Abstract
The nucleoside analog 5-aza-2′-deoxycytidine (Decitabine, DAC) is one of several drugs in clinical use that inhibit DNA methyltransferases, leading to a decrease of 5-methylcytosine in newly replicated DNA and subsequent transcriptional activation of genes silenced by cytosine methylation. In addition to methyltransferase inhibition, DAC has demonstrated toxicity and potential mutagenicity, and can induce a DNA-repair response. The mechanisms accounting for these events are not well understood. DAC is chemically unstable in aqueous solutions, but there is little consensus between previous reports as to its half-life and corresponding products of decomposition at physiological temperature and pH, potentially confounding studies on its mechanism of action and long-term use in humans. Here we have employed a battery of analytical methods to estimate kinetic rates and to characterize DAC decomposition products under conditions of physiological temperature and pH. Our results indicate that DAC decomposes into a plethora of products, formed by hydrolytic opening and deformylation of the triazine ring, in addition to anomerization and possibly other changes in the sugar ring structure. We also discuss the advantages and problems associated with each analytical method used. The results reported here will facilitate ongoing studies and clinical trials aimed at understanding the mechanisms of action, toxicity, and possible mutagenicity of DAC and related analogs.
Introduction
DAC1 (5-aza-2′-deoxycytidine, Decitabine) is a nucleoside analog that is converted intracellularly to the corresponding 5′-triphosphate and serves as a substrate for DNA replication (1, 2). The most pharmacologically interesting property of DAC is its ability to form a complex with DNA-dependent cytosine methyltransferases (3), resulting in methyltransferase inhibition and decreased 5-methylcytosine (5mC) in newly replicated DNA (4). The inhibition of the methyltransferase with subsequent decreased 5mC content results in loosening of the local chromatin structure and transcriptional activation of genes silenced by cytosine methylation (5, 6). DAC is currently used clinically for the treatment of myelodysplastic syndromes (7) and is in clinical trial for other human cancers (8-10) and sickle cell anemia (11). The primary mechanism for the activity of DAC is believed to be methyltransferase inhibition (4), although DAC has demonstrated toxicity (12) and some reports suggest that it could be mutagenic (13). DAC is also known to induce upregulation of DNA repair genes, including double-strand break repair proteins (14) and p53 (15). It has been suggested that the cytotoxicity and DNA repair-inducing properties of DAC result from the DNA-enzyme suicide complex formed when DAC in DNA covalently traps the methyltransferase (12). However, a recent study indicates that DAC may induce degradation of methyltransferases without covalent bond formation between DAC-containing DNA and the enzyme, and in the absence of DNA replication (16), leading to the question of whether DAC or its decomposition products may have biological activity even without being incorporated into DNA.
One of the primary challenges with DAC is its chemical instability. It is known from several studies that DAC and related nucleoside analogs such as azaCyd (5-azacytidine) decompose within hours at physiological temperature and pH (17-22). According to previous reports, the degradation products of DAC result from hydrolytic opening of the triazine ring, deformylation, and anomerization (17, 23). Not only does the decomposition result in reduced drug concentration, but the degradation products themselves likely have pharmacological and toxic properties independent of the parent compound. It is as yet unknown how the degradation products of DAC might account for its toxicity, potential mutagenicity, and DNA repair-inducing capacity.
The decomposition of DAC has been studied previously, but there is little consensus about the identification of the degradation products (17, 18, 23), and there is a wide range of values reported for the half-life of DAC (3.5 to 21 h) under conditions of physiological temperature and neutral pH (17-21). Each of the previous studies focused on a particular degradation pathway or applied a specific analytical method. In this paper, we present results on DAC degradation obtained with a battery of methods including gas chromatograpy/mass spectrometry, UV spectrophotometry, NMR spectroscopy, and HPLC/UV/mass spectrometry. These results provide a substantially more comprehensive examination of DAC decomposition specifically under conditions of physiological temperature and pH. Using our results, we are able to resolve disagreements between previous studies, and we indicate the advantages and problems associated with the various methods we and others have applied. The results presented here will facilitate ongoing pharmacological studies and clinical trials aimed at understanding the mechanisms of action, toxicity and potential mutagenicity of DAC and related analogs.
Experimental Procedures
Synthesis of compounds
The α- and β-anomers of DAC were synthesized according to the synthetic strategy of Liu et al (24) with modifications. Briefly, 2′-deoxyuridine was converted to 3′-5′-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-2′-deoxyuridine and transglycosylation was performed with the trimethylsilylated derivative of azaC in the presence of the catalyst trimethylsilyl trifluoromethanesulfonate to yield the α- and β-anomers of 3′,5′-O-(1,1,3,3-tetraisopropyldisiloxane-1,3-diyl)-5-aza-2′-deoxycytidine. These protected anomers were separated and purified by open column silica gel chromatography, and deprotected by incubation with tetrabutylammonium fluoride in THF. After deprotection, compounds were further purified by silica gel flash chromatography and recrystallization.
The β-anomer of DAC (β-DAC) was > 96% pure by HPLC, and was analyzed by 1H NMR (d6-DMSO): δ 8.50 (s, 1, H-6), 7.49 (d, 2, NH2), 6.03 (t, 1, H-1′), 5.23 (d, 1, OH-3′), 5.03 (t, 1, OH-5′), 4.23 (m, 1, H-3′), 3.81 (m, 1, H-4′), 3.57 (m, 2, H-5′, H-5″), 2.17 (m, 2, H-2′, H-2″). The α-anomer of DAC was ∼ 80% pure by HPLC (contained some β-anomer), and was analyzed by 1H NMR (d6-DMSO): δ 8.30 (s, 1, H-6), 7.41 (d, 2, NH2), 5.97 (dd, 1, H-1′), 5.24 (s, 1, OH-3′), 4.88 (s, 1, OH-5′), 4.23 (m, 2, H-3′, H-4′), 3.28 (m, H-5′, H-5″, partially underneath H2O peak), 2.48 (m, H-2′, H-2″, partially underneath DMSO peak). Chemical shifts for both anomers compared well with previous literature values (25). The α-anomer of DAC (α-DAC) was HPLC-purified using C-18 stationary phase and isocratic 5% methanol/H2O mobile phase prior to its use in decomposition experiments.
Stable-isotope-labeled [6-13C]-5-azacytosine (13C-azaC) was synthesized according to the method of Hartenstein and Fridovich (26). The compound was HPLC-purified and was > 94% pure by HPLC. Isotopic purity was 99% by GC/MS.
Decomposition of β-DAC measured by GC/MS
A solution (9.2 mM) of β-DAC was prepared in 100 mM potassium phosphate buffer (pH 7.4). The solution was heated at 37 °C for 24 h and aliquots (10 μL) were removed at selected time intervals and immediately frozen at -80 °C until all aliquots were taken. Samples were thawed, diluted 10-fold in water, and 10 μL (9.2 × 10-9 mol) were added to 5.5 × 10-9 mol stable-isotope labeled 13C-azaC internal standard, after which samples were dried by vacuum centrifugation for ∼ 20 min. The time from aliquot thawing to immediately before drying was < 5 min per sample, in order to minimize DAC degradation during sample processing. Next, 200 μL formic acid (88%) were added to each vial, vials were capped, and heated at 100 °C for 30 min in order to hydrolyze the glycosidic bond. Vials were uncapped and formic acid evaporated by vacuum centrifugation, after which samples were derivatized to the TBDMS derivatives of azaC using the following conditions: 100 μL dry acetonitrile and 100 μL MTBSTFA + 1% TBDMCS added to each vial, vials capped and heated at 140 °C for 30 min. Samples (1 μL) were injected splitless onto a 6890 GC coupled with a 5973 inert-MSD and a 7683B series autoinjector (Agilent Technologies, Wilmington, DE) using electron impact (EI) mode, analyzed by scanning over a range of 35-450 m/z. Injector and interface temperatures were 250 °C and 280 °C, respectively. Oven conditions were as follows: 100 °C for 2 min, 10 °C/min to 180 °C, 30 °C/min to 260 °C, hold for 5 min. Hydrolyzed and derivatized DAC samples yielded azaC compounds containing two TBDMS groups, with the predominant peak representing fragmentation loss of a tert-butyl group ([M-C(CH3)3], or [M-57]). Unenriched azaC yielded m/z = 283 and enriched 13C-azaC, m/z = 284, both corresponding to the respective [M-57] peaks. Samples were quantitated by comparing the peak area of unenriched azaC to 13C-azaC (1 mass unit heavier). The contribution of unenriched azaC to 13C-azaC due to the natural abundance isotope envelope was subtracted to get an accurate value of the heavier internal standard. Control experiments with formic acid were performed on azaC and β-DAC to determine the conditions that would minimize decomposition of the free base, yet efficiently cleave the glycosidic bond of β-DAC, leading to our choice of 100 °C for 30 min.
Decomposition of β-DAC estimated by UV spectrophotometry
Aliquots (10 μL) of the above- mentioned solution of β-DAC (9.2 mM) were removed at selected time intervals and placed in ice, diluted 100-fold in 200 mM potassium phosphate buffer (pH 7.0), and measured on a Cary-100 UV-Vis spectrophotometer (Varian Inc., Walnut Creek, CA) using a range of 200-300 nm. In order to minimize DAC degradation during sample processing, the time from aliquot removal to measurement was < 5 min for each sample. In a previous study with the ribonucleoside, azaCyd, it was noted that hydrolytic opening of the triazine ring results in the molar absorptivity at 240 nm increasing from 6900 to 18700 M-1 cm-1 (measured at pH 7.0), whereas the guanylurea derivative (resulting from irreversible loss of formic acid after hydrolytic ring-opening) lacks substantial absorbance at 240 nm (27). Therefore we measured and quantitated absorbance at 240 nm as an approximation of the sum of DAC and its ring-open form, normalized according to the ratio of the two extinction coefficients noted above for quantitation.
Decomposition of α-DAC and β-DAC measured by NMR spectroscopy
Studies were performed with a 500 MHz NMR (Bruker Biospin, Fremont, CA) equipped with a variable temperature controller (Eurotherm, BVT 3000). The temperature was calibrated with a sample of 80% ethylene glycol in d6-DMSO by relating temperature to chemical shift difference and constructing a calibration curve (28). A water suppression double gradient echo WATERGATE pulse program was used to obtain spectra (29). Solutions of β-DAC (13 mM) and α-DAC (15 mM) were prepared separately in 100 mM potassium phosphate buffer (pH 7.4), containing 10% D2O, and DSS for spectral calibration. The solution was heated at 37 °C for ∼ 24 h and spectra (128 scans per spectrum) were recorded approximately every hour. Solutions were made fresh and kept on ice for a brief time before starting the NMR experiments. Before beginning the time course, a spectrum of the sample was collected at approximately 283 K (10 °C), after which the sample was heated to 310.15 K (37 °C) inside the NMR. The time course for the reaction was said to begin when the temperature controller reached 310.15 K. The pH of samples was tested before and after the experiment. Both α-DAC and β-DAC samples had a marginal decrease in pH (∼ 0.05 units) from start to finish, likely due to the formation of formic acid. Peak areas of the H-6 protons of each species and of the formate proton were integrated and quantitated for use in determining apparent rate constants of the decomposition process. A standard of formic acid (10 mM) in the same buffer was also analyzed to verify the chemical shift of the CH formate proton under our conditions (δ = 8.445 ppm).
Decomposition of β-DAC measured by HPLC/UV and QTOF/MS
For quantitation of β-DAC decomposition analyzed by HPLC, aliquots (10 μL) of the above-mentioned solution of β-DAC (9.2 mM, used for GC/MS and UV measurements) were removed at selected time intervals and immediately frozen at -80 °C until all samples were collected. When ready for injection, aliquots were thawed, and 2 μL (1.8 × 10-8 mol) of each were added to 10 μL of 2′-deoxyuridine (dU) as an internal standard (2.8 × 10-9 mol), after which the mixtures were injected into a 1050 HPLC (Agilent) connected to a Surveyor PDA detector (Thermo Finnigan, Waltham, MA). In order to minimize degradation of DAC during sample processing, the time from aliquot thawing to injection on the HPLC was < 10 min for each sample. The mobile phase was isocratic 20 mM ammonium acetate (pH 6.8) with a 1 mL/min flow rate and the stationary phase was a 25 cm × 4.6 mm, 5 μm C-18 analytical column (Supelco #58928-U, Sigma-Aldrich, St. Louis, MO). Peak areas of β-DAC and dU were quantitated at 260 nm and the ratios of β-DAC to dU were calculated and normalized.
Separate aliquots (20 μL) were injected into a 2795 HPLC connected to a 2996 PDA detector and a quadrupole-time of flight (QTOF) API-US mass spectrometer (Waters Corp., Milford, MA). The mobile phase and stationary phase were as mentioned above. After UV detection, the flow was split to 0.3 mL/min before entering the QTOF/MS. The first 2.5 min of each run was discarded before attaching the flow to the QTOF/MS in order to avoid entry into the mass spectrometer of the potassium and phosphate ions present in the sample. For UV-detection, samples were monitored at 240 and 220 nm. For mass-spectral detection, samples were monitored in positive ion mode, resulting in species of 1 mass unit higher than the neutral molecules. Data were visualized by extracting the ion chromatograms at m/z = 229 (β-DAC and α-DAC, both singly-charged), 247 (ring-open-formylated derivatives, singly-charged), and 437 (guanylurea derivatives, singly-charged dimeric species). Extracted ion chromatograms at m/z = 219 (guanylurea derivatives, singly-charged monomeric species) were not useful for discrimination, as compounds with m/z = 229 and 247 also produced fragments with m/z = 219 when visualized by the mass spectrometer.
Mathematical models of DAC decomposition
The simplest model for the decomposition of DAC used in this study is a first-order exponential decay equation:
| (1) |
where [I] represents the concentration of DAC at time t, and k is the apparent rate constant. The half-life of the process is obtained by solving for t when the ratio of [I]/[I]0 equals 0.5:
| (2) |
Scheme 1 presents a more sophisticated model of DAC decomposition (17), but does not account for anomerization. The system of linear, first-order differential equations can be written for the decomposition of DAC shown in Scheme 1 as follows:
Scheme 1.
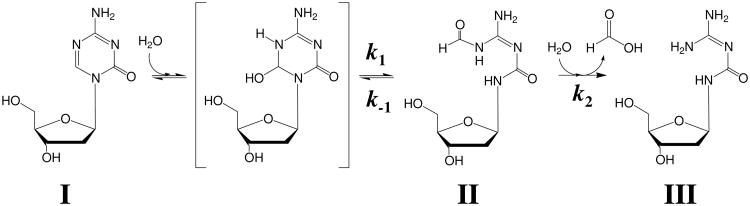
Simple model of DAC decomposition in aqueous solution. β-DAC (I) is in equilibrium with its ring-open-formylated derivative (II), followed by irreversible deformylation and formation of the guanylurea derivative (III). The intermediate ring-closed hydrated intermediate is also shown. The forward and reverse rate constants, k1 and k-1, although displayed here between the ring-closed hydrated intermediate and II, are intended to represent the overall observed rate constants of conversion between I and II.
| (3) |
| (4) |
| (5) |
where [I], [II], and [III] represent the concentration of each species at time t, and kn are the apparent rate constants for the process shown in Scheme 1. The equations were solved analytically (30):
| (6) |
| (7) |
| (8) |
where A and B are the roots of the following quadratic equation:
| (9) |
Scheme 2 presents our more complex model of DAC decomposition, which accounts for anomerization. The system of differential equations can be written for this scheme as follows:
Scheme 2.
Model of DAC decomposition based on NMR and HPLC analyses. Either β-DAC (I) or α-DAC (IV) are converted into their corresponding ring-open-formylated derivatives (IIa and IIc, respectively), followed by equilibration between the furanoside (5-membered-ring sugars) and pyranoside (6-membered-ring sugars) derivatives of II and irreversible deformylation into the guanylurea derivatives (III), which are also in a similar equilibrium. The H-6 proton of compounds I, II, and IV, as well as the CH proton of formic acid are bolded and italicized, as these were quantified by NMR. The ring-close hydrated intermediates corresponding to that seen in Scheme 1 are not shown here for the sake of clarity.
| (10) |
| (11) |
| (12) |
| (13) |
where [I], [II], [III], and [IV] represent the concentration of each species at time t, and kn are the apparent rate constants for the decomposition process shown in Scheme 2. We used numerical solutions for this more complex scheme (see below), instead of analytical solutions.
The data from GC/MS and HPLC/UV experiments were fit to theoretical curves generated using Equation 1 and the half-lives of β-DAC loss were calculated from Equation 2. The data from UV experiments were modeled according to Scheme 1 using Equations 6-9, and fit to a theoretical curve representing the sum of I and II weighted for their relative extinction coefficient values (27). The individual contributions of I, II, and III were calculated separately and the half-life of β-DAC loss was calculated by obtaining the value of t at which I had declined to 50% of its initial value. Data-sets from the two separate NMR experiments starting with α-DAC or β-DAC were modeled according to Scheme 2, and simultaneously fit to theoretical curves using Equations 10-13, generated by approximating solutions using the Euler method of numerical integration with a Δt = 1 min. For the NMR data, a solution was forced that would solve both sets of data using the same rate constants, with the half-life of β-DAC loss calculated by obtaining the value of t at which I had declined to 50% of its initial value.
All of the above data were fit in Microsoft Excel using the solver function with the apparent rate constants as variable parameters. The solver function minimized the sum of the squared deviations between experimental and theoretical values, each deviation being normalized to its respective experimental value so as to give equal weight to all data points during the fit. For the kinetic rates determined with our more complex models (UV and NMR data), multiple, various, initial values were tested, with the solver nearly always yielding the same rate values. The few cases in which different rate constants were obtained resulted in much higher values of the sum of the squared deviations and obviously inaccurate visual fit to the data. The values we report represent the only solutions with appreciably low sum of the squared deviations and visually accurate fit, indicating that our solutions result in a global minimum of the error and not just a local minimum.
To estimate the error of our apparent rate constant values and the associated half-lives of DAC decomposition, we generated confidence intervals for these values using a model comparison method as previously described (31). For each data set, the value of each rate constant was independently allowed to vary from that of the optimized value while keeping all other rate constants at their optimized values, such that the resulting sum of the squared deviations was higher by the amount accounted for when using a P-value of 0.05 (95% confidence interval). This variation always yielded a noticeable worsening of the fit between the data and their theoretical lines. The error values were nearly symmetrical around the optimized value, and in all cases, the larger of the two values was chosen in order to allow simple presentation of the confidence intervals as symmetric error bars.
Results
In this paper, we studied the chemical decomposition of DAC under physiological conditions of pH and temperature using multiple analytical techniques. The data obtained from each separate method of analysis are instructive, but the complete picture of DAC degradation is most clear when the results from all methods are considered together, as shown below.
Rate of DAC loss estimated by stable isotope dilution GC/MS
The first method we employed to measure the decomposition of DAC was GC/MS, which required acid hydrolysis to release the free bases and chemical derivatization with volatile functional groups prior to analysis. DAC is prone to hydrolytic ring-opening at its C-6 position in aqueous solutions at increased pH (17), but under acidic conditions, its glycosidic bond is labile, releasing the free base azaC and deoxyribose (32). We measured the time-dependent decomposition of the triazine ring of β-DAC in potassium phosphate buffer (pH 7.4) at 37 °C for 24 h, removing aliquots at selected time intervals and processing for GC/MS analysis. Peak area ratios of the derivatized azaC relative to the stable isotope labeled internal standard were quantitated and normalized, shown in Figure 1a. Data were fit to a first-order exponential decay equation (Equation 1), an apparent rate constant for the process was determined, and a half-life of 12.9 ± 0.9 h was calculated (Equation 2) for the decomposition of β-DAC.
Figure 1.
GC/MS analysis of β-DAC decomposition at 37 °C in 100 mM potassium phosphate buffer (pH 7.4). (A) Graph of GC/MS data versus time. Theoretical line was generated as described in Experimental Procedures. Rate constant and half-life are presented in Table 1. (B) Electron impact (EI) mass spectrum of the 6-h time point seen in A, representing a mixture of derivatized azaC and the internal standard. The structure of the derivatized internal standard, 13C-5-azacytosine-TBDMS2, is shown, and the proposed fragmentation pattern is indicated. The 13C stable-isotope label at C-6 of the triazine ring is indicated by *.
While azaC is more stable to alkali than DAC, it can decompose in acid at high temperature (32, 33), resulting in 5-azauracil (azaU) and further breakdown products (33). In order to obtain an accurate rate of DAC decomposition by our GC/MS method, we had to carefully choose our formic acid hydrolysis conditions so as to simultaneously maximize cleavage of the glycosidic bond of DAC, but minimize breakdown of the azaC base. Control experiments were performed with DAC and azaC, leading us to choose 100 °C for 30 min in 88% formic acid for our hydrolysis conditions. Under these conditions, DAC glycosidic bond cleavage was complete, and azaC degraded less than 15% when compared to thymine as an internal standard. Additionally, any degradation of azaC occurring during acid-hydrolysis and derivatization were accounted for, since we report the normalized ratio between azaC and the stable-isotope labeled internal standard 13C-azaC, both of which would degrade to an equivalent extent.
The compounds we detected correspond to azaC containing two TBDMS groups [M], fragments from loss of a methyl group ([M-CH3] or [M-15]), and fragments from loss of a tert-butyl group ([M-C(CH3)3], or [M-57]), the latter of which is the most abundant ion. The unenriched azaC fragment has m/z = 283 and the enriched 13C-azaC, m/z = 284, both of which correspond to the [M-57] peak. Figure 1b shows the mass spectrum of the 6-h data-point from Figure 1a. With our method, we observed peaks with m/z corresponding to ring-closed azaC, but not any ring-open derivatives, nor azaU. It is possible that the ring-open forms of DAC are unstable to the hydrolysis and/or derivatization conditions, explaining their lack of detection by our GC/MS method. Alternatively, our formic acid hydrolysis conditions could potentially force the ring-open-formylated derivatives to ring-close, or could even cause the guanylurea derivatives to reformylate followed by ring-closure, in both cases potentially regenerating DAC and leading to an artificially long half-life of DAC decomposition.
Rates of the ring-opening equilibrium and deformylation of DAC determined by UV spectrophotometry
Although the data analyzed by GC/MS fit relatively well to a model described by Equation 1 as shown in Figure 1a, this model does not account for the previously described ring-opening equilibrium of DAC (17). During the same decomposition experiment mentioned above, we also removed aliquots at selected time intervals to measure the UV absorbance at 240 nm. Data seen in Figure 2 were fit to solutions of a system of differential equations (Equations 6-9) that describe a scheme previously determined for β-DAC (17) under alkaline conditions (Scheme 1). In this scheme, β-DAC (I) becomes hydrated at the C-6 position and ring-opens to produce a ring-open-formylated derivative, N-(formylamidino)-N′-β-d-2′-deoxyribofuranosylurea (II), followed by loss of formic acid, producing a guanylurea derivative, 1-β-d-2′-deoxyribofuranosyl-3-guanylurea (III). The base-catalyzed opening of the triazine ring results in an initial increase in UV absorbance, due to the increased molar absorptivity of the ring-open-formylated derivative (27). The UV measurements here represent a combination of I and II, since both compounds have maximal absorbance near 240 nm, whereas III has neglible absorbance at this wavelength. The lines in Figure 2 represent the theoretical individual contributions of I, II and III, as well as the normalized combination of I and II, the latter of which fits the measured data. The apparent rate constants for the process were determined, and the half-life of β-DAC decomposition was calculated to be 10.2 ± 1.2 h, modestly lower than the half-life of 12.9 h measured by GC/MS, a difference which would be expected if the GC/MS method overestimates the half-life of DAC, as discussed above. Table 1 presents the apparent rate constants and corresponding half-life of β-DAC loss for each analytical technique used.
Figure 2.
UV-spectrophotometric analysis of β-DAC decomposition at 37 °C in 100 mM potassium phosphate buffer (pH 7.4). Theoretical lines were generated as described in Experimental Procedures. Rate constants and half-life of β-DAC decomposition are presented in Table 1. Solid line – theoretical line representing the sum of I and II normalized for relative extinction coefficient values, which fits the values from the UV measurements. Dashed line, dotted line, and x's – theoretical contribution of I, II, and III, respectively, as calculated by the equations using the determined rate constants.
Table 1.
Apparent rate constants and corresponding half-lives for the time-dependent decomposition of β-DAC analyzed by multiple techniques. All decomposition experiments were conducted in 100 mM potassium phosphate buffer (pH 7.4) at 37 °C. Rate constants and half-lives corresponding to each analytical method were determined using equations as described in Experimental Procedures.
| GC/MS | UV | NMR | HPLC | |
|---|---|---|---|---|
| Apparent Rate Constants (min-1 × 103) | k = 0.893 ± 0.059 |
k1 = 5.89 ± 0.54 k-1 = 19.9 ± 2.4 k2 = 3.95 ± 0.23 |
k1 = 1.46 ± 0.08 k-1 = 2.44 ± 0.20 k2 = 6.01 ± 0.32 k3 = 0.537 ± 0.033 k-3 = 2.71 ± 0.23 |
k = 1.26 ± 0.10 |
| t1/2 (min) | 776 ± 51 | 613 ± 71 | 603 ± 33 | 549 ± 43 |
| t1/2 (h) | 12.9 ± 0.9 | 10.2 ± 1.2 | 10.1 ± 0.6 | 9.15 ± 0.72 |
DAC decomposition and anomerization determined by NMR spectroscopy
The decomposition of DAC involves ring-opening and deformylation (17), but according to a previous report, DAC can also anomerize in neutral aqueous solution (23). Therefore, we measured DAC decomposition as a function of time by 1H-NMR spectroscopy, a technique that would distinguish between anomers. The two anomers of DAC were synthesized, characterized, and separately dissolved in potassium phosphate buffer, and the time-dependent decomposition of each was analyzed by NMR at 37 °C and pH 7.4 for approximately 24 h. Figure 3 shows representative NMR spectra and graphical data from experiments performed with β-DAC. The corresponding data for α-DAC are available as supporting information (Figure S1). The resonances corresponding to the H-6 protons of the DAC species are singlets and are furthest downfield, in a region of the spectrum where there is minimal to no overlap of peaks. This allowed quantitation of the peak areas for determining the kinetic rate constants. NMR data were fit to numerical solutions of a system of differential equations as modeled by Scheme 2, which was devised based on our NMR experiments and the HPLC/UV/MS experiments discussed below. In this scheme, β-DAC (I) becomes hydrated at the C-6 position and ring-opens (IIa). Compound IIa can epimerize at the anomeric 1′-carbon and recyclize, resulting in attachment of the base in 2 different ways (α- or β-anomeric configuration) to 2 different sugar conformations – the furanoside (5-membered ring) and pyranoside (6-membered ring) derivatives. Any one of these 4 compounds (IIa, IIb, IIc, and IId) could deformylate, producing the corresponding guanylurea derivatives (III). Additionally, compound IIc could also recyclize and dehydrate, reforming the triazine ring to produce α-DAC (IV).
Figure 3.

NMR analysis of β-DAC decomposition at 37 °C in 100 mM potassium phosphate buffer (pH 7.4), 10% D2O. (A) 1H-NMR spectrum of β-DAC after 35 min at 37 °C. (B) 1H-NMR spectrum of β-DAC after 12.6 h at 37 °C. (C) Points represent data obtained from integrated peak areas of the H-6 protons and CH proton of formate shown in A and B. Theoretical lines were obtained as described in Experimental Procedures. Rate constants and half-life of β-DAC decomposition are presented in Table 1. See Scheme 2 for identification of each species.
As displayed in Figure 3 and Figure S1, we were able to assign chemical shifts for β-DAC (I, δ = 8.515 ppm) and α-DAC (IV, 8.455 ppm) from the spectra of our synthesized compounds. Based on the time-dependent initial increase and subsequent slow decrease of the peak at δ = 8.61 ppm, we identified this as the most abundant ring-open-formylated derivative of DAC, and the much smaller peaks at 8.64 and 8.66 ppm are possibly derivatives containing the same base with a different sugar conformation (all derivatives of II). The broad peak shape at δ = 8.61 ppm and shift downfield of ∼ 0.1 ppm from β-DAC follows the same trend as that noted in the literature for the character of the ring-open-formylated derivative of azaCyd, N-(formylamidino)-N′-β-d-ribofuranosylurea (27). Deformylation of any of the ring-open-formylated derivatives (II) into the guanylurea derivatives (III) results in loss of the H-6 proton as formic acid. Since the proton resonances of the guanylurea derivatives were in regions of the spectra with significant overlap, these analogs were instead measured indirectly as a sum, by quantitating the singlet resonance of the formate CH proton (8.445 ppm).
Differential equations for the scheme were written (Equations 10-13), numerical methods were used to obtain solutions as described in Experimental Procedures, and both data sets starting with α-DAC and β-DAC were fit simultaneously to the solutions, with apparent rate constants presented in Table 1. The resulting half-life for β-DAC decomposition was 10.1 ± 0.6 h, shorter than the value obtained by GC/MS and almost identical to that obtained by UV analysis. The parallel half-life of α-DAC was obtained by extrapolating the theoretical curve for α-DAC decomposition (Figure S1), resulting in 30.2 ± 2.0 h, approximately three times that of β-DAC.
Separation and characterization of β-DAC decomposition products by HPLC/UV and QTOF/MS
Although our NMR method provides clear evidence of anomerization and allows for accurate quantitation, it is limited to the compounds with a proton at the H-6 position, which excludes the guanylurea derivatives (III). Therefore, we developed an HPLC method with simultaneous UV and QTOF/MS detection in order to verify the existence of the proposed structures found in Scheme 2. Aliquots were removed from the same sample of β-DAC used above for GC/MS and UV measurements, and were analyzed, resulting in at least 9 compounds separable by HPLC.
Figure 4 shows HPLC/UV chromatograms of the β-DAC sample after a 12-h incubation (UV spectra of representative compounds are available as supporting information, Figure S2). Four of the observed peaks (Figure 4, peaks 1, 2, 3, and 7) absorb strongly in the UV at 220 nm and very little at 240 nm. When analyzed by QTOF/MS, these same peaks have mass spectra containing m/z = 219 as the monomeric species and 437 as the dimeric species (data available as supporting information, Figure S3). Together, these data identify the 4 peaks as guanylurea derivatives (III) of DAC. Three separate peaks seen in Figure 4 (peaks 5, 8, and 9) absorb more at 240 than at 220 nm, and have mass spectra containing m/z = 247 as the monomeric and 269 as the monomeric-Na+-adduct species (Figure S3). These data confirm the identity of these 3 peaks to be ring-open-formylated derivatives (II) of DAC. The remaining two peaks absorb equally at 240 and 220 nm, and have mass spectra containing predominantly m/z = 229 as the monomeric species and 457 as the dimeric species (Figure S3), indicating that these are ring-closed derivatives, α-DAC (peak 4 in Figure 4) and β-DAC (peak 6).
Figure 4.
HPLC/UV chromatograms of the 12-h time point from β-DAC decomposition at 37 °C in 100 mM potassium phosphate buffer (pH 7.4). Upper chromatogram, absorbance at 240 nm. Lower chromatogram, absorbance at 220 nm. A total of 9 peaks were visualized, represented in Scheme 2 and explained in the Results and Table S1. Peaks 1 (6.48), 2 (8.35), 3 (11.01), and 7 (17.01 min) have m/z = 219 and 437, corresponding to the guanylurea derivatives (III); peaks 4 (12.28) and 6 (15.41 min) have m/z = 229 and 457, corresponding to ring-closed α-DAC (IV) and β-DAC (I), respectively; peaks 5 (13.08), 8 (19.27), and 9 (22.34 min) have m/z = 247 and 269, corresponding to the ring-open-formylated derivatives (II).
Together with our NMR results, these data confirm our decomposition scheme (Scheme 2). We propose that the four peaks corresponding to the UV and mass spectra of the guanylurea derivative (III) are the furanoside (5-membered ring) and pyranoside (6-membered ring) forms of the deoxyribose sugar attached to guanylurea in the α- and β-anomeric configurations. In a similar fashion, the three peaks identified as ring-open-formylated derivatives are 3 of the 4 possible combinations of anomers and forms of the sugar attached to the base. It is likely that the fourth derivative is in too low abundance to be detected under these conditions, or it may co-elute with another peak on the chromatogram. Scheme 3 is a proposed mechanism by which conversion between the different sugar conformations may occur. Although it is not possible using our techniques to say with certainty which peak corresponds to which specific anomer or sugar derivative, we propose a preliminary identification of each peak, available in a table as supporting information (Table S1).
Scheme 3.
Proposed mechanism for anomerization and interconversion between the different deoxyribose sugar conformations of ring-opened DAC. After the deoxyribose sugar ring-opens, free rotation about the C-1′ carbon positions the nucleobase (denoted by R) in either of the α- or β-configurations before hydroxyl attack at C-1′ and ring closure. Likewise, free rotation about the C-4′ carbon places one of two hydroxyl groups in position for attack of C-1′ and formation of either the furanoside (5-membered ring) or pyranoside (6-membered ring) conformations of the deoxyribose sugar.
In order to quantitatively compare the rate of β-DAC decomposition obtained by HPLC with the previous analytical methods, aliquots from the above-mentioned time-course (used for GC/MS and UV analyses) were added to constant amounts of dU as an internal standard, injected into an HPLC, and peak areas were quantitated at a fixed wavelength. The internal standard, dU, was found to be appropriate as it elutes from the HPLC column > 1 min after the last DAC degradation product. Data are shown in Figure 5, fit to Equation 1 to obtain an apparent rate constant, and the half-life was calculated (Equation 2) to be 9.15 ± 0.72 h for β-DAC loss (Table 1).
Figure 5.
HPLC/UV quantitation of β-DAC decomposition at 37 °C in 100 mM potassium phosphate buffer (pH 7.4). Points represent the normalized peak area ratios of β-DAC to the internal standard, dU. Theoretical line was generated as described in Experimental Procedures. Rate constant and half-life of β-DAC decomposition are presented in Table 1.
A control experiment was performed in which an aliquot of the 24-h time-point of β-DAC decomposition was added to an equimolar amount of azaC free base. Then samples of the 24-h aliquot with and without added azaC were injected into the HPLC and compared. Formation of azaC after 24-h did occur, and the compound had a retention time of 4.15 min, but formation was negligible, accounting for less than 0.5% of the original starting amount of β-DAC. This suggests that hydrolysis of the glycosidic bond of DAC is a real but negligible side-reaction under conditions of physiological pH and temperature, and would not contribute substantially to our analysis of rates. Furthermore, due to the lack of other peaks present on HPLC, it is unlikely that deamination of DAC to 5-aza-2′-deoxyuridine (azadU) occurred to any appreciable extent under our conditions.
Discussion
Interest in DAC as a potential chemotherapy agent derives primarily from its demonstrated capacity to inhibit enzymatic methylation of cytosine residues in DNA (4) with subsequent transcriptional activation of surrounding genes that had been silenced by normal or aberrant cytosine methylation (5, 6). The instability of DAC in aqueous solution under physiological conditions creates a significant challenge to its use in humans. Furthermore, the degradation products derived from DAC likely have independent and as yet unknown pharmacological properties that may contribute to the observed toxicity (12) and possible mutagenicity (13) of DAC. The results presented in this paper are intended to enhance our understanding of the pathways and kinetics of DAC degradation and to help understand its toxicity.
In the first approach utilized here, a GC/MS assay was developed to measure DAC decomposition in aqueous buffered solution. Central to this method is the use of a 13C-enriched standard of azaC prepared in this laboratory by the method of Hartenstein and Fridovich (26). We developed this method to measure the stability of DAC in solution, but it could also be used to study the stability of phosphorylated DAC or when DAC is incorporated into oligonucleotides or cellular DNA. The net loss of the triazine ring of DAC at physiological temperature and pH was measured by our GC/MS method, and data were fit to a simple exponential decay equation and the apparent half-life was calculated to be 12.9 ± 0.9 h (Figure 1, Table 1).
We next measured the UV spectrum of DAC in aqueous buffered solution as a function of time, shown in Figure 2. In this scheme (Scheme 1), the triazine ring hydrates and ring-opens reversibly at C-6, whereas the loss of formic acid is irreversible. Equations were derived to explain the concentrations of the possible species as a function of time and three rate constants (Table 1). Our data are consistent with the ring-opened and ring-closed forms of DAC being in equilibrium, with the ring-closed favored by a factor of approximately 3.4 (k-1/k1). The rate-limiting step for DAC degradation, according to our data, would be the loss of formic acid (k2). The corresponding half-life for DAC under these conditions was calculated to be 10.2 ± 1.2 h. This apparent half-life is approximately 20% lower than determined when using the above-described GC/MS method. The UV-based method would be more accurate than the GC/MS method, as it accounts for the ring-opening equilibrium. When analyzing by GC/MS, ring-opened intermediates may be forced back into the ring-closed structure during sample preparation, and thus mask the contribution of this equilibrium to the overall loss of DAC.
We then turned to a method that utilizes NMR spectroscopy to examine the composition of solutions containing DAC and its degradation products. When examined as a function of time, the resonance corresponding to the H-6 proton of β-DAC (I) declines in magnitude and the resonances belonging to the formate CH proton and the H-6 proton of DAC when found as the α-anomer (IV) both increase with time (Figure 3). Multiple smaller and broad peaks are observed in the spectrum, attributable to the ring-open-formylated intermediates (II). The chemical synthesis of DAC yields both the α and β-anomers, which are chromatographically separable. We therefore had the authentic α-anomer for use as a standard and to measure the reverse anomerization of the α- to β-anomer (Figure S1).
The complexity of the scheme that corresponded to observations obtained from our NMR experiments was too great to solve analytically, and therefore, a numerical method was applied to estimate the rate constants. The good agreement between the experimental and theoretical results is shown in Figure 3c and the corresponding rate constants are presented in Table 1. The advantage of this method over the previous methods is that it allows examination of the anomerization of β-DAC to α-DAC, as well as the reverse pathway. The apparent rate constant we measure for ring-opening of the β-anomer (k1) is approximately three times that for the α-anomer (k3), whereas the rate of ring-closing is slightly greater for the α-anomer (k-3 compared to k-1). The α-anomer is therefore more likely to be found in the ring-closed form than the β-anomer. Thus, as seen in Figure 3c, with time α-DAC could become more predominant than β-DAC under conditions of physiological temperature and pH. The apparent half-life of β-DAC using this method, 10.1 ± 0.6 h, is virtually identical to that measured with the previous method (10.2 ± 1.2 h), but it accounts for the net degradation of DAC that results from the known anomerization of β-DAC.
The final method we examined was HPLC separation followed by UV detection and mass spectral analysis. Our above-described NMR method only allowed detection of the guanylurea derivatives indirectly, by observing the time-dependent increase of the resonance associated with the CH proton of formate. We anticipated that HPLC analysis might allow separation and assessment of these products. We expected to observe peaks corresponding to β- and α-DAC, absorbing strongly at 240 nm, as well as a series of minor peaks with absorbance at 240 nm (ring-open-formylated analogs) and a series of peaks with only end absorbance (guanylurea analogs). We observed nine peaks with the expected UV absorbance (Figure 4, Figure S2) and mass spectra (Figure S3). The multiplicity for the number of ring-open-formylated and guanylurea peaks results from another equilibrium in the degradation of DAC, conversion of the 5-membered deoxyribofuranoside to the 6-membered deoxyribopyranoside sugars (Scheme 3). Both α- and β-anomers can exist for each sugar configuration. This conversion between anomers and 5- and 6-membered sugar rings has been previously observed during synthesis of a closely related compound, 2′-deoxyribosylurea (34), which is itself known to be both a lethal and mutagenic lesion when in DNA (35). As we can separate β-DAC from the series of at least eight degradation products, it is possible to directly determine the rate of β-DAC decay. The observed half-life of β-DAC was measured with this method to be 9.15 ± 0.72 h, modestly shorter than that determined by all the preceding methods.
Previous literature values for the decomposition of β-DAC at 37 °C and neutral pH range from 3.5 to 21 h. One study reports the half-life of DAC to be approximately 11 h when incubated at 37 °C, pH 7.0, analyzed by HPLC with UV detection (17). Two recent studies report half-lives of 20 to 21 h at 37 °C, one using a neutral buffer with unreported pH (20), and the other using a buffer at pH 7.4 (19). One of these studies (20) used capillary electrophoresis with UV detection, but as their exact pH is unreported, it is difficult to directly compare their results with ours. Investigators conducting the other study (19) obtained their half-life by measuring the decline of UV-absorbance of DAC, but they did not provide a mathematical formulation to account for the ring-opening equilibrium, as they were only comparing the stability of DAC with a DAC-containing dinucleotide. Thus, their apparent half-life for DAC (20 h) would be expected to be higher than our UV-determined half-life (10.2 h), as our UV study accounts for the ring-opening equilibrium. Indeed, analyzing our UV data without using an equation to account for the ring-opening equilibrium would result in a similar half-life of approximately 21 h, the time at which the data reach 50% of the initial absorbance (Figure 2). Two more studies report the half life of β-DAC at 37 °C and neutral pH to be approximately 3.5 to 4 h (18, 21). However, in one of the studies, DAC was incubated in cell culture media supplemented with fetal calf serum (21) and in the other, DAC was incubated in human plasma (18). It is likely that enzymatic degradation in addition to chemical decomposition occurred in each case. Indeed, enzymes such as cytidine deaminase can rapidly convert DAC into the corresponding uracil derivative, azadU (36). Our study provides the basis for chemical decomposition of DAC, upon which future studies can build complexity when analyzing DAC in biological systems that involve both chemical and enzymatic degradation. In our study, in spite of the range of analytical techniques and models with equations of increasing complexity, the average of the half-lives determined for β-DAC degradation among our methods was 10.6 ± 1.6 h.
In addition to the range of half-lives reported for DAC, previous studies of the products of DAC decomposition under conditions of neutral pH disagree. In 1981, Lin et al (17) analyzed the chemical stability of β-DAC and azaCyd by HPLC at different temperatures and in buffer solutions of varying pH. Both compounds had similar rates and products in alkaline solution, but DAC decomposition differed substantially from azaCyd at neutral pH, with several products left unidentified. In 1984, Vesely and Piskala (23) synthesized both anomers of DAC and noted that incubation of either anomer of DAC in water at room temperature for 72 h resulted in formation of the opposite anomer, with α-DAC showing more stability than β-DAC. This suggested that anomerization to α-DAC accounts for at least one of the previously unidentified decomposition products. More recently, a study was published characterizing the decomposition products of DAC in water and plasma by HPLC/MS methods (18). The authors proposed a scheme showing DAC in equilibrium with several isomers of the ring-open-formylated derivatives, which then deformylate into two possible isomers of the guanylurea derivatives. All of their proposed structures are structural isomers of the triazine ring attached to the deoxyribofuranoside sugar in the β-anomeric configuration exclusively. Anomerization to α-DAC is not discussed, despite previous literature findings of spontaneous anomerization under neutral pH conditions (23). Furthermore, a major peak was observed on HPLC with identical mass and tandem mass spectral fragmentation to that of β-DAC, but with a shorter retention time. They proposed this molecule to be “cyclodecitabine”, a bicyclic analog formed by intramolecular addition of the 5′-hydroxyl group to the C-6 position of the triazine ring of β-DAC (18). However, cyclodecitabine would likely be less polar than β-DAC due to loss of the 5′-OH group. When separating compounds on a C-18 column (reverse-phase HPLC), this would result in cyclodecitabine eluting later than β-DAC, not earlier. Based on our results and other studies (23), we find it much more likely that the compound they observed is actually α-DAC.
The mechanism we propose in Scheme 3 for anomerization has been previously eluded to (23, 37), but to our knowledge, has not previously been explicitly presented. As reported by Lin et al (17), decomposition of β-DAC at high pH (10.4) resulted in observation of only 3 species by HPLC: β-DAC (I) and its corresponding ring-open-formylated (II) and guanylurea derivative (III) (see Scheme 1). As the pH was lowered, this rate of decomposition became slower, indicating that the hydrolytic ring-opening of DAC is base-catalyzed. At neutral pH, the decomposition profile was more complex, and the authors were unable to identify the additional products (17), which we propose here to be the α- and β-anomers of furanoside and pyranoside derivatives. The mechanism we propose in Scheme 3 requires a proton and is acid-catalyzed, such that anomerization would not occur to an appreciable extent at higher pH, in agreement with the previous study as mentioned above (17). Thus it is only at neutral pH, where both base-catalyzed events (hydrolytic ring-opening) and acid-catalyzed events (sugar ring-opening followed by anomerization and re-closure) can take place simultaneously, that DAC decomposition will result in such a complex mixture. This increases the importance of our study, as the chemotherapeutic use of DAC in humans results in its exposure to aqueous, neutral pH environments.
As is clear from our NMR studies (Figure S1, Figure 3), α-DAC has a significantly longer half-life than does β-DAC under physiological conditions, 30.2 ± 2.0 h compared to 10.1 ± 0.6 h, respectively. This agrees with previous literature on the differential stability of these anomers (21, 23). Using the rate constants obtained from our NMR experiments (Table 1), we calculate that when starting with 100% β-DAC, α-DAC would reach a maximum of 17.4% at approximately 26 h before declining due to hydrolytic ring-opening and deformylation. Alternatively, when starting with 100% α-DAC, the same rate constants allow us to calculate that β-DAC would reach a maximum of 5.7% at approximately 26 h before declining. One major structural difference between the α- and β-anomers of DAC is the position of the 5′-OH, which is in closer proximity to the triazine ring in β-DAC than in α-DAC. It is possible that intramolecular interactions between the 5′-OH and the triazine ring of β-DAC facilitate hydrolytic ring-opening, resulting in a shorter half-life for the β-anomer.
The ability of β-DAC to undergo anomerization to the α-anomer might in part explain some of the observed toxicity associated with β-DAC. Although we do not provide experimental evidence here to support this possibility, several previous studies have been done on the ability of α-DAC to act as an antitumor compound and as a demethylating agent (21, 23, 37). Although the mechanism is unclear, the activities are several-fold to two orders of magnitude less than β-DAC, which was tested in parallel in each study. It may be that α-DAC shows activity in these studies simply because of its partial conversion to the β-anomer, which then exerts its activity. This could allow α-DAC to act as a reservoir of β-DAC, and our prediction of a less than 6% maximum conversion to the β-anomer over 26 h would explain the lower biological activity of α-DAC. Alternatively, the α-anomer may be enzymatically converted to the triphosphate and incorporated into DNA, as has been previously reported for another α-anomer nucleoside analogue, α-2′-deoxythioguanosine (38), although there are very few α-anomers reported to be incorporated into DNA by polymerases. As a still further possibility, α-DAC could itself have activity without the need for DNA incorporation (proposed in ref 37), just as a recent study on β-DAC showed its ability to selectively degrade DNMT1 in the absence of DNA replication (16). α-DAC may act as a formylating agent via its unstable triazine ring moiety under aqueous conditions, much like β-DAC (4), leading to inactivation of enzymes such as DNMT1 or others. These considerations are highly relevant to determining the mechanism of DAC antitumor activity, given our data showing that anomerization of β-DAC to α-DAC is a major side-reaction of its chemical decomposition.
Our results suggest that a plethora of compounds are generated during the chemical decomposition of DAC at physiological pH and temperature. This raises questions about which derivatives are pharmacologically active, and which will be incorporated into DNA. Besides the decomposition products generated at the nucleoside level, it is likely that the mono-, di-, and triphosphates of DAC are also chemically labile and may decompose even more quickly than the deoxynucleoside, as has been observed of the 5′-phosphorylated derivatives of azaCyd (39). It is unknown whether the corresponding degradation products of DAC-triphosphate would be incorporated by polymerases into DNA. Furthermore, there is also potential for hydrolysis, ring-opening, and anomerization of DAC after incorporation of the ring-closed triphosphate into DNA. The products of DAC decomposition in DNA may generate substrates for DNA-repair enzymes, or could possibly lead to miscoding, as has been suggested for the guanylurea derivative (4, 13). These questions are the subject of ongoing investigation in our laboratory.
The various methods presented here to examine the degradation of DAC have identified a complex scheme containing multiple pathways. Each of the methods utilized independently support branches within the overall scheme. While the activity of α-DAC has been studied (21, 23, 37), the activities of the other various degradation products are as yet unknown. It is possible that some of the products described here account for the toxic or potentially mutagenic activities of DAC. Future studies are required to determine the consequences of these products.
Supplementary Material
Acknowledgments
We thank Dr. Kangling Zhang for his assistance with the QTOF/MS studies. We also thank the reviewers for their very helpful suggestions, which we have incorporated into our manuscript. This study was supported by the National Institutes of Health. D.K.R. is supported in part by the Loma Linda University School of Medicine Medical Scientist Training Program.
Footnotes
Abbreviations: DAC, 5-aza-2′-deoxycytidine; 5mC, 5-methylcytosine; α-DAC, α-anomer of 5-aza-2′-deoxycytidine; β-DAC, β-anomer of 5-aza-2′-deoxycytidine; azaC, 5-azacytosine; 13C-azaC, [6-13C]-5-azacytosine; azaCyd, 5-azacytidine; azaU, 5-azauracil; azadU, 5-aza-2′-deoxyuridine; QTOF/MS, quadrupole-time of flight/mass spectrometry; MTBSTFA, N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide; TBDMCS, tert-butyldimethylchlorosilane; TBDMS, tert-butyldimethylsilyl; d6-DMSO, 2H6-dimethyl sulfoxide; D2O, deuterium oxide; DSS, 2,2-dimethyl-2-silapentane-5-sulfonic acid; PDA, photodiode array; DNMT1, DNA (cytosine-5-)-methyltransferase 1.
Supporting Information Available: Table S1 provides a preliminary identification of all the observed decomposition products of DAC based on HPLC/UV/MS and NMR analyses. Figure S1 shows NMR spectra and graphical data of the decomposition of α-DAC, analogous to Figure 3. Figures S2 and S3 provide an HPLC/MS chromatogram of the separation of β-DAC from its products of degradation, along with characteristic UV spectra and mass spectra of representative products. This material is available free of charge vie the internet at http://pubs.acs.org.
References
- 1.Momparler RL, Derse D. Kinetics of phosphorylation of 5-aza-2′-deoxycytidine by deoxycytidine kinase. Biochem Pharmacol. 1979;28:1443–1444. doi: 10.1016/0006-2952(79)90454-4. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard J, Momparler RL. Incorporation of 5-aza-2′-deoxycytidine-5′-triphosphate into DNA. Interactions with mammalian DNA polymerase α and DNA methylase. Mol Pharmacol. 1983;24:109–114. [PubMed] [Google Scholar]
- 3.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 5.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 6.Daskalakis M, Nguyen TT, Nguyen C, Guldberg P, Kohler G, Wijermans P, Jones PA, Lubbert M. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-aza-2′-deoxycytidine (decitabine) treatment. Blood. 2002;100:2957–2964. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 7.Kantarjian H, Issa JPJ, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C, Ravandi F, Helmer R, 3rd, Shen L, Nimer SD, Leavitt R, Raza A, Saba H. Decitabine improves patient outcomes in myelodysplastic syndromes. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 8.Kantarjian HM, O'Brien S, Cortes J, Giles FJ, Faderl S, Issa JP, Garcia-Manero G, Rios MB, Shan J, Andreeff M, Keating M, Talpaz M. Results of decitabine (5-aza-2′-deoxycytidine) therapy in 130 patients with chronic myelogenous leukemia. Cancer. 2003;98:522–528. doi: 10.1002/cncr.11543. [DOI] [PubMed] [Google Scholar]
- 9.Schrump DS, Fischette MR, Nguyen DM, Zhao M, Xinmin L, Kunst TF, Hancox A, Hong JA, Chen GA, Pishchik V, Figg WD, Murgo AJ, Steinberg SM. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res. 2006;12:5777–5785. doi: 10.1158/1078-0432.CCR-06-0669. [DOI] [PubMed] [Google Scholar]
- 10.Appleton K, Mackay HJ, Judson I, Plumb JA, McCormick C, Strathdee G, Lee C, Barrett S, Reade S, Jadayel D, Tang A, Bellenger K, Mackay L, Setanoians A, Schatzlein A, Twelves C, Kaye SB, Brown R. Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J Clin Oncol. 2007;25:4603–4609. doi: 10.1200/JCO.2007.10.8688. [DOI] [PubMed] [Google Scholar]
- 11.DeSimone J, Koshy M, Dorn L, Lavelle D, Bressler L, Molokie R, Talischy N. Maintenance of elevated fetal hemoglobin levels by decitabine during dose interval treatment of sickle cell anemia. Blood. 2002;99:3905–3908. doi: 10.1182/blood.v99.11.3905. [DOI] [PubMed] [Google Scholar]
- 12.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci USA. 1997;94:4681–4685. doi: 10.1073/pnas.94.9.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karpf AR, Moore BC, Ririe TO, Jones DA. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol Pharmacol. 2001;59:751–757. [PubMed] [Google Scholar]
- 16.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Lin KT, Momparler RL, Rivard GE. High-performance liquid chromatographic analysis of chemical stability of 5-aza-2′-deoxycytidine. J Pharm Sci. 1981;70:1228–1232. doi: 10.1002/jps.2600701112. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Marcucci G, Byrd JC, Grever M, Xiao J, Chan KK. Characterization of decomposition products and preclinical and low dose clinical pharmacokinetics of decitabine (5-aza-2′-deoxycytidine) by a new liquid chromatography/tandem mass spectrometry quantification method. Rapid Commun Mass Spectrom. 2006;20:1117–1126. doi: 10.1002/rcm.2423. [DOI] [PubMed] [Google Scholar]
- 19.Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, Tang C, Redkar S, Jones PA. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 2007;67:6400–6408. doi: 10.1158/0008-5472.CAN-07-0251. [DOI] [PubMed] [Google Scholar]
- 20.Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 21.Hajek M, Votruba I, Holy A, Krecmerova M, Tloustova E. Alpha anomer of 5-aza-2′-deoxycytidine down-regulates hTERT mRNA expression in human leukemia HL-60 cells. Biochem Pharmacol. 2008;75:965–972. doi: 10.1016/j.bcp.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 22.Chan KK, Giannini DD, Staroscik JA, Sadee W. 5-Azacytidine hydrolysis kinetics measured by high-pressure liquid chromatography and 13C-NMR spectroscopy. J Pharm Sci. 1979;68:807–812. doi: 10.1002/jps.2600680705. [DOI] [PubMed] [Google Scholar]
- 23.Vesely J, Piskala A. Effects of the α-d-anomer of 5-aza-2′-deoxycytidine on L1210 mouse leukemic cells in vitro and in vivo. Cancer Res. 1984;44:5165–5168. [PubMed] [Google Scholar]
- 24.Liu MC, Luo MZ, Mozdziesz DE, Lin TS, Dutschman GE, Cheng YC, Sartorelli AC. Synthesis of 2′-methylene-substituted 5-azapyrimidine, 6-azapyrimidine, and 3-deazaguanine nucleoside analogues as potential antitumor/antiviral agents. Nucleosides Nucleotides. 1999;18:55–72. doi: 10.1080/07328319908045594. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Hattar J, Jiricny J. An improved synthesis of 2′-deoxy-5-azacytidine by condensation of a 9-fluorenylmethoxycarbonyl-protected sugar onto the silylated base. J Org Chem. 1986;51:3211–3213. [Google Scholar]
- 26.Hartenstein R, Fridovich I. Amidinourea formate, a precursor of 2-amino-4-hydroxy-s-triazine. J Org Chem. 1967;32:1653–1654. doi: 10.1021/jo01280a095. [DOI] [PubMed] [Google Scholar]
- 27.Beisler JA. Isolation, characterization, and properties of a labile hydrolysis product of the antitumor nucleoside, 5-azacytidine. J Med Chem. 1978;21:204–208. doi: 10.1021/jm00200a012. [DOI] [PubMed] [Google Scholar]
- 28.Braun S, Kalinowski HO, Berger S. 150 and more basic NMR experiments. Wiley-VCH; Weinheim, Germany: 1998. pp. 140–143. [Google Scholar]
- 29.Liu M, Mao X, Ye C, Huang H, Nicholson JK, Lindon JC. Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J Magn Reson. 1998;132:125–129. [Google Scholar]
- 30.Rodiguin NM, Rodiguina EN. Consecutive chemical reactions. D. Van Nostrand; Princeton, NJ.: 1964. pp. 29–31. [Google Scholar]
- 31.Motulsky H, Christopoulos A. Fitting models to biological data using linear and nonlinear regression. GraphPad Software Inc.; San Diego, CA: 2005. pp. 109–117. [Google Scholar]
- 32.Tomankova H, Zyka J. Study of the time dependence of the stability of 5-aza-2′-deoxycytidine in acid medium. Microchem J. 1980;25:281–288. [Google Scholar]
- 33.Notari RE, DeYoung JL. Kinetics and mechanisms of degradation of the antileukemic agent 5-azacytidine in aqueous solutions. J Pharm Sci. 1975;64:1148–1157. doi: 10.1002/jps.2600640704. [DOI] [PubMed] [Google Scholar]
- 34.Dubey I, Pratviel G, Robert A, Meunier B. Convenient method for the preparation of 2′-deoxyribosylurea by thymidine oxidation and NMR study of both anomers. Nucleosides Nucleotides Nucleic Acids. 2001;20:1463–1471. doi: 10.1081/NCN-100105241. [DOI] [PubMed] [Google Scholar]
- 35.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 36.Chabot GG, Bouchard J, Momparler RL. Kinetics of deamination of 5-aza-2′-deoxycytidine and cytosine arabinoside by human liver cytidine deaminase and its inhibition by 3-deazauridine, thymidine or uracil arabinoside. Biochem Pharmacol. 1983;32:1327–1328. doi: 10.1016/0006-2952(83)90293-9. [DOI] [PubMed] [Google Scholar]
- 37.Fojtova M, Piskala A, Votruba I, Otmar M, Bartova E, Kovarik A. Efficacy of DNA hypomethylating capacities of 5-aza-2′-deoxycytidine and its alpha anomer. Pharmacol Res. 2007;55:16–22. doi: 10.1016/j.phrs.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Peery A, LePage GA. Nucleotide formation from α- and β-2′-deoxythioguanosine in extracts of murine and human tissues. Cancer Res. 1969;29:617–623. [PubMed] [Google Scholar]
- 39.Zielinski WS, Sprinzl M. Chemical synthesis of 5-azacytidine nucleotides and preparation of tRNAs containing 5-azacytidine in its 3′-terminus. Nucleic Acids Res. 1984;12:5025–5036. doi: 10.1093/nar/12.12.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.