Abstract
Multi-isotope imaging mass spectrometry (MIMS) is the quantitative imaging of stable isotope labels in cells with a new type of secondary ion mass spectrometer (NanoSIMS). The power of the methodology is attributable to (i) the immense advantage of using non-toxic stable isotope labels, (ii) high resolution imaging that approaches the resolution of usual transmission electron microscopy and (iii) the precise quantification of label down to 1 part-per-million and spanning several orders of magnitude. Here we review the basic elements of MIMS and describe new applications of MIMS to the quantitative study of metabolic processes including protein and nucleic acid synthesis in model organisms ranging from microbes to humans.
Keywords: Stable isotope, Quantitative imaging, Stem cell, Human, Metabolism, Cell division
1. Introduction
Radiolabeled nucleotides [1] and halogenated nucleotide analogs [2] have been extensively used to label and track nucleic acids during chromosome replication and cell division. These approaches, however, are associated with pitfalls related to reagent toxicity [3,4] and the fidelity of techniques used for label measurement. Indeed, numerous active controversies in cell biology can be traced to limitations inherent to the use of label detection methods such as autoradiography or immunofluorescence to answer questions of cell turnover and fate.
One such area of active debate is whether some stem cells non-randomly segregate chromosomes during mitosis [5,6]. The resultant preservation of DNA template strands may influence the genetic stability of the stem cell lineage or impact daughter cell fate by the asymmetric inheritance of epigenetic gene-regulatory elements. The mere fact that the debate over the existence of so-called “immortal DNA template strands” is unresolved after nearly four decades of investigation [7] illustrates the limitations of the methods used to test the hypothesis.
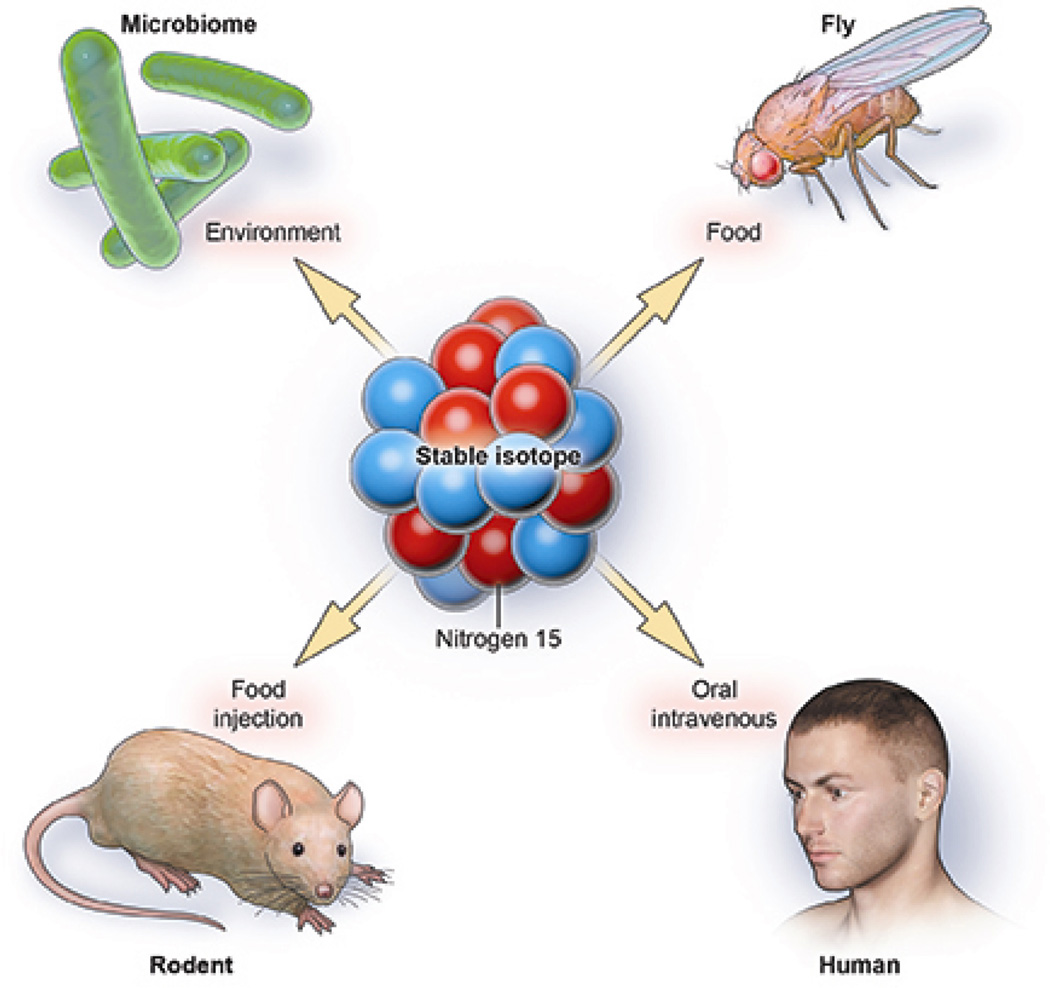
Here we review a new approach to quantitatively study intracellular metabolic processes, including DNA synthesis, a methodology that we call multi-isotope imaging mass spectrometry (MIMS) [8]. MIMS is a synergy of two methods pioneered in the first half of the last century: the vision of using stable isotope tracers (Figs. 1 and 2) to quantitatively study metabolism as shown by Schoenheimer [9–12] and the extension by Leblond [13] to use autoradiography with electron microscopy to visualize radiotracers intracellularly. With MIMS, stable isotope tracers can be quantitatively imaged and measured in domains at least 10-fold smaller than a micron cubed. Here we summarize the important components of MIMS, including (i) stable isotopes and their advantages as metabolic tracers, (ii) a new-generation secondary ion mass spectrometer (Fig. 3) that emerged after crucial advances in ion optics, and (iii) an evolving computational interface that enables efficient data analysis [8]. We conclude by illustrating the power of the methodology, with particular emphasis on the study of immortal strands and the immense potential for human translation.
Fig. 1.
Stable isotopes as metabolic tracers. An immense advantage of stable isotope tracers is that they do not decay and are natural in the environment. When delivered in enriched form as a component of tracer molecules, they seamlessly incorporate into the metabolic pathway under study, and they are detectable with mass spectrometers, including NanoSIMS. The innocuous nature of stable isotopes has led to their widespread use as tracers in organisms ranging from microbes to humans. Since Urey [18] discovered deuterium and Schoenheimer [9–12] pioneered the use of stable isotopes as tracers to study metabolism, they have been increasely used in human studies.
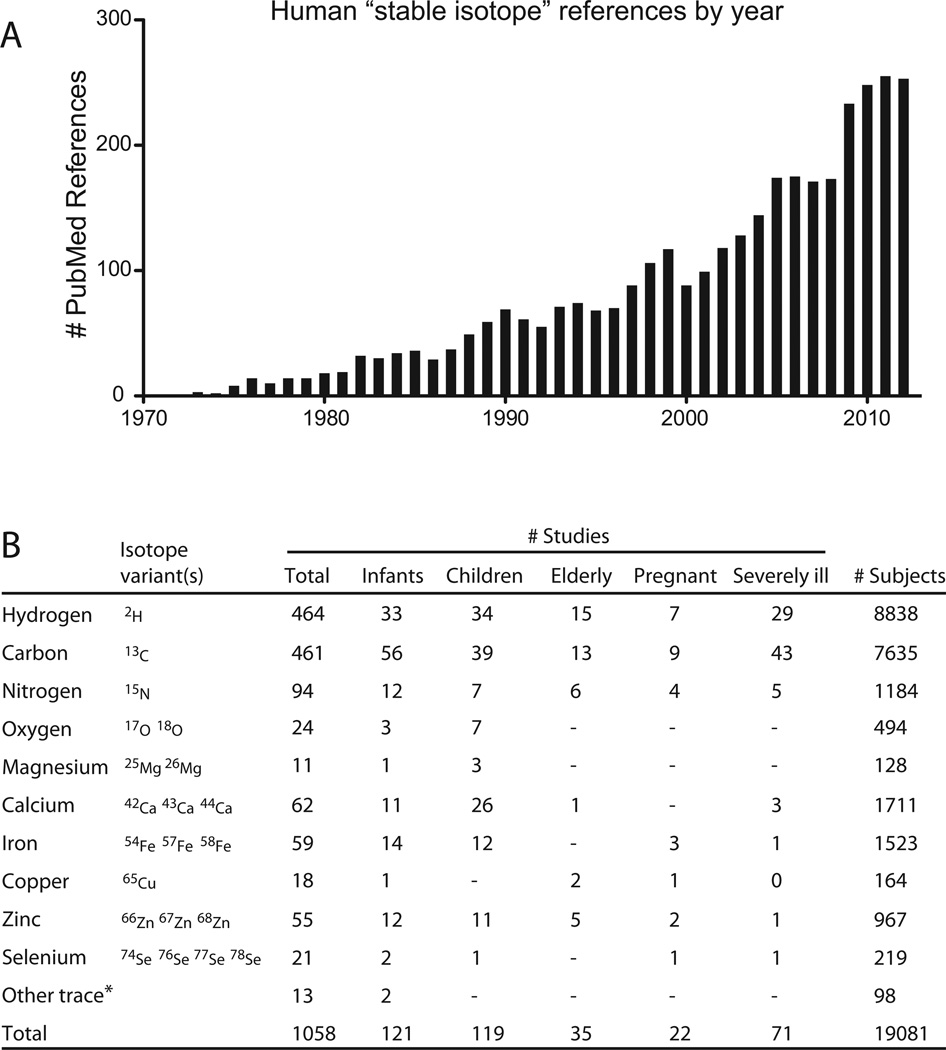
Fig. 2.
Precedent for administration of stable isotopes to humans. This figure and table illustrate the extensive history of safe stable isotope administration to humans, including the most vulnerable populations such as pregnant women, infants, and the severely ill. (A) These data are derived from a Pubmed search with search term “stable isotope” then limited to “human” studies. The search (4/10/2013) yielded 3507 references, which when parsed by year show a dramatic increase in stable isotope use with time. (B) The references were then reviewed to assess key pieces of data, including whether the study indeed administered stable isotopes to humans, details regarding the subject population, and the isotope(s) administered. Patient populations were defined as follows: infant < 12 mos, child = 1–18 yrs, elderly > 65 yrs, severely ill = occupying intensive care unit. These data certainly underestimate the cumulative human experience, in part because the 13C–urea breath test is widely used in clinical practice. While most of the low abundance stable isotope variants are heavier than the most common form, selenium is an example of a trace mineral where the most common form (80Se = 49.61%) is heavier than 3 other selenium stable isotopes. Note: total values are often smaller than the sum of the column because many studies employed multiple stable isotope tracers. For full list of references: http://nrims.partners.org/HumanStableIsotopeReferences.xls. *Uncommonly used stable isotope tracers in human studies included sulfur, nickel, chromium, molybdenum, and cadmium.
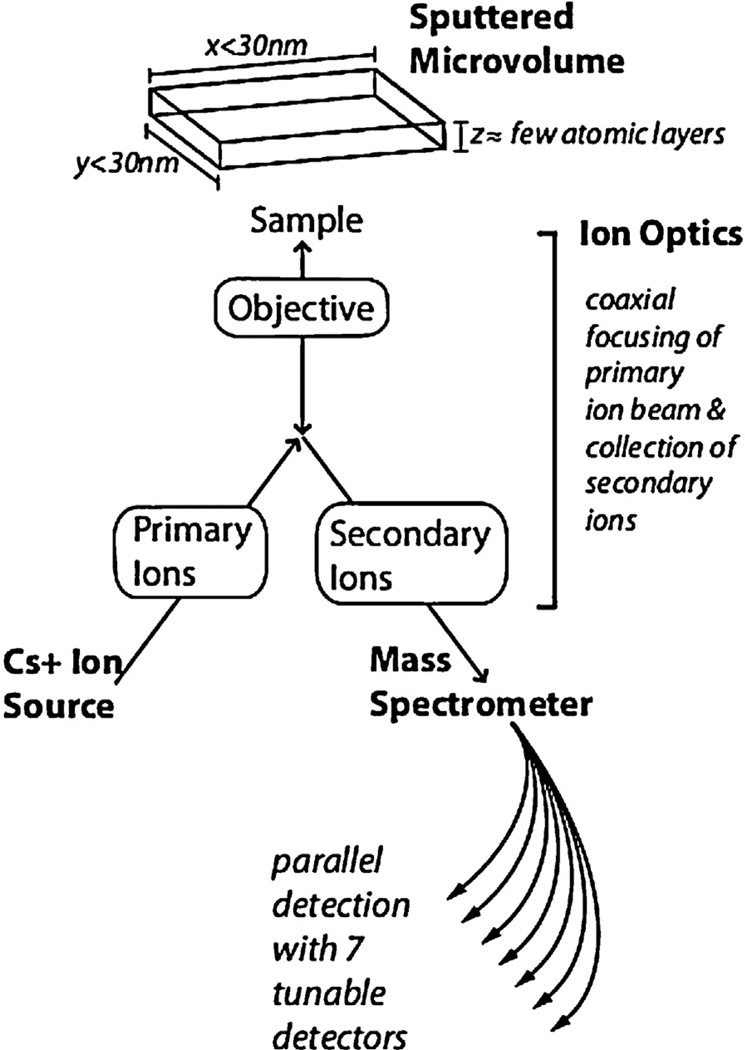
Fig. 3.
Ion optics and the NanoSIMS instrument. In a NanoSIMS instrument, an ion optical system directs a primary ion beam (Cs+) to bombard the sample surface. The sample is sputtered with release of atoms and atomic clusters, of which a small fraction is ionized. This ionized fraction (secondary ions), representative of the composition of the sample, is efficiently collected by the immersion objective. The ion optical system shapes and transports negatively charged secondary ions to the mass spectrometer, where 7 tunable detectors simultaneously count 7 masses of interest.
2. Stable isotope tracers
Stable isotopes are ideal metabolic tracers, because they are easily detectable and topographically mappable with high precision, they seamlessly integrate in the biochemical and physiologic processes of cells and organisms, and there is extensive precedent of safety in model organisms and humans (Fig. 2). Although radiolabels, fluorescent compounds, and halogenated analogs have been invaluable tools for biological studies, they each have the potential for toxicity and direct influence on the pathway under study. The toxicity of radiolabels includes the induction of DNA damage and modification of cell cycle activity [14]. The covalent attachment of a reporter, such as a fluorescent protein or halogen atoms, may alter the structure and biochemical properties of the parent molecule, resulting in cytotoxicity or cell cycle alterations [4,15–17].
In contrast to radioisotopes, stable isotopes do not decay and simply are isotopic variants that differ in atomic mass due to an alternative number of neutrons [18,19]. Many elements have more than one stable isotope form, but the term “stable isotope” often denotes the heavier, less abundant variants. Stable isotopes exist in animate and inanimate matter in constant ratios; therefore, incorporation of stable isotope tracers in a domain of interest is detectable by a measurable change in the isotope ratio. Schoenheimer pointed to their existence in equivalent ratios in both animate and inanimate matter as evidence that living organisms must not distinguish the few molecules containing the heavier variant from those containing the lighter variant [11].
Deuterium is the only stable isotope with evident toxicity when administered in high doses, several orders of magnitude above “tracer” doses. Inhibitory effects on the growth of organisms ranging from microbes to mammals generally emerge when >15% of all hydrogen atoms comprising the animal have been replaced by deuterium, a substitution that requires prolonged exposure to high concentration deuterium [20–23]. This effect is generally attributed to an isotopic effect on the kinetics of biochemical reactions due to the addition of a neutron, which results in the doubling of the atom’s atomic weight. Minor isotopic effects may also exist for stable isotopes of the heavier elements, such as carbon and nitrogen, for which an extra neutron has a smaller relative effect on the atomic weight. While these isotopic effects may account for subtle changes in the kinetics of biochemical reactions and minute naturally occurring differences in isotopic ratio can occur over paleontological time [24,25], there is no evidence that such subtle isotopic effects impact the global physiologic function of cells or organisms, even after replacement of a substantial fraction of organism-wide atoms with the heavier isotope. For example, replacing approximately 20% of a rodent’s carbon atoms with 13C or 60% of oxygen atoms with 18O incurs no discernable physiologic effect [26,27]. Most importantly, the safety of stable isotopes, including “tracer” doses of deuterium, is born out by decades of animal and human studies, in which no harmful signals have emerged, even from studies including the most vulnerable research subjects, such as pregnant, neonatal, or critically ill patients [23,28,29] (see table contained in Fig. 2).
3. NanoSIMS™
The fundamental concept of SIMS developed in the 1960s remains largely unchanged with new generation NanoSIMS™ instruments (Fig. 3) [30,31]. The surface of a sample is sputtered with an ion beam (e.g. cesium ions), triggering a cascade of atomic collisions and resulting in the ejection of atoms, molecular fragments, and atomic clusters. A small fraction of these emitted secondary particles are ionized, and it is this fraction that is transmitted and shaped through an ion optical system to a mass spectrometer, where ions of a given mass are counted. Images are generated after registering the counts of a given ionic particle to the region on the sample from where the ions were sputtered. The fundamental goal of MIMS – to quantitatively localize stable isotope tags in subcellular domains within a reasonable amount of time – is now possible because of innovations in ion optics and particle discrimination.
With NanoSIMS, image resolution is in part dictated by the statistical power of inter-pixel comparisons, and therefore improved efficiency of ion capture and transport decreases the size of the analyzed microvolume required to generate a statistically meaningful pixel on a mass image. The immersion objective increases the fraction of ions collected by exerting an electrostatic accelerating field in close proximity to the sample surface to collect projectile ions with disparate energies and trajectories. The secondary ions, which are representative of the sample composition, are shaped by ionic lenses and ultimately separated by mass charge along the focal plane of the magnet. The ion optical system, contained in NanoSIMS instruments, efficiently delivers up to 80% of all ions released from a sputtered microvolume to the mass spectrometer [32]. This efficiency makes high resolution imaging possible within a practical timeframe.
Secondary ions are delivered from the optical column to a mass spectrometer with high mass resolution, a feature that broadens the range of potential analytes, including nitrogen. The direct analysis of nitrogen is rendered inefficient by the low electron affinity of the native atom, resulting in rare emission of nitrogen ions. Instead, the analysis requires resolution of a surrogate molecule, such as CN, which is challenging due to the similar molecular weights of 13C14N, 12C15N, and 12C14N1H all of which have a reported molecular weight of 27. The mass spectrometer, however, can resolve the subtle differences in mass between these molecules [8,33]. Indeed, the ability to analyze samples containing 15N-tagged tracers is invaluable to biology, where many molecules of high relevance, such as proteins and nucleic acids contain nitrogen.
It is the use of multiple detectors in parallel that makes possible the precise quantification of stable isotope tracers [8]. With MIMS, stable isotope incorporation is measured by an increase above natural occurrence of the stable isotope tag relative to its common variant. Because both ionic species are measured simultaneously from the same sputtered microvolume, the measured isotope ratio is unaffected by regional differences in ionization or instrumental drift. This built-in control makes the isotope ratio measurements more precise and efficient than if the two components of the isotope ratio were measured serially.
4. Computation
An underappreciated aspect of imaging mass spectrometry is the analytic challenge posed by large data files, which can routinely exceed one gigabyte. A number of software platforms are available to analyze the massive quantities of data produced by NanoSIMS analyses [34], some of which are freely available. At the National Resource for Imaging Mass Spectrometry (NRIMS), we use a custom-built “plugin” to ImageJ, called OpenMIMS (http://www.nrims.harvard.ed/software.php).
A central theme in software development for MIMS applications is to improve the efficiency and sophistication of identification and analysis of regions of interest (ROI). NanoSIMS data analyses generally entail manually outlining ROI, before extracting the quantitative data for each ROI or group of ROI. The process of manual ROI selection and classification is often the most time-consuming aspect of nanoSIMS experiments, while also introducing potential for observer bias. Ongoing software development aims to facilitate reproducible drawing of ROIs via interactive thresholding or semi-automated boundary detection [34,35]. Such programs may still require some degree of qualitative “expert” validation of the auto-generated ROIs, but can greatly accelerate experimental analysis.
While NanoSIMS images are most easily rendered in 2 dimensions, each pixel is representative of a microvolume of matter, in which the vertical dimension is on the order of a few atomic layers. Thus, the vertical resolution far exceeds the lateral resolution. Accessing the vertical-resolving capabilities opens the possibility of using successively sputtered planes for 3D reconstruction [36]. Further software development will enable efficient selection and quantitation of volumetric ROI in 3D images.
Software innovation may also facilitate synergy between NanoSIMS analyses and other commonly used methods of phenotypic or genetic characterization, such as detection of proteins, nuclei acids, or genetic reporters by in situ hybridization or immunohistochemistry. Although there is immense potential to directly measure probes or antibodies, as has been established for electron microscopy with heavy-metal conjugated antibodies, there is also a role for computational cross-talk between MIMS images and data derived in parallel with other modalities. Such approaches have been used in the study of bacteria, where fluorescent signals obtained with fluorescent in situ hybridization can be registered with NanoSIMS images and used to generate ROI in a semi-automated manner [34]. Continued improvements in automated interaction with other methodologic platforms holds the potential to greatly accelerate the discovery process, while also removing operator bias from the analysis process.
5. Applying MIMS to biology
One way to think of MIMS is that it merges the analytical and quantitative power of mass spectrometry as envisioned by Schoenheimer [11] with imaging at a lateral resolution equivalent to that of usual transmission electron microscopy, over exquisitely thin depth – a few atomic layers – much smaller than the thinnest electron microscopy sections. The power of MIMS relies on this dynamic interplay between the high-resolution qualitative imaging of cells and tissues with the quantitative data that can be extracted for any region of interest within an image.
5.1. Illuminating complex tissues with high-resolution mass images
Quantitative mass images are reconstructed into a gray-scale image in which the pixel intensity is derived from the total number of counts of a given secondary ion within the area represented by a given pixel (Fig. 4). We have found that mass images of unlabeled tissues often provide striking regional contrasts in mass intensity, illuminating complex tissue architectures with clarity reminiscent of electron micrographs. CN− images provide an excellent first-pass histologic view of biological tissues, often revealing striking subcellular detail [37]. Nuclear borders are easily resolved and subnuclear structures, such as nucleoli are often visible. Submicron cellular projections, including lamillapodia and the cilia of the intestinal brush border are identifiable [8].
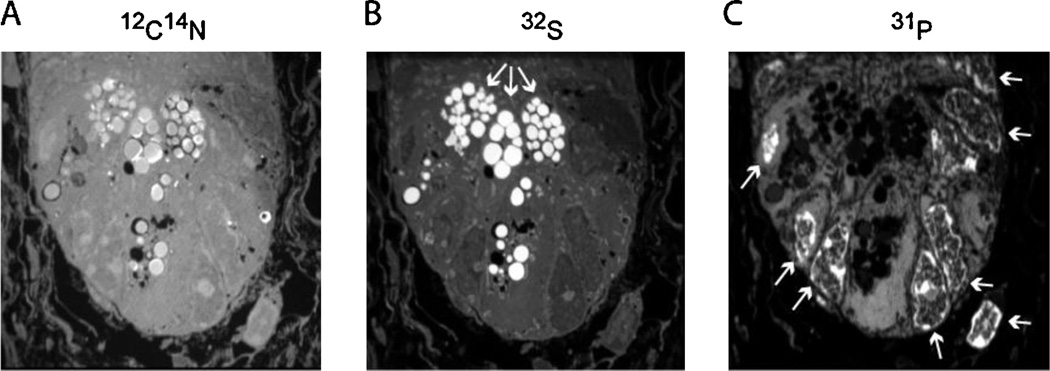
Fig. 4.
Mass images of the small intestinal crypt. The base of the small intestinal crypt is shown (A) 12C14N mass image (surrogate for 14N). (B) 32S mass image shows intense signal in the granules found in the apical cytoplasm of Paneth cells (white arrows). (C) 31P mass image showing intense signal in the nuclei (white arrows). Tissue embedded in LR white; sectioned to 0.5 µm. Image acquisition: raster size = 30 µm, 512 × 512 pixels, 0.750 ms/pixel, 92 consecutive planes summed.
Regional differences in mass intensity can often be accounted for based on known differences in elemental composition within different tissues or subcellular domains. A bright 31P− signal is observed in nuclei in a pattern that resembles chromatin, and which is attributable to the high phosphate content of DNA [38,39]. Similarly, cytoplasmic granules often contain high relative concentrations of elements like sulfur or zinc, likely accounting for the dramatic illumination of Paneth cell granules in the small intestine by 32S− images [39]. Melanin granules in human hair are seen brightly in 32S− images due to variations in sulfur-content [40].
5.2. Measuring stable isotopes in subcellular domains
The exceptional power of MIMS is the confluence of imaging at high resolution with measurement of isotope incorporation in micro-domains (sub-micron cubed) of interest. Experiments are designed so that stable isotopes have been incorporated in cells via a metabolic or biosynthetic pathway of interest. Examples for which MIMS has already been used include nitrogen fixation by bacteria [41–45], fatty acid transport by adipocytes [46], protein synthesis by the hair cells of the inner ear [36], and DNA synthesis [39,47].
We recently applied MIMS in the small intestinal stem cell niche to test the “immortal strand hypothesis” [7], which holds that with each round of stem cell division, chromosomes containing older template strands are segregated to the daughter cell destined to remain a stem cell. The resultant indefinite maintenance of a full complement of DNA template strands was proposed to explain the phenomenon of the “label-retaining” stem cell, in which DNA label remains undiluted despite ongoing cell division during label-free chase. With a series of pulse-chase experiments with stable isotope (15N)-tagged thymidine, we showed that dividing cells in the stem cell compartment of the small intestine randomly segregated chromosomes and thus do not display evidence of “immortal strands”, at least under normal homeostatic conditions [39].
This study showcased many of the key aspects of MIMS for in vivo biology. First, pulse-chase experiments designed to detect “label-retaining” stem cells entailed label administration for periods spanning in utero through post-natal development. Such extensive labeling duration is made possible by the use of non-toxic stable isotope labels. Second, we quantified the magnitude of label dilution in dividing cells, which showed that the degree of label dilution was consistent with random distribution of labeled DNA strands. Our conclusion that label was equally distributed to daughter cells in a pattern consistent with random segregation was greatly strengthened by the precise measurement of label in domains as small as the segregating chromosomes (Fig. 5) [39,48]. This study left unanswered whether small intestinal stem cells are stimulated to undergo asymmetric division after injury, as described in both the intestine [49,50] and skeletal muscle [51,52], and it does not definitively exclude very subtle asymmetry of chromosomal segregation as suggested in the large intestine [53]; however, it provides a template for quantitatively studying DNA synthesis, chromosome fate, and cell turnover in a broad range of experimental settings.
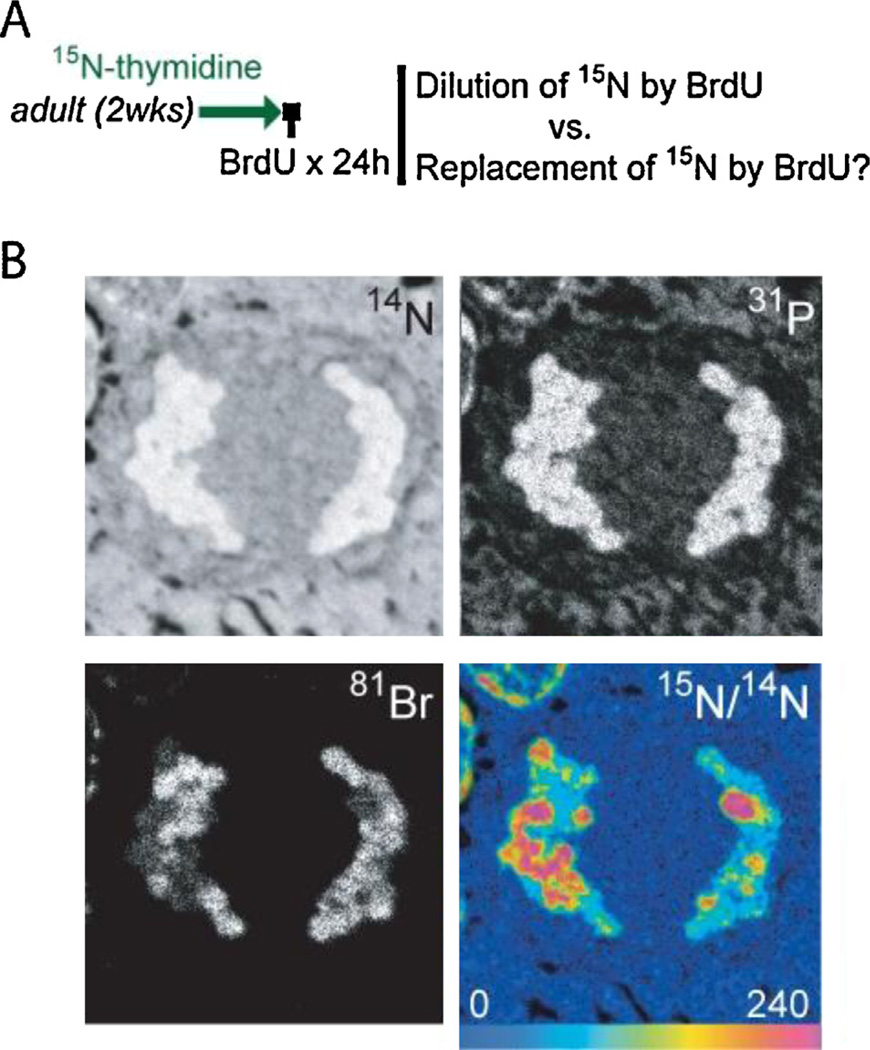
Fig. 5.
MIMS images of mitotic cell in the small intestinal crypt. (A) Label administered to adult mice for 2 wks, followed by BrdU for 24 h during 15N-thymidine-free chase. The “immortal strand hypothesis” would predict complete replacement of 15N by BrdU, rather than the dilution of 15N signal by approximately 50% with each division. There were no examples of the complete replacement of 15N by BrdU, including in mitotic figures. Note: BrdU was administered at the end of the experiment; BrdU is quantitatively imaged by directly analyzing 81Br−. (B) Small intestine from mouse after sequential administration of 15N-thymidine and BrdU (see A). Segregating chromosomes are visible showing BrdU (bottom left) and 15N-label (bottom right) in both segregating chromosomal complements. Bottom right: Hue saturation intensity (HSI) image mapping the 15N/14N ratio. The rainbow scale ranges from blue, where the ratio is equivalent to natural ratio (0.37%, expressed as 0% above natural ratio), to red, where the ratio is several fold above natural ratio (240% above natural ratio). Although this image (representative sampling of the larger 3-dimensional mitotic nucleus) shows a slight imbalance in the 15N distribution, the frequency of signal imbalance in mitotic figures was similar to that predicted by random chance (n = 232 mitotic figures in late stages of mitosis). Image acquisition: raster size = 9 µm, resolution 512 × 512 pixels, 0.750 ms/pixel, 100 consecutive planes summed.
5.3. Human translation
Methods of bulk tissue analyses of stable isotope tracers [10,54] and recent advances in molecular imaging [55] allow the study of human metabolism and immune and inflammatory processes on the tissue or organ level. Now MIMS opens the possibility of studying such diverse biological processes on the cellular and subcellular level with stable isotope tracers. We recently performed a proof-of-principle study, in which we identified rare 15N-labeled lymphocytes in peripheral blood samples taken from a healthy human volunteer after pulse-chase administration of intravenous 15N-thymidine [39].
A central issue with such translational human experiments is that they be conducted safely. Though stable isotopes are inherently safe and tracer studies generally involve administering naturally occurring molecules, most stable isotope tagged compounds are unavailable in pharmaceutical grade. While specific quality control metrics vary amongst different regulatory agencies, a series of quality control tests can facilitate safe label administration. Thus, prior to administering 15N-thymidine to human subjects, we performed stability testing of 15N-thymidine after suspension in 0.9% NaCl infusate, verifying that it was stable in solution at room temperature over 14 days. In addition, sterility testing, endotoxin screens, and the use of sterile packaging techniques ensure protection from complications, such as bacterial infections or systemic inflammatory responses. We anticipate that our experience with 15N-thymidine coupled with the decades of experience using other stable isotope tagged compounds in human studies will provide a template to study cell turnover and a range of metabolic processes on the cellular and subcellular level in humans with MIMS.
6. Summary
MIMS combines the use of innocuous stable isotope tracers with the quantitative imaging power of modern nanoSIMS instrumentation. MIMS has illuminated a wide range of cellular processes including fatty acid transport and storage [46], protein synthesis [36], nitrogen fixation [41], and DNA synthesis [39] in model organisms ranging from bacteria to humans. The methodology has provided answers to controversial questions in cell biology, including the mechanism of cytoskeletal turnover in inner ear hair cells [36], the regenerative potential of mammalian cardiac myocytes [47], and the “immortal strand hypothesis” in the small intestine [39]. Just as initial demonstrations of the power of MIMS to show nitrogen fixation by bacteria [41] led to a proliferation of its use in the field of microbiology [56,57], we anticipate heightened interest in the broader biomedical research community to quantitatively image subcellular biochemical processes with MIMS in organisms ranging from single microbes to humans.
References
- 1.Walker BE, Leblond CP. Sites of nucleic acid synthesis in the mouse visualized by radioautography after administration of C14-labelled adenine and thymidine. Experimental Cell Research. 1958;14:510–531. doi: 10.1016/0014-4827(58)90158-7. [DOI] [PubMed] [Google Scholar]
- 2.Latt SA. Microfluorometric detection of deoxyribonucleic acid replication in human metaphase chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:3395–3399. doi: 10.1073/pnas.70.12.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Littlefield JW, Gould EA. The toxic effect of 5-bromodeoxyuridine on cultured epithelial cells. Journal of Biological Chemistry. 1960;235:1129–1133. [PubMed] [Google Scholar]
- 4.Matsuoka K, Nomura K, Hoshino T. Mutagenic effects of brief exposure to bromodeoxyuridine on mouse FM3A cells. Cell and Tissue Kinetics. 1990;23:495–503. doi: 10.1111/j.1365-2184.1990.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 5.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Rando TA. The immortal strand hypothesis: segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 8.Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, et al. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. Journal of Biology. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenheimer R, Rittenberg D, Foster GL, Keston AS, Ratner S. The application of the nitrogen isotope N15 for the study of protein metabolism. Science. 1938;88:599–600. doi: 10.1126/science.88.2295.599. [DOI] [PubMed] [Google Scholar]
- 10.Schoenheimer R, Rittenberg D. The application of isotopes to the study of intermediary metabolism. Science. 1938;87:221–226. doi: 10.1126/science.87.2254.221. [DOI] [PubMed] [Google Scholar]
- 11.Schoenheimer R. The investigation of intermediary metabolism with the aid of heavy hydrogen: Harvey lecture, January 21, 1937. Bulletin of the New York Academy of Medicine. 1937;13:272–295. [PMC free article] [PubMed] [Google Scholar]
- 12.Schoenheimer R, Rittenberg D. Deuterium as an indicator in the study of intermediary metabolism. Science. 1935;82:156–157. doi: 10.1126/science.82.2120.156. [DOI] [PubMed] [Google Scholar]
- 13.Belanger LF, Leblond CP. A method for locating radioactive elements in tissues by covering histological sections with a photographic emulsion. Endocrinology. 1946;39:8–13. doi: 10.1210/endo-39-1-8. [DOI] [PubMed] [Google Scholar]
- 14.Hu VW, Black GE, Torres-Duarte A, Abramson FP. 3H-thymidine is a defective tool with which to measure rates of DNA synthesis. FASEB Journal. 2002;16:1456–1457. doi: 10.1096/fj.02-0142fje. [DOI] [PubMed] [Google Scholar]
- 15.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 16.Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is green fluorescent protein toxic to the living cells? Biochemical and Biophysical Research Communications. 1999;260:712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 17.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 18.Urey HC. The separation and properties of the isotopes of hydrogen. Science. 1933;78:566–571. doi: 10.1126/science.78.2034.566. [DOI] [PubMed] [Google Scholar]
- 19.Sturup S, Hansen HR, Gammelgaard B. Application of enriched stable isotopes as tracers in biological systems: a critical review. Analytical and Bioanalytical Chemistry. 2008;390:541–554. doi: 10.1007/s00216-007-1638-8. [DOI] [PubMed] [Google Scholar]
- 20.Lester W, Jr, Sun SH, Seber A. Observations on the influence of deuterium on bacterial growth. Annals of the NewYork Academy of Sciences. 1960;84:667–677. doi: 10.1111/j.1749-6632.1960.tb39099.x. [DOI] [PubMed] [Google Scholar]
- 21.Lewis GN. The biology of heavy water. Science. 1934;79:151–153. doi: 10.1126/science.79.2042.151. [DOI] [PubMed] [Google Scholar]
- 22.Barbour HG. The basis of the pharmacological action of heavy water in mammals. Yale Journal of Biology and Medicine. 1937;9:551–565. [PMC free article] [PubMed] [Google Scholar]
- 23.Schellekens RC, Stellaard F, Woerdenbag HJ, Frijlink HW, Kosterink JG. Applications of stable isotopes in clinical pharmacology. British Journal of Clinical Pharmacology. 2011;72:879–897. doi: 10.1111/j.1365-2125.2011.04071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand WA, Coplen TB. Stable isotope deltas: tiny, yet robust signatures in nature. Isotopes in Environmental and Health Studies. 2012 doi: 10.1080/10256016.2012.666977. [DOI] [PubMed] [Google Scholar]
- 25.Snider MJ, Reinhardt L, Wolfenden R, Cleland WW. 15N kinetic isotope effects on uncatalyzed and enzymatic deamination of cytidine. Biochemistry. 2002;41:415–421. doi: 10.1021/bi011410i. [DOI] [PubMed] [Google Scholar]
- 26.Gregg CT, Hutson JY, Prine JR, Ott DG, Furchner JE. Substantial replacement of mammalian body carbon with carbon-13. Life Sciences. 1973;13:775–782. doi: 10.1016/0024-3205(73)90068-4. [DOI] [PubMed] [Google Scholar]
- 27.Samuel D, Wof A, Meshorer A, Wasserman I. The effect of 18O on the growth and reproduction of mice; Stable Isotopes: Proceedings of the Third International Conference; 1973. pp. 353–360. [Google Scholar]
- 28.Koletzko B, Demmelmair H, Hartl W, Kindermann A, Koletzko S, Sauerwald T, et al. The use of stable isotope techniques for nutritional and metabolic research in paediatrics. Early Human Development. 1998;53(Suppl.):S77–S97. doi: 10.1016/s0378-3782(98)00067-x. [DOI] [PubMed] [Google Scholar]
- 29.Koletzko B, Sauerwald T, Demmelmair H. Safety of stable isotope use. European Journal of Pediatrics. 1997;156(Suppl. 1):S12–S17. doi: 10.1007/pl00014267. [DOI] [PubMed] [Google Scholar]
- 30.Castaing R, Slodzian GJ. Optique corpusculaire-premiers essais de microanalyse par emission ionique secondaire. Microscopie. 1962;1:395–399. [Google Scholar]
- 31.Liebl H. Ion microprobe mass analyzer. Journal of Applied Physics. 1967;38:5277–5283. [Google Scholar]
- 32.Slodzian G, Daigne B, Girard F, Boust F, Hillion F. Scanning secondary ion analytical microscopy with parallel detection. Biologie Cellulaire. 1992;74:43–50. doi: 10.1016/0248-4900(92)90007-n. [DOI] [PubMed] [Google Scholar]
- 33.McMahon G, Saint-Cyr HF, Lechene C, Unkefer CJ. CN-secondary ions form by recombination as demonstrated using multi-isotope mass spectrometry of 13C- and 15N-labeled polyglycine. Journal of the American Society for Mass Spectrometry. 2006;17:1181–1187. doi: 10.1016/j.jasms.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Polerecky L, Adam B, Milucka J, Musat N, Vagner T, Kuypers MM. Look@NanoSIMS – a tool for the analysis of nanoSIMS data in environmental microbiology. Environmental Microbiology. 2012;14:1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x. [DOI] [PubMed] [Google Scholar]
- 35.Gormanns P, Reckow S, Poczatek JC, Turck CW, Lechene C. Segmentation of multi-isotope imaging mass spectrometry data for semi-automatic detection of regions of interest. PLoS ONE. 2012;7:e30576. doi: 10.1371/journal.pone.0030576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang DS, Piazza V, Perrin BJ, Rzadzinska AK, Poczatek JC, Wang M, et al. Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature. 2012;481:520–524. doi: 10.1038/nature10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peteranderl R, Lechene C. Measure of carbon and nitrogen stable isotope ratios in cultured cells. Journal of the American Society for Mass Spectrometry. 2004;15:478–485. doi: 10.1016/j.jasms.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Guerquin-Kern JL, Hillion F, Madelmont JC, Labarre P, Papon J, Croisy A. Ultra-structural cell distribution of the melanoma marker iodobenzamide: improved potentiality of SIMS imaging in life sciences. Biomedical Engineering Online. 2004;3:10. doi: 10.1186/1475-925X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinhauser ML, Bailey AP, Senyo SE, Guillermier C, Perlstein TS, Gould AP, et al. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481:516–519. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallegot P, Peteranderl R, Lechene C. In-situ imaging mass spectrometry analysis of melanin granules in the human hair shaft. Journal of Investigative Dermatology. 2004;122:381–386. doi: 10.1046/j.0022-202X.2004.22217.x. [DOI] [PubMed] [Google Scholar]
- 41.Lechene CP, Luyten Y, McMahon G, Distel DL. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science. 2007;317:1563–1566. doi: 10.1126/science.1145557. [DOI] [PubMed] [Google Scholar]
- 42.Popa R, Weber PK, Pett-Ridge J, Finzi JA, Fallon SJ, Hutcheon ID, et al. Carbon and nitrogen fixation and metabolite exchange in and between individual cells of Anabaena oscillarioides. ISME Journal. 2007;1:354–360. doi: 10.1038/ismej.2007.44. [DOI] [PubMed] [Google Scholar]
- 43.Dekas AE, Poretsky RS, Orphan VJ. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science. 2009;326:422–426. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- 44.Halm H, Musat N, Lam P, Langlois R, Musat F, Peduzzi S, et al. Co-occurrence of denitrification and nitrogen fixation in a meromictic lake, Lake Cadagno (Switzerland) Environmental Microbiology. 2009;11:1945–1958. doi: 10.1111/j.1462-2920.2009.01917.x. [DOI] [PubMed] [Google Scholar]
- 45.Finzi-Hart JA, Pett-Ridge J, Weber PK, Popa R, Fallon SJ, Gunderson T, et al. Fixation and fate of C and N in the cyanobacterium trichodesmium using nanometer-scale secondary ion mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6345–6350. doi: 10.1073/pnas.0810547106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleinfeld AM, Kampf JP, Lechene C. Transport of 13C-oleate in adipocytes measured using multi imaging mass spectrometry. Journal of the American Society for Mass Spectrometry. 2004;15:1572–1580. doi: 10.1016/j.jasms.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 47.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabin-Flaman A, Monnier AF, Coffinier Y, Audinot JN, Gibouin D, Wirtz T, et al. Combed single DNA molecules imaged by secondary ion mass spectrometry. Analytical Chemistry. 2011;83:6940–6947. doi: 10.1021/ac201685t. [DOI] [PubMed] [Google Scholar]
- 49.Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 50.Quyn AJ, Appleton PL, Carey FA, Steele RJ, Barker N, Clevers H, et al. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6:175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell Biology. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 52.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biology. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falconer E, Chavez EA, Henderson A, Poon SS, McKinney S, Brown L, et al. Identification of sister chromatids by DNA template strand sequences. Nature. 2010;463:93–97. doi: 10.1038/nature08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neese RA, Misell LM, Turner S, Chu A, Kim J, Cesar D, et al. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15345–15350. doi: 10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuypers MM. Microbiology. Sizing up the uncultivated majority. Science. 2007;317:1510–1511. doi: 10.1126/science.1148538. [DOI] [PubMed] [Google Scholar]
- 57.Musat N, Foster R, Vagner T, Adam B, Kuypers MM. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiology Review. 2012;36:486–511. doi: 10.1111/j.1574-6976.2011.00303.x. [DOI] [PubMed] [Google Scholar]