523/JNEUROSCI.4680-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu M, Bose R, Pye M, Zhang L, Miller B, Ching P, Sakuma R, Luga V, Roncari L, Attisano L, Wrana JL. Regulation of Planar Cell Polarity by Smurf Ubiquitin Ligases. Cell. 2009;137:295–307. doi: 10.1016/j.cell.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Niswander L, Martin GR. FGF-4 and BMP-2 have opposite effects on limb growth. Nature. 1993;361:68–71. doi: 10.1038/361068a0. [DOI] [PubMed] [Google Scholar]
- Okuda H, Miyata S, Mori Y, Tohyama M. Mouse Prickle1 and Prickle2 are expressed in postmitotic neurons and promote neurite outgrowth. FEBS Letters. 2007;581:4754–4760. doi: 10.1016/j.febslet.2007.08.075. [DOI] [PubMed] [Google Scholar]
- Oldridge M, M Fortuna A, Maringa M, Propping P, Mansour S, Pollitt C, DeChiara TM, Kimble RB, Valenzuela DM, Yancopoulos GD, Wilkie AOM. Dominant mutations in ROR2, encoding an orphan receptor tyrosine kinase, cause brachydactyly type B. Nat Genet. 2000;24:275–278. doi: 10.1038/73495. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov D. Alcian Blue/Alizarin Red Staining of Cartilage and Bone in Mouse. Cold Spring Harbor Protocols. 2009 doi: 10.1101/pdb.prot5170. 2009:pdb.prot5170. [DOI] [PubMed] [Google Scholar]
- Randall RM, Shao YY, Wang L, Ballock RT. Activation of Wnt Planar cell polarity (PCP) signaling promotes growth plate column formation in vitro. Journal of Orthopaedic Research. 2012;30:1906–1914. doi: 10.1002/jor.22152. [DOI] [PubMed] [Google Scholar]
- Rath N, Wang Z, Lu MM, Morrisey EE. LMCD1/Dyxin Is a Novel Transcriptional Cofactor That Restricts GATA6 Function by Inhibiting DNA Binding. Molecular and Cellular Biology. 2005;25:8864–8873. doi: 10.1128/MCB.25.20.8864-8873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe GC, Tinschert S, Buschow C, Meinecke P, Wolff G, Gillessen-Kaesbach G, Oldridge M, Wilkie AOM, Kömec R, Mundlos S. Distinct Mutations in the Receptor Tyrosine Kinase Gene ROR2 Cause Brachydactyly Type B. The American Journal of Human Genetics. 2000;67:822–831. doi: 10.1086/303084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe GC, Trepczik B, Süring K, Brieske N, Tucker AS, Sharpe PT, Minami Y, Mundlos S. Ror2 knockout mouse as a model for the developmental pathology of autosomal recessive Robinow syndrome. Developmental Dynamics. 2004;229:400–410. doi: 10.1002/dvdy.10466. [DOI] [PubMed] [Google Scholar]
- Sheth R, Marcon L, Bastida MF, Junco M, Quintana L, Dahn R, Kmita M, Sharpe J, Ros MA. Hox Genes Regulate Digit Patterning by Controlling the Wavelength of a Turing-Type Mechanism. Science. 2012;338:1476–1480. doi: 10.1126/science.1226804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo M, Hersh LB. REST/NRSF-Interacting LIM Domain Protein, a Putative Nuclear Translocation Receptor. Molecular and Cellular Biology. 2003;23:9025–9031. doi: 10.1128/MCB.23.24.9025-9031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo M, Hersh LB. Characterization of the REST/NRSF-interacting LIM domain protein (RILP): localization and interaction with REST/NRSF. Journal of Neurochemistry. 2006;96:1130–1138. doi: 10.1111/j.1471-4159.2005.03608.x. [DOI] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- Song M-R, Shirasaki R, Cai C-L, Ruiz EC, Evans SM, Lee S-K, Pfaff SL. T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development. 2006;133:4945–4955. doi: 10.1242/dev.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes & Development. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabin CJ, McMahon AP. Developmental biology. Grasping limb patterning. Science. 2008;321:350–352. doi: 10.1126/science.1162474. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Nakabayashi J, Sakaguchi T, Yamamoto TS, Takahashi H, Takeda H, Ueno N. The prickle-Related Gene in Vertebrates Is Essential for Gastrulation Cell Movements. Current Biology. 2003;13:674–679. doi: 10.1016/s0960-9822(03)00245-8. [DOI] [PubMed] [Google Scholar]
- Tao H, Inoue K-i, Kiyonari H, Bassuk AG, Axelrod JD, Sasaki H, Aizawa S, Ueno N. Nuclear localization of Prickle2 is required to establish cell polarity during early mouse embryogenesis. Developmental Biology. 2012;364:138–148. doi: 10.1016/j.ydbio.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Manak JR, Sowers L, Mei X, Kiyonari H, Abe T, Dahdaleh NS, Yang T, Wu S, Chen S, Fox MH, Gurnett C, Montine T, Bird T, Shaffer LG, Rosenfeld JA, McConnell J, Madan-Khetarpal S, Berry-Kravis E, Griesbach H, Saneto RP, Scott MP, Antic D, Reed J, Boland R, Ehaideb SN, El-Shanti H, Mahajan VB, Ferguson PJ, Axelrod JD, Lehesjoki A-E, Fritzsch B, Slusarski DC, Wemmie J, Ueno N, Bassuk AG. Mutations in Prickle Orthologs Cause Seizures in Flies, Mice, and Humans. The American Journal of Human Genetics. 2011;88:138–149. doi: 10.1016/j.ajhg.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Suzuki M, Kiyonari H, Abe T, Sasaoka T, Ueno N. Mouse prickle1, the homolog of a PCP gene, is essential for epiblast apical-basal polarity. Proceedings of the National Academy of Sciences. 2009;106:14426–14431. doi: 10.1073/pnas.0901332106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Expression of planar cell polarity genes during development of the mouse CNS. European Journal of Neuroscience. 2006;23:597–607. doi: 10.1111/j.1460-9568.2006.04596.x. [DOI] [PubMed] [Google Scholar]
- Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent {beta}-catenin degradation. J. Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torban E, Wang H-J, Groulx N, Gros P. Independent Mutations in Mouse Vangl2 That Cause Neural Tube Defects in Looptail Mice Impair Interaction with Members of the Dishevelled Family. Journal of Biological Chemistry. 2004;279:52703–52713. doi: 10.1074/jbc.M408675200. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Celli J, Kayserili H, van Beusekom E, Balci S, Brussel W, Skovby F, Kerr B, Percin EF, Akarsu N, Brunner HG. Mutation of the gene encoding the ROR2 tyrosine kinase causes autosomal recessive Robinow syndrome. Nat Genet. 2000;25:423–426. doi: 10.1038/78113. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish Prickle, a Modulator of Noncanonical Wnt/Fz Signaling, Regulates Gastrulation Movements. Current Biology. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of Rate of Cartilage Differentiation by Indian Hedgehog and PTH-Related Protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Wang B, Sinha T, Jiao K, Serra R, Wang J. Disruption of PCP signaling causes limb morphogenesis and skeletal defects and may underlie Robinow syndrome and brachydactyly type B. Human Molecular Genetics. 2011;20:271–285. doi: 10.1093/hmg/ddq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-KL, Omi M, Ferrari D, Cheng H-C, Lizarraga G, Chin H-J, Upholt WB, Dealy CN, Kosher RA. Function of BMPs in the apical ectoderm of the developing mouse limb. Developmental Biology. 2004;269:109–122. doi: 10.1016/j.ydbio.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Witte F, Chan D, Economides AN, Mundlos S, Stricker S. Receptor tyrosine kinase-like orphan receptor 2 (ROR2) and Indian hedgehog regulate digit outgrowth mediated by the phalanx-forming region. Proceedings of the National Academy of Sciences. 2010;107:14211–14216. doi: 10.1073/pnas.1009314107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- Yin H, Copley CO, Goodrich LV, Deans MR. Comparison of Phenotypes between Different vangl2 Mutants Demonstrates Dominant Effects of the Looptail Mutation during Hair Cell Development. PLoS ONE. 2012;7:e31988. doi: 10.1371/journal.pone.0031988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi Y, Sakiyama J, Kameda T, Iba H, Suzuki A, Ueno N, Kuroiwa A. BMP-2/-4 mediate programmed cell death in chicken limb buds. Development. 1996;122:3725–3734. doi: 10.1242/dev.122.12.3725. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Zhao Y. The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein—protein interaction. Biology of the Cell. 2007;99:489–502. doi: 10.1042/BC20060126. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Zhu J, Wang Y, Zhou J, Ren K, Ding X, Zhang J. LIM domain protein TES changes its conformational states in different cellular compartments. Molecular and Cellular Biochemistry. 2009;320:85–92. doi: 10.1007/s11010-008-9901-7. [DOI] [PubMed] [Google Scholar]