Abstract
Mesenchymal stem cells (MSCs) in different anatomic locations possess diverse biological activities. Maintaining the pluripotent state and differentiation depend on the expression and regulation of thousands of genes, but it remains unclear which molecular mechanisms underlie MSC diversity. Thus, potential MSC applications are restricted. Long noncoding RNAs (lncRNAs) are implicated in the complex molecular circuitry of cellular processes. We investigated differences in lncRNA and mRNA expression profiles between bone marrow stem cells (BMSCs) and periodontal ligament stem cells (PDLSCs) with lncRNA microarray assays and bioinformatics analysis. In PDLSCs, numerous lncRNAs were significantly upregulated (n = 457) or downregulated (n = 513) compared to BMSCs. Furthermore, 1,578 mRNAs were differentially expressed. These genes implicated cellular pathways that may be associated with MSC characteristics, including apoptosis, MAPK, cell cycle, and Wnt signaling pathway. Signal-net analysis indicated that phospholipase C beta 4, filamin B beta, calcium/calmodulin-dependent protein kinase II gamma, and the ionotropic glutamate receptor, AMPA 1, had the highest betweenness centrality among significant genes in the differential gene profile network. A comparison between the coding-noncoding gene coexpression networks of PDLSCs and BMSCs identified chemokine (C-X-C motif) ligand 12 as a core regulatory factor in MSC biology. These results provided insight into the mechanisms underlying MSC biology.
1. Introduction
Stem cells are undifferentiated cells that can either self-renew or differentiate to produce mature progeny cells [1, 2]. The two major categories are embryonic and adult stem cells. Adult stem cells are undifferentiated cells found in specialized tissues and organs of adults. Compared to embryonic stem cells, adult stem cells that exist in various organs of the body are easily accessible, and their use is less controversial in terms of ethics [3, 4]. Mesenchymal stem cells (MSCs) have been identified as mesoderm-derived stromal cells that can differentiate into various mesoderm-type cell lineages. MSCs hold significant promise for tissue regeneration, due to their potential for self-renewal and multilineage differentiation [5–7]. Humans have abundant adult MSCs available for use in cell-based tissue engineering. MSCs from various tissues, including bone marrow, periosteum, skeletal muscle, and adipose tissue, have similar epitope profiles, but significant differences have been observed in MSC properties; that is, MSCs vary in their differentiation, proliferation, and migration potentials according to the tissue source [8–12]. Traditionally, bone-marrow-derived MSCs (BMSCs) have been studied for bone regeneration applications. BMSCs are a population of multipotent, nonhematopoietic marrow-derived cells that are easily expanded in culture and differentiate into cells with an osteogenic phenotype [13, 14]. BMSC transplantations have enhanced periodontal tissue regeneration and bone formation [15, 16]. Interestingly, Hu and colleagues investigated whether BMSCs might give rise to different types of epithelial cells, and they tested their potential for serving as a source of ameloblasts. Those results showed, for the first time, that BMSCs could be reprogrammed to become ameloblast-like cells [17]. Thus, BMSCs offered a novel approach for tooth-tissue engineering; they could be induced to become both mesenchymal and epithelial cells in tooth applications [17]. However, scientists disagree on whether BMSCs are ideal seeding cells for tooth engineering. Jing pointed out that the differentiation ability of BMSCs decreases significantly with increasing age of the donor [18]. In the past few decades, several new populations of MSCs have been isolated from dental and craniofacial tissues on the basis of their stem cell properties. These new populations included stem cells derived from the periodontal ligament (PDLSCs), from dental pulp, and from apical papilla, among others [19–24]. When transplanted into animals, these dental tissue-derived stem cells could generate bone/dentin-like mineralized tissue, and they were capable of repairing tooth defects and regenerating periodontal tissue [21, 25, 26]. In contrast to BMSCs, these cells were easily accessible, and they were more intimately associated with dental tissues [3]. Although dental tissue-derived MSCs and BMSCs are regulated by similar factors and share a common protein expression profile, these populations differ significantly in their proliferative ability and developmental potentials in vitro. Furthermore, importantly, they differ in their ability to develop into distinct tissues representative of the microenvironments from which they were derived in vivo. For example, BMSCs formed only bone tissue in the mouse model when treated in the same manner as the dental tissue-derived stem cells [19, 27]. However, the chondrogenic and adipogenic potentials of dental tissue-derived MSCs appeared to be weaker than those of BMSCs [22, 28]. Conversely, the neurogenicity of dental tissue-derived stem cells may be more potent than that of BMSCs, probably due to their neural crest origin [22, 28].
From the time that dental stem cells were first identified, they have been spotlighted in the dental tissue engineering field. Recently, numerous investigators have attempted to use these cells for dental tissue regeneration and assess their potential in preclinical applications [26, 29]. However, little is known about the characteristics of dental stem cells and the molecular mechanism underlying their diverse biological activities; thus, their potential application is restricted. Clues on the molecules that control MSC biology can be obtained by comparing molecular expression in MSCs with different biological activities. The development of microarray methods for large-scale analyses of mRNA gene expression has made it possible to search systematically for key molecules [30, 31]. With the introduction of these genome-wide research techniques, various groups have attempted to describe and compare the gene expression patterns of specialized adult stem cells [32–34]. Long, noncoding RNAs (lncRNAs) are transcribed RNA molecules longer than 200 nucleotides. LncRNAs have been shown to have comprehensive functions in both normal development and disease states [35]. Many studies have revealed that lncRNAs exert important roles in biological processes, including roles in cell differentiation, transcription, imprinting, chromatin modification, and others [36, 37]. Specifically, previous studies have demonstrated that lncRNAs are extremely important for controlling cell or tissue differentiation [38–40].
In this study, we investigated differences in lncRNA and mRNA expression profiles between PDLSCs and BMSCs with microarray assays and bioinformatics analyses. Our results provided useful information for elucidating the different mechanisms that govern MSCs derived from different tissues.
2. Materials and Methods
2.1. Cell Culture
All research involving human stem cells complied with the International Society for Stem Cell Research “Guidelines for the Conduct of Human Embryonic Stem Cell Research.” We collected impacted, third molars with immature roots from 3 healthy male patients (18–20 years old) under approved guidelines set by the Beijing Stomatological Hospital, Capital Medical University, after obtaining informed patient consent. Molars were removed, disinfected with 75% ethanol, and then washed with PBS. PDLSCs were isolated from each sample, cultured, and identified as previously described [21]. Briefly, PDLSCs were separated from the periodontal ligament in the middle one-third of the root. Then, the tissue was digested in a solution of 3 mg/mL collagenase type I (Worthington-Biochem, USA) and 4 mg/mL dispase (Roche, Germany) for 1 h at 37°C. Single-cell suspensions were obtained by passing the cells through a 70 μm strainer (Falcon, BD Labware, USA). Three separate PDLSC cultures were grown in a humidified, 5% CO2 incubator at 37°C in alpha-modified Eagle's medium (α-MEM; Invitrogen, California, USA) supplemented with 15% fetal bovine serum (FBS; Invitrogen), 2 mmol/L glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen).
BMSCs derived from 18–20-year-old males (n = 3) were obtained from Cyagen Biosciences (Guangzhou, China). Three separate BMSC cultures were grown in a humidified, 5% CO2 incubator at 37°C, in Dulbecco's MEM (Invitrogen), supplemented with 15% FBS (Invitrogen), 2 mmol/L glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). The culture medium was changed every 3 days. All MSCs were used in subsequent experiments after 3–5 passages.
2.2. Microarray Detection
MSCs were grown to 90% confluence; then, the BMSCs (n = 3) and PDLSCs (n = 3) were briefly rinsed with PBS, lysed, and total RNA was isolated with Trizol reagents (Invitrogen). rRNA was removed from total RNA and purified RNA was amplified and transcribed to produce fluorescent cRNA. Reverse transcription was performed along the entire length of the transcripts, without the 3′ bias, with a random priming method. cDNA was labeled and hybridized to the GeneChip Human Gene 2.0 ST Array (Affymetrix), according to the manufacturer's protocol. After hybridization, washing, and staining, the chip was scanned according to the manufacturer's instructions. Microarray experiments were performed at Genminix Informatic Ltd. (Shanghai, China), a microarray service certified by Affymetrix.
2.3. Real-Time RT-PCR Analysis
Real-time, reverse transcription-PCR (RT-PCR) was used to verify the differential expression of genes that were detected on the microarray. Total RNA was isolated from MSCs with Trizol reagents (Invitrogen). For real-time RT-PCR, 2 μg aliquots of RNA as template were combined with random hexamers and reverse transcriptase, according to the manufacturer's protocol (Invitrogen). Real-time PCR reactions were performed with the QuantiTect SYBR Green PCR kit (Qiagen, Germany) and an iCycler iQ Multicolor Real-Time PCR Detection System. The relative level of gene expression was calculated with the 2−ΔΔCT method, as previously described [41]. Primers used for amplifying specific genes are shown in Table 1.
Table 1.
Primer sequences used in the real-time RT-PCR validation of microarray analyses.
| Target gene symbol | Primer sequences (5′-3′) | Target size (bp) | T m (°C) |
|---|---|---|---|
| NR_045555-F | GTTGCAAGGAAACCTTTGGA | 96 | 60 |
| NR_045555-R | CTGCATGCTGTTGACCTTGT | ||
| NR_027621-F | CTGCGTGGATTGCTACAAGA | 102 | 60 |
| NR_027621-R | CCTTCATAGGCCACCACACT | ||
| XR_111050-F | ATGGCCAGTTCGTTTCTCAC | 60 | |
| XR_111050-R | AAGACACGTCCTTGGTTTGG | ||
| NR_037595-F | CCCTGTGCAAGAGCACATAA | 60 | |
| NR_037595-R | TGCCAGCTCATACAAGATGC | ||
| NR_033651-F | CCCCTTGGTATTCTCCCAAT | 60 | |
| NR_033651-R | CAGCCTTTTGTTGGGTGTTT | ||
| NR_037182-F | CTTCTGCAGGAGGAATCCAG | 60 | |
| NR_037182-R | TCCCAGTTTTTGGTGACTCC | ||
| GAPDH-F | CGGACCAATACGACCAAATCCG | 83 | 60 |
| GAPDH-R | AGCCACATCGCTCAGACACC | ||
| HOXA9-F | CGGTTATGGCATTAAACCTGAACCG | 67 | 60 |
| HOXA9-R | GTGAGTGTCAAGCGTGGGACAG | ||
| HOXC8-F | CGGTAAGTTCCAAGGTCTGATACCG | 99 | 60 |
| HOXC8-R | CGTCTCCCAGCCTCATGTTTC | ||
| WNT2B-F | CTTTCCTTTGCACCAGCTTC | 52 | 60 |
| WNT2B-R | TACCCTTCCTCTTGCACACC | ||
| BARX1-F | CGCTTCGAGAAGCAGAAGTA | 111 | 60 |
| BARX1-R | CTTCATCCTCCGATTCTGGT | ||
| IGFBP5-F | GCACCTGAGATGAGACAGGA | 139 | 60 |
| IGFBP5-R | TGTAGAATCCTTTGCGGTCA | ||
| S100A4-F | GTACTTGGTGTCCACCTTCCACAAGTAC | 60 | |
| S100A4-R | CCGGGTCAGCAGCTCCTTTAG |
2.4. Bioinformatics Analysis
Differentially expressed genes were selected with the TwoClassDif method [9, 42, 43]. Gene ontology (GO) analysis was applied to analyze the main functions of differentially expressed genes. Gene ontology is the key functional classification method used at NCBI. GO can organize genes into hierarchical categories and uncover gene regulatory networks on the basis of biological processes and molecular functions [17, 44]. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, significantly changed pathways were identified and connected in a pathway network (Path-net), where connections were based on the relationship between these pathways. This approach was previously used to summarize the pathway interactions among genes that were differentially expressed under the influence of disease, and it revealed why certain pathways were activated [45].
Based on the GO and KEGG pathway analyses, we established an interactions repository (Signal-net) derived from KEGG to show the core genes that played an important role in this MSC gene network [46, 47]. To determine the interactions among genes, we constructed a coding-noncoding gene coexpression network (CNC network), which has also been called a gene coexpression network. This CNC network was based on a correlation analysis that evaluated associations between differentially expressed lncRNAs and mRNAs [45]. We calculated the Pearson correlation for each pair of genes and used the most significantly correlated pairs to construct the network [48]. The purpose of network structure analysis was to locate core regulatory factors (genes). In the network, the core regulatory factors were those connected to large numbers of adjacent genes, and, thus, they exhibited the greatest degrees of connectivity. In considering different networks, we evaluated the core regulatory factors by the degree of difference they showed in their roles in the PDLSC and BMSC networks [49], which was measured with the variable Diffk (difference in normalized connectivities).
2.5. Statistics
All statistical calculations were performed with SPSS10 statistical software. Statistical analyses included comparisons with the t-test, Fisher's exact test, χ 2test, and the Pearson correlation, as appropriate; P values less than 0.05 were considered statistically significant.
3. Results
3.1. Comparison of lncRNA and mRNA Expression Profiles between PDLSCs and BMSCs
To reveal the molecular mechanisms underlying MSCs derived from different tissues, we screened the gene expression patterns in PDLSCs and BMSCs with the human GeneChip microarray method. Because we included only three samples in each group, we applied the RVM t-test, which can effectively raise the degrees of freedom in analyses of small sample sizes to filter the genes that were differentially expressed in PDLSCs and BMSCs. After determining significant differences and the false discovery rate (FDR) in the analysis, the differentially expressed genes were selected according to the P value threshold. Hierarchical clustering showed systematic variations in the expression of lncRNAs and mRNAs between PDLSCs and BMSCs. From the microarray data, a comparison of lncRNA expression levels between PDLSCs and BMSCs identified an average of 970 lncRNAs that were significantly differentially expressed (see Supplementary Table 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2014/317853); of those, 457 were upregulated and 513 were downregulated in the PDLSCs compared to the BMSCs. In addition, a total of 1,578 mRNAs were differentially expressed in the PDLSCs and BMSCs (Supplementary Table 2); of those, 862 were upregulated and 716 were downregulated in the PDLSCs compared to the BMSCs.
To confirm the reliability of the microarray data, we randomly selected six lncRNAs among the 970 differentially expressed lncRNAs and analyzed their expression with real-time RT-PCR. These data confirmed that, compared to BMSCs, PDLSCs showed increased expression of the lncRNAs coded as NR_045555, NR_027621, and NR_033651, and decreased expression of the lncRNAs coded as NR_037182, NR_037595, and XR_111050 (Figure 1). Similarly, we randomly selected six mRNAs among the 1,578 differentially expressed mRNAs and analyzed their expression with real-time RT-PCR. These data confirmed that the mRNAs BARX1, S100A4, WNT2B, and IGFBP5 were increased and that the mRNAs HOXA9 and HOXC8 were decreased in PDLSCs compared to BMSCs (Figure 2). The expression levels of these 12 genes were consistent with the microarray results; thus, these results confirmed the reliability of the microarray data.
Figure 1.
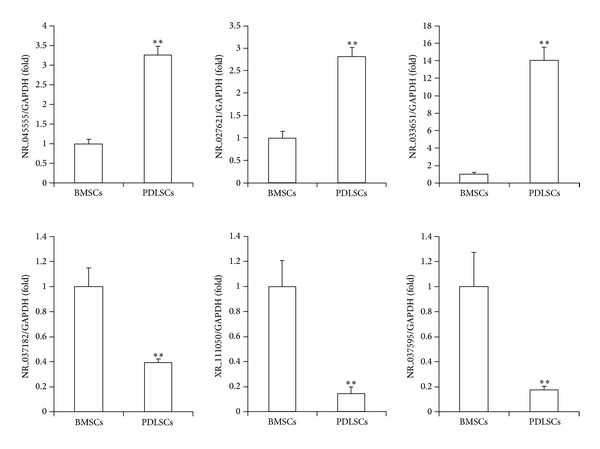
Real-time RT-PCR results show differential lncRNA expression levels in stem cells derived from bone marrow (BMSCs) or periodontal ligament tissue (PDLSCs). The lncRNAs coded as NR_045555, NR_027621, and NR_033651 showed increased expression in PDLSCs, and the lncRNAs coded as NR_037182, XR_111050, and NR_037595 showed decreased expression in PDLSCs compared to BMSCs. GAPDH was used as an internal control. Student's t-test was performed to determine statistical significance; all error bars represent s.d. (n = 3 tissue samples); **P < 0.01.
Figure 2.
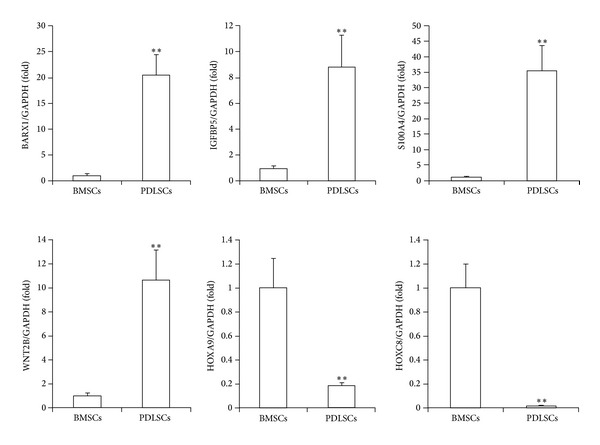
Real-time RT-PCR results show differential mRNA expression levels in stem cells derived from bone marrow (BMSCs) or periodontal ligament tissue (PDLSCs). The mRNAs BARX1, IGFBP5, S100A4, and WNT2B showed increased expression, and the mRNAs HOXA9 and HOXC8 showed decreased expression in PDLSCs compared to BMSCs. GAPDH was used as an internal control. Student's t-test was performed to determine statistical significance; all error bars represent s.d. (n = 3 tissue samples); **P < 0.01.
3.2. Bioinformatics Analysis of BMSC and PDLSC Microarray Data
Next, we performed a bioinformatics analysis to discover the key factors that controlled MSC functions. First, a GO analysis was applied to analyze the main functions of the differentially expressed genes according to gene ontology, which is the key functional classification used by NCBI. According to the threshold, the analysis determined which GOs were significantly differently regulated between PDLSCs and BMSCs with a P value and FDR < 0.05. The negative logarithm of the P value (-LgP) was used to represent the correlation between gene expression and the relevant biological process. The GO analysis identified 166 genes that were significantly upregulated and 104 that were downregulated among all differentially expressed genes in PDLSCs (data not shown). The results clearly showed which important functions were involved with the differentially expressed genes. The top five upregulated GO functions (upGOs) were related to the response to the mitotic cell cycle, the M phase of the mitotic cell cycle, mitotic prometaphase, the cell cycle checkpoint, and mitotic sister chromatid segregation (Supplementary Figure 1). The top five downregulated GO functions (downGOs) were related to the anterior/posterior pattern, embryonic skeletal system morphogenesis, signal transduction, cochlea morphogenesis, and blood vessel remodeling (Supplementary Figure 2).
Based on the KEGG database, we identified the pathways that mediated the functions of the differentially expressed genes. We identified a total of 67 pathways that showed significant differences due to differential gene expression; changes in 31 pathways involved upregulated genes and changes in 36 pathways involved downregulated genes (Supplementary Figures 3 and 4). We performed Path-net analysis to generate an interaction network that included these significantly changed pathways (Figure 3). The top 3 upregulated pathways were apoptosis, MAPK, and cell cycle signaling. The top 3 downregulated pathways were focal adhesion, Wnt, and adherens junction signaling. In addition, cytokine-cytokine receptor interactions and pathways related to cancer were up-/downregulated. These data suggested that these pathways may play key roles in the different core epigenetic mechanisms of PDLSCs and BMSCs.
Figure 3.
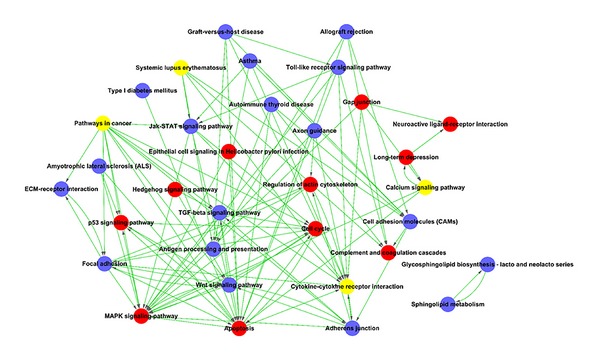
The interaction network of significant pathways (Path-net) in stem cells. Pathways that were significantly different between PDLSCs and BMSCs were connected in a Path-net diagram to show the relationships between these pathways. The role of each pathway in the network was measured by counting its connections to upstream and downstream pathways, known as in-degree (upstream connections), out-degree (downstream connections), or degree (all connections). A high degree pathway indicated that it regulated or was regulated by many other pathways, which implied an important role in the signaling network. The circles represent the pathways; blue represents downregulated pathways, red represents upregulated pathways, and yellow represents up- and downregulated pathways. The lines indicate interactions between pathways.
We performed a Signal-net analysis to further investigate the global network, based on the significantly regulated GOs and pathways. With Signal-net, we screened important candidate genes involved in the differences between PDLSCs and BMSCs (Figure 4). In the Signal-net analysis, the genes are characterized by measuring their “betweenness centrality,” the number of times a node is located in the shortest path between 2 other nodes. This measure reflects the importance of a node in a graphic network relative to other nodes. The four most important differentially expressed genes were identified in the network (Supplementary Table 3); these were phospholipase C beta 4 (PLCβ4), filamin B beta (FLNB), calcium/calmodulin-dependent protein kinase II gamma (CAMK2G), and the ionotropic glutamate receptor, AMPA 1 (GRIA1).
Figure 4.
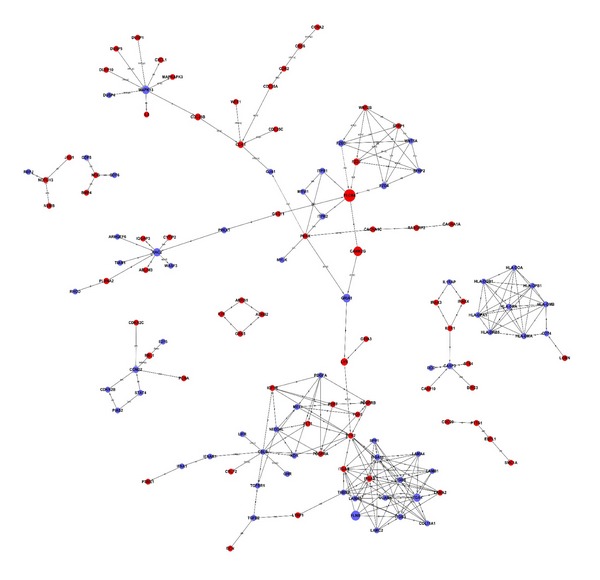
The interaction network of differentially expressed genes (Signal-net). The circles represent important functional genes in PDLSCs (red: upregulated genes; blue: downregulated genes); the circle size represents the degree of interaction (betweenness centrality), and lines indicate the interactions.
Finally, we used a coding-noncoding gene coexpression (CNC) network to evaluate the interactions among genes and identify the core regulatory genes in the network. Based on our previous results, we built CNC networks to identify the interactions among the differentially expressed lncRNAs and mRNAs in PDLSCs and BMSCs [45]. We used 65 lncRNAs and 208 mRNAs to build the CNC network for PDLSCs and 75 lncRNAs and 187 mRNAs to build the network for BMSCs. In the CNC networks, each mRNA could correlate with one to tens of lncRNAs and vice versa. We used the CNC networks to implicate the interregulation of lncRNAs and mRNAs in the different molecular mechanisms of PDLSCs and BMSCs (Supplementary Figures 5 and 6). In the CNC network of PDLSCs, 17 genes showed a degree ≥ 59 and a clustering coefficient ≥ 0.6. This indicated that these genes, including 4 lncRNAs and 13 mRNAs (Table 2), played important roles in the network. In the CNC network of BMSCs, 20 mRNAs showed a degree ≥ 29 and a clustering coefficient ≥ 0.7. This indicated that (Table 3) these genes played important roles in the network. According to the Diffk values (|Diffk| ≥ 0.75) for these networks, 16 genes (Table 4), including 2 lncRNAs and 14 mRNAs, showed different connectivities between PDLSCs and BMSCs, indicating that their roles were different in core pathways that governed MSC functions. The top three mRNAs were chemokine (C-X-C motif) ligand 12 (CXCL12), integrin alpha 2 (ITGA2, CD49B), and cell division cycle 20 homolog (CDC20), which were upregulated. The two lncRNAs identified (|Diffk| ≥ 0.75) were FR020479 and FR191603; the former was downregulated and the latter was upregulated.
Table 2.
Seventeen genes identified in the PDLSC CNC network with high degrees of connectivity and clustering coefficients (degree ≥59, clustering coefficient ≥0.6).
| Gene symbol | Description | Clustering coefficient | Degree | Style | Type |
|---|---|---|---|---|---|
| FLNB | Filamin B, beta | 0.67332309 | 63 | Down | mRNA |
| PTTG1 | Pituitary tumor-transforming factor-1 | 0.72021858 | 61 | Up | mRNA |
| GNG11 | Guanine nucleotide binding protein (G protein), gamma 11 | 0.72021858 | 61 | Up | mRNA |
| IGF1R | Insulin-like growth factor-1 receptor | 0.67431694 | 61 | Up | mRNA |
| ITGA2 | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 0.66994536 | 61 | Up | mRNA |
| ENPP1 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | 0.73107345 | 60 | Down | mRNA |
| CDC20 | Cell division cycle 20 homolog (S. cerevisiae) | 0.70734463 | 60 | Up | mRNA |
| COL11A1 | Collagen, type XI, alpha 1 | 0.70734463 | 60 | Down | mRNA |
| DBF4 | DBF4 homolog (S. cerevisiae) | 0.70734463 | 60 | Up | mRNA |
| NR_040093 | gi∣338968843∣ref∣NR_040093.1∣ | 0.76446523 | 59 | Down | lncRNA |
| XR_112964 | gi∣310115154∣ref∣XR_112964.1∣ | 0.74868498 | 59 | Down | lncRNA |
| XR_108725 | gi∣310119896∣ref∣XR_108725.1∣ | 0.74868498 | 59 | Down | lncRNA |
| XR_110624 | gi∣310118206∣ref∣XR_110624.1∣ | 0.74868498 | 59 | Down | lncRNA |
| CCNB2 | Cyclin B2 | 0.68322618 | 59 | Up | mRNA |
| GSTM5 | Glutathione S-transferase mu 5 | 0.68322618 | 59 | Up | mRNA |
| HLA-DMA | Major histocompatibility complex, class II, DM alpha | 0.68322618 | 59 | Down | mRNA |
| WASF3 | WAS protein family, member 3 | 0.68264173 | 59 | Down | mRNA |
Table 3.
Twenty genes identified in the BMSC CNC network with high degrees of connectivity and clustering coefficients (degree ≥29, clustering coefficient ≥0.7).
| Gene symbol | Description | Clustering coefficient | Degree | Style | Type |
|---|---|---|---|---|---|
| CXCL12 | Chemokine (C-X-C motif) ligand 12 | 0.75568182 | 33 | Up | mRNA |
| PRIM1 | Primase, DNA, polypeptide 1 (49 kDa) | 0.87931034 | 29 | Up | mRNA |
| LIFR | Leukemia inhibitory factor receptor alpha | 0.87931034 | 29 | Down | mRNA |
| MAD2L1 | MAD2 mitotic arrest deficient-like 1 (yeast) | 0.87931034 | 29 | Up | mRNA |
| TGFBR1 | Transforming growth factor, beta receptor 1 | 0.87931034 | 29 | Down | mRNA |
| PARP1 | Poly(ADP-ribose) polymerase 1 | 0.87931034 | 29 | Up | mRNA |
| FGF5 | Fibroblast growth factor-5 | 0.87931034 | 29 | Up | mRNA |
| CCNE2 | Cyclin E2 | 0.87931034 | 29 | Up | mRNA |
| TTK | TTK protein kinase | 0.87931034 | 29 | Up | mRNA |
| RBL1 | Retinoblastoma-like 1 (p107) | 0.87931034 | 29 | Up | mRNA |
| POLE2 | Polymerase (DNA directed), epsilon 2 (p59 subunit) | 0.87931034 | 29 | Up | mRNA |
| CDK1 | Cyclin-dependent kinase 1 | 0.87931034 | 29 | Up | mRNA |
| MCM3 | Minichromosome maintenance complex component 3 | 0.87931034 | 29 | Up | mRNA |
| CDK2 | Cyclin-dependent kinase 2 | 0.87931034 | 29 | Up | mRNA |
| BMPR1B | Bone morphogenetic protein receptor, type IB | 0.87931034 | 29 | Down | mRNA |
| HIST1H2BO | Histone cluster 1, H2bo | 0.80295567 | 29 | Up | mRNA |
| F10 | Coagulation factor X | 0.80295567 | 29 | Up | mRNA |
| BDKRB1 | Bradykinin receptor B1 | 0.80295567 | 29 | Up | mRNA |
| GSTM5 | Glutathione S-transferase mu 5 | 0.80295567 | 29 | Up | mRNA |
| PRPH2 | Peripherin 2 (retinal degeneration, slow) | 0.80295567 | 29 | Down | mRNA |
Table 4.
Sixteen genes with different pathway connectivities (identified with Diffk) in PDLSCs and BMSCs (∣Diffk∣ ≥ 0.75).
| Gene symbol | Description | Style | Type | ∣Diffk∣ |
|---|---|---|---|---|
| CXCL12 | Chemokine (C-X-C motif) ligand 12 | Up | mRNA | 1 |
| ITGA2 | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | Up | mRNA | 0.968254 |
| CDC20 | Cell division cycle 20 homolog (S. cerevisiae) | Up | mRNA | 0.952381 |
| WASF3 | WAS protein family, member 3 | Down | mRNA | 0.9365079 |
| CAMK4 | Calcium/calmodulin-dependent protein kinase IV | Up | mRNA | 0.8571429 |
| SEMA3C | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C | Down | mRNA | 0.8571429 |
| CCNA2 | Cyclin A2 | Up | mRNA | 0.8253968 |
| POLA1 | Polymerase (DNA directed), alpha 1, catalytic subunit | Up | mRNA | 0.8253968 |
| FR020479 | AB209345, AC006512, U47924 | Down | lncRNA | 0.7878788 |
| FR191603 | AJ609445, AK128061, AP001273 | Up | lncRNA | 0.7532468 |
| SLK | STE20-like kinase | Up | mRNA | 0.7460317 |
| PDGFA | Platelet-derived growth factor alpha polypeptide | Down | mRNA | 0.7388167 |
| PTK2 | PTK2 protein tyrosine kinase 2 | Up | mRNA | 0.7142857 |
| ENPP1 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | Down | mRNA | 0.7099567 |
| PRKCE | Protein kinase C, epsilon | Up | mRNA | 0.7056277 |
| BDKRB1 | Bradykinin receptor B1 | Up | mRNA | 0.7041847 |
4. Discussion
The presence of different MSCs in dental and craniofacial tissues has encouraged clinical studies to investigate tissue regeneration in orofacial and periodontal regions [50, 51]. In the past few decades, MSC-mediated tissue regeneration has made surprising progress [25, 26, 52]. However, bone marrow has remained the principal source of MSCs for most preclinical and clinical applications. Interestingly, the MSCs from different anatomic locations possess diverse biological activities [8–12]. The challenge lies in identifying the specific genes that are associated with distinct MSC functions. To that end, in the present study, we identified lncRNAs and mRNAs that were differentially expressed in PDLSCs and BMSCs.
We identified 970 differentially expressed lncRNAs and 1,578 differentially expressed mRNAs in BMSCs and dental tissue-derived MSCs. This information may be useful for further studies on gene functions and regulation mechanisms in MSCs. Furthermore, we found that several of the upregulated genes in PDLSCs may be associated with PDLSC characteristics. For instance, BARX1, a transcription factor expressed in the mesenchyme of molar primordia, is involved in the regulation of tooth morphogenesis, in the development of tooth and craniofacial mesenchyme that originates from the neural crest [53–55], and possibly, in the regulation of MSC differentiation.
To identify the key factors that regulated MSC functions, we applied bioinformatics analyses to classify the microarray data. The GO analysis revealed specific functional pathways that were enriched in the genes responsible for the divergent features of PDLSCs and BMSCs. These differentially expressed genes were subsequently organized into hierarchical categories based on pertinent biological processes. A high degree pathway interacted with a high number of other pathways, which implied an important role in cell biological features. Further pathway analyses indicated that apoptosis, MAPK, cytokine-cytokine receptor interaction, focal adhesion, pathways in cancer, Wnt, cell cycle, and adherens junctions signaling pathways were involved in the diverse biological activities of PDLSCs and BMSCs. It is well known that these pathways play an important role in regulating cellular apoptosis, survival, and differentiation.
To identify important genes involved in the different epigenetic mechanisms of PDLSCs and BMSCs, we performed Signal-net analysis on the significantly regulated GOs and pathways. This analysis revealed that PLCβ4, FLNB, CAMK2G, and GRIA1 exhibited the most betweenness centrality. PLCβ4 and CAMK2G were upregulated in PDLSCs. It was reported that PLCβ4 was highly expressed in the retina and the cerebellum, where calcium plays an important role in the transduction of extracellular signals [56–58]. Moreover, CAMK2G is activated by intracellular calcium/calmodulin [59]. Thus, the Signal-net analysis results suggested that these genes were important in calcium-sensitive signaling cascades that regulate cell function. In addition, FLNB regulates intracellular communication and signaling by linking the protein actin to the cell membrane. This activity allows direct communication between the cell membrane and the cytoskeletal network, which provides a means to control and guide proper skeletal development [60, 61].
The CNC network comparisons indicated that CXCL12 was a core regulatory factor, which may be involved in the diverse biological activities of PDLSCs and BMSCs. CXCL12, also known as stromal cell-derived factor-1, stimulates migration by rearranging the actin cytoskeleton, increasing focal adhesion, and stimulating matrix metalloproteinase production in MSCs [62, 63]. Thus, CXCL12 can recruit MSC to participate in the regeneration of injured tissues [64]. Presumably, MSC migration is mediated through an intracellular pathway, for example, the MAPK/ERK signaling pathways [62]. Our results were consistent with previous reports and may also be applicable to the differentiation mechanisms previously described in MSCs.
Additionally, we identified some lncRNAs that were differentially expressed in PDLSCs and BMSCs, for example, FR020479 and FR191603. Previous studies demonstrated that lncRNAs may function by controlling the transcriptional regulation of neighboring coding genes [65, 66]. Identifying differentially expressed nearby coding mRNAs may enhance our understanding of the function of lncRNAs in MSCs. However, further studies must be performed to investigate that hypothesis.
5. Conclusion
This study provided comprehensive profiles of mRNA and lncRNA expression in PDLSCs and BMSCs, two tissue-derived MSCs. In addition, potential regulatory mechanisms were identified with bioinformatics analyses. Although more studies are required to demonstrate the precise role and mechanisms of these lncRNAs and mRNAs, the genomic data we identified with microarray analyses may increase our understanding of MSC biology.
Supplementary Material
Expression profiles of lncRNA and mRNA between bone marrow stem cells (BMSCs) and periodontal ligament stem cells (PDLSCs) were investigated with lncRNA microarray assays and bioinformatics analysis. In PDLSCs, 970 lncRNAs that were significantly differentially expressed compared to BMSCs (Supplementary Table 1). Furthermore, 1,578 mRNAs were differentially expressed in the PDLSCs and BMSCs (Supplementary Table 2). For bioinformatics analysis, the results of the GO analysis showed which important functions were involved with the differentially expressed genes, including the top upregulated and downregulated GO functions (upGOs and downGOs) (Supplementary Figures 1 and 2). Then based on the KEGG database, there were identified 67 pathways that showed significant differences due to differential gene expression (Supplementary Figures 3 and 4), which play key roles in the different core epigenetic mechanisms of PDLSCs and BMSCs. To further investigate the global network, differentially expressed genes were identified by a Signal-net analysis (Supplementary Table 3). Finally, coding-noncoding gene coexpression (CNC) networks were used to implicate the interregulation of lncRNAs and mRNAs in the different molecular mechanisms of PDLSCs and BMSCs (Supplementary Figures 5 and 6).
Acknowledgments
This work was supported by Grants from the National Natural Science Foundation of China (81271101 to Zhipeng Fan, 81271100 to Rui Dong, and 81170931 to Juan Du), the Program for New Century Excellent Talents in the University (NCET-12-0611 to Zhipeng Fan), High-Level Talents of the Beijing Health System (2013-3-035 to Zhipeng Fan and 2013-3-034 to Rui Dong), the Beijing Natural Science Foundation (7112057 to Rui Dong), and the Beijing Nova Program (2011083 to Rui Dong).
Conflict of Interests
The authors declare that there is no potential conflict of interests.
References
- 1.Cai J, Weiss ML, Rao MS. In search of “stemness’. Experimental Hematology. 2004;32(7):585–598. doi: 10.1016/j.exphem.2004.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 2004;116(5):639–648. doi: 10.1016/s0092-8674(04)00208-9. [DOI] [PubMed] [Google Scholar]
- 3.Morsczeck C, Schmalz G, Reichert TE, Völlner F, Galler K, Driemel O. Somatic stem cells for regenerative dentistry. Clinical Oral Investigations. 2008;12(2):113–118. doi: 10.1007/s00784-007-0170-8. [DOI] [PubMed] [Google Scholar]
- 4.Mauron A, Jaconi ME. Stem cell science: current ethical and policy issues. Clinical Pharmacology and Therapeutics. 2007;82(3):330–333. doi: 10.1038/sj.clpt.6100295. [DOI] [PubMed] [Google Scholar]
- 5.Jahagirdar BN, Verfaillie CM. Multipotent adult progenitor cell and stem cell plasticity. Stem Cell Reviews. 2005;1(1):53–59. doi: 10.1385/SCR:1:1:053. [DOI] [PubMed] [Google Scholar]
- 6.Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair—current views. Stem Cells. 2007;25(11):2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 7.Feng J, Mantesso A, de Bari C, Nishiyama A, Sharp PT. Dual origin of mesenchymal stem cells contributing to organ growth and repair. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(16):6503–6508. doi: 10.1073/pnas.1015449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Gao LN, An Y, et al. Comparison of mesenchymal stem cells derived from gingival tissue and periodontal ligament in different incubation conditions. Biomaterials. 2013;34(29):7033–7047. doi: 10.1016/j.biomaterials.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Yu S, Long J, Yu J, et al. Analysis of differentiation potentials and gene expression profiles of mesenchymal stem cells derived from periodontal ligament and Wharton’s jelly of the umbilical cord. Cells Tissues Organs. 2013;197(3):209–223. doi: 10.1159/000343740. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi O, Katsube Y, Hirose M, Ohgushi H, Ito H. Comparison of osteogenic ability of rat mesenchymal stem cells from bone marrow, periosteum, and adipose tissue. Calcified Tissue International. 2008;82(3):238–247. doi: 10.1007/s00223-008-9112-y. [DOI] [PubMed] [Google Scholar]
- 11.Sakaguchi Y, Sekiya I, Yagishita K, Muneta T. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis and Rheumatism. 2005;52(8):2521–2529. doi: 10.1002/art.21212. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Li C-D, Jiang X-X, Li H-L, Tang P-H, Mao N. Comparison of mesenchymal stem cells from human placenta and bone marrow. Chinese Medical Journal. 2004;117(6):882–887. [PubMed] [Google Scholar]
- 13.Caplan AI. Mesenchymal stem cells: cell-based reconstructive therapy in orthopedics. Tissue Engineering. 2005;11(7-8):1198–1211. doi: 10.1089/ten.2005.11.1198. [DOI] [PubMed] [Google Scholar]
- 14.Bruder SP, Kraus KH, Goldberg VM, Kadiyala S. The effect of implants loaded with autologous mesenchymal stem cells on the healing of canine segmental bone defects. Journal of Bone and Joint Surgery A. 1998;80(7):985–996. doi: 10.2106/00004623-199807000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Lee YM, Seol YJ, Lim YT, et al. Tissue-engineered growth of bone by marrow cell transplantation using porous calcium metaphosphate matrices. Journal of Biomedical Materials Research. 2001;54:216–223. doi: 10.1002/1097-4636(200102)54:2<216::aid-jbm8>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi H, Hirachi A, Hasegawa N, et al. Enhancement of periodontal tissue regeneration by transplantation of bone marrow mesenchymal stem cells. Journal of Periodontology. 2004;75(9):1281–1287. doi: 10.1902/jop.2004.75.9.1281. [DOI] [PubMed] [Google Scholar]
- 17.Hu B, Unda F, Bopp-Kuchler S, et al. Bone marrow cells can give rise to ameloblast-like cells. Journal of Dental Research. 2006;85(5):416–421. doi: 10.1177/154405910608500504. [DOI] [PubMed] [Google Scholar]
- 18.Jing W, Wu L, Lin Y, Liu L, Tang W, Tian W. Odontogenic differentiation of adipose-derived stem cells for tooth regeneration: necessity, possibility, and strategy. Medical Hypotheses. 2008;70(3):540–542. doi: 10.1016/j.mehy.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(25):13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura M, Gronthos S, Zhao M, et al. SHED: stem cells from human exfoliated deciduous teeth. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):5807–5812. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo B-M, Miura M, Gronthos S, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. The Lancet. 2004;364(9429):149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 22.Sonoyama W, Liu Y, Yamaza T, et al. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. Journal of Endodontics. 2008;34(2):166–171. doi: 10.1016/j.joen.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.d’Aquino R, Graziano A, Sampaolesi M, et al. Human postnatal dental pulp cells co-differentiate into osteoblasts and endotheliocytes: a pivotal synergy leading to adult bone tissue formation. Cell Death and Differentiation. 2007;14(6):1162–1171. doi: 10.1038/sj.cdd.4402121. [DOI] [PubMed] [Google Scholar]
- 24.Laino G, D’Aquino R, Graziano A, et al. A new population of human adult dental pulp stem cells: a useful source of living autologous fibrous bone tissue (LAB) Journal of Bone and Mineral Research. 2005;20(8):1394–1402. doi: 10.1359/JBMR.050325. [DOI] [PubMed] [Google Scholar]
- 25.Ding G, Liu Y, Wang W, et al. Allogeneic periodontal ligament stem cell therapy for periodontitis in swine. Stem Cells. 2010;28(10):1829–1838. doi: 10.1002/stem.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Zheng Y, Ding G, et al. Periodontal ligament stem cell-mediated treatment for periodontitis in miniature swine. Stem Cells. 2008;26(4):1065–1073. doi: 10.1634/stemcells.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batouli S, Miura M, Brahim J, et al. Comparison of stem-cell-mediated osteogenesis and dentinogenesis. Journal of Dental Research. 2003;82(12):976–981. doi: 10.1177/154405910308201208. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Walboomers XF, Shi S, Fan M, Jansen JA. Multilineage differentiation potential of stem cells derived from human dental pulp after cryopreservation. Tissue Engineering. 2006;12(10):2813–2823. doi: 10.1089/ten.2006.12.2813. [DOI] [PubMed] [Google Scholar]
- 29.Kim S-H, Kim K-H, Seo B-M, et al. Alveolar bone regeneration by transplantation of periodontal ligament stem cells and bone marrow stem cells in a canine peri-implant defect model: a pilot study. Journal of Periodontology. 2009;80(11):1815–1823. doi: 10.1902/jop.2009.090249. [DOI] [PubMed] [Google Scholar]
- 30.Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nature Genetics. 1999;21(1):10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 31.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nature Genetics. 1999;21(1):33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 32.Shi S, Robey PG, Gronthos S. Comparison of human dental pulp and bone marrow stromal stem cells by cDNA microarray analysis. Bone. 2001;29(6):532–539. doi: 10.1016/s8756-3282(01)00612-3. [DOI] [PubMed] [Google Scholar]
- 33.Inomata N, Tomita H, Ikezawa Z, Saito H. Differential gene expression profile between cord blood progenitor-derived and adult progenitor-derived human mast cells. Immunology Letters. 2005;98(2):265–271. doi: 10.1016/j.imlet.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Schilling T, Küffner R, Klein-Hitpass L, Zimmer R, Jakob F, Schütze N. Microarray analyses of transdifferentiated mesenchymal stem cells. Journal of Cellular Biochemistry. 2008;103(2):413–433. doi: 10.1002/jcb.21415. [DOI] [PubMed] [Google Scholar]
- 35.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosal S, Das S, Chakrabarti J. Long noncoding RNAs: new players in the molecular mechanism for maintenance and differentiation of pluripotent stem cells. Stem Cells and Development. 2013;22(16):2240–2253. doi: 10.1089/scd.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Reports. 2012;13(11):971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes and Development. 2012;26(4):338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bustin SA. Absolute quantification of mrna using real-time reverse transcription polymerase chain reaction assays. Journal of Molecular Endocrinology. 2000;25(2):169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 42.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19(18):2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 43.Clarke R, Ressom HW, Wang A, et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nature Reviews Cancer. 2008;8(1):37–49. doi: 10.1038/nrc2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pujana MA, Han J-DJ, Starita LM, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nature Genetics. 2007;39(11):1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 46.Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24(9):1175–1182. doi: 10.1093/bioinformatics/btn081. [DOI] [PubMed] [Google Scholar]
- 47.Zhang JD, Wiemann S. KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25(11):1470–1471. doi: 10.1093/bioinformatics/btp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prieto C, Risueño A, Fontanillo C, de Las Rivas J. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS ONE. 2008;3(12) doi: 10.1371/journal.pone.0003911.e3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson MRJ, Zhang B, Fang Z, Mischel PS, Horvath S, Nelson SF. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics. 2006;7, article 40 doi: 10.1186/1471-2164-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silvério KG, Benatti BB, Casati MZ, Sallum EA, Nociti FH., Jr. Stem cells: potential therapeutics for periodontal regeneration. Stem Cell Reviews. 2008;4(1):13–19. doi: 10.1007/s12015-008-9011-7. [DOI] [PubMed] [Google Scholar]
- 51.Rimondini L, Mele S. Stem cell technologies for tissue regeneration in dentistry. Minerva Stomatologica. 2009;58(10):483–500. [PubMed] [Google Scholar]
- 52.Giuliani A, Manescu A, Langer M, et al. Three years after transplants in human mandibles, histological and in-line holotomography revealed that stem cells regenerated a compact rather than a spongy bone: biological and clinical implications. Stem Cells Translational Medicine. 2013;2(4):316–324. doi: 10.5966/sctm.2012-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sperber SM, Dawid IB. barx1 is necessary for ectomesenchyme proliferation and osteochondroprogenitor condensation in the zebrafish pharyngeal arches. Developmental Biology. 2008;321(1):101–110. doi: 10.1016/j.ydbio.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miletich I, Buchner G, Sharpe PT. Barx1 and evolutionary changes in feeding. Journal of Anatomy. 2005;207(5):619–622. doi: 10.1111/j.1469-7580.2005.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitsiadis TA, Mucchielli M-L, Raffo S, Proust J-P, Koopman P, Goridis C. Expression of the transcription factors Otlx2, Barx1 and Sox9 during mouse odontogenesis. European Journal of Oral Sciences. 1998;106(supplement 1):112–116. doi: 10.1111/j.1600-0722.1998.tb02161.x. [DOI] [PubMed] [Google Scholar]
- 56.Taylor CW. Controlling calcium entry. Cell. 2002;111(6):767–769. doi: 10.1016/s0092-8674(02)01197-2. [DOI] [PubMed] [Google Scholar]
- 57.Kido Y, Gordon CT, Sakazume S, et al. Further characterization of atypical features in auriculocondylar syndrome caused by recessive PLCB4 mutations. American Journal of Medical Genetics A. 2013;161(9):2339–2346. doi: 10.1002/ajmg.a.36066. [DOI] [PubMed] [Google Scholar]
- 58.Lyon AM, Tesmer JJ. Structural insights into phospholipase C-beta function. Molecular Pharmacology. 2013;84(4):488–500. doi: 10.1124/mol.113.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mattingsdal M, Brown AA, Djurovic S, et al. Pathway analysis of genetic markers associated with a functional MRI faces paradigm implicates polymorphisms in calcium responsive pathways. Neuroimage. 2013;70:143–149. doi: 10.1016/j.neuroimage.2012.12.035. [DOI] [PubMed] [Google Scholar]
- 60.Lu J, Lian G, Lenkinski R, et al. Filamin B mutations cause chondrocyte defects in skeletal development. Human Molecular Genetics. 2007;16(14):1661–1675. doi: 10.1093/hmg/ddm114. [DOI] [PubMed] [Google Scholar]
- 61.Stossel TP, Condeelis J, Cooley L, et al. Filamins as integrators of cell mechanics and signalling. Nature Reviews Molecular Cell Biology. 2001;2(2):138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 62.Gao H, Priebe W, Glod J, Banerjee D. Activation of signal transducers and activators of transcription 3 and focal adhesion kinase by stromal cell-derived factor 1 is required for migration of human mesenchymal stem cells in response to tumor cell-conditioned medium. Stem Cells. 2009;27(4):857–865. doi: 10.1002/stem.23. [DOI] [PubMed] [Google Scholar]
- 63.Hsieh JY, Huang TS, Cheng SM, et al. miR-146a-5p circuitry uncouples cell proliferation and migration, but not differentiation, in human mesenchymal stem cells. Nucleic Acids Research. 2013;41(21):9753–9763. doi: 10.1093/nar/gkt666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ponomaryov T, Peled A, Petit I, et al. Induction of the chemokine stromal-derived factor-1 following DNA damage improves human stem cell function. Journal of Clinical Investigation. 2000;106(11):1331–1339. doi: 10.1172/JCI10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biology. 2010;7(5):582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma H, Hao Y, Dong X, et al. Molecular mechanisms and function prediction of long noncoding RNA. TheScientificWorldJOURNAL. 2012;2012:11 pages. doi: 10.1100/2012/541786.541786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profiles of lncRNA and mRNA between bone marrow stem cells (BMSCs) and periodontal ligament stem cells (PDLSCs) were investigated with lncRNA microarray assays and bioinformatics analysis. In PDLSCs, 970 lncRNAs that were significantly differentially expressed compared to BMSCs (Supplementary Table 1). Furthermore, 1,578 mRNAs were differentially expressed in the PDLSCs and BMSCs (Supplementary Table 2). For bioinformatics analysis, the results of the GO analysis showed which important functions were involved with the differentially expressed genes, including the top upregulated and downregulated GO functions (upGOs and downGOs) (Supplementary Figures 1 and 2). Then based on the KEGG database, there were identified 67 pathways that showed significant differences due to differential gene expression (Supplementary Figures 3 and 4), which play key roles in the different core epigenetic mechanisms of PDLSCs and BMSCs. To further investigate the global network, differentially expressed genes were identified by a Signal-net analysis (Supplementary Table 3). Finally, coding-noncoding gene coexpression (CNC) networks were used to implicate the interregulation of lncRNAs and mRNAs in the different molecular mechanisms of PDLSCs and BMSCs (Supplementary Figures 5 and 6).


