Abstract
Inflammation-mediated reactive molecules can damage DNA by oxidation and chlorination. The biological consequences of this damage are as yet incompletely understood. In this paper, we have constructed oligonucleotides containing 5-chlorouracil (ClU), one of the known inflammation damage products. The thermodynamic stability, base pairing configuration and duplex conformation of oligonucleotides containing ClU paired opposite adenine have been examined. NMR spectra reveal that the ClU-A base pair adopts a geometry similar to that of the T-A base pair, and the ClU-A containing duplex adopts a normal B-form conformation. The linewidth of the imino proton of the ClU residue is substantially greater than that of the corresponding T imino proton; however, this difference is not attributed to a reduced thermal or thermodynamic stability or to increased proton exchange with solvent. While the NMR studies reveal increased chemical exchange for the ClU imino proton of the ClU-A base pair, the ClU residue is not a target for removal by the Escherichia coli mispaired uracil glycosylase, which senses damage-related helix instability. The results of this study are consistent with previous reports indicating that the DNA of replicating cells can tolerate substantial substitution with CIU. The fraudulent, pseudo-Watson-Crick ClU-A base pair is sufficiently stable to avoid glycosylase removal and, therefore, might constitute a persistent form of cellular DNA damage.
Introduction
Emerging studies indicate that the DNA of mammalian cells can be damaged by reactive molecules generated from activated neutrophils (1–5). The damaging agents include hydrogen peroxide and hypochlorous acid resulting in both oxidized and chlorinated bases. Among these damage products is 5-chlorouracil (ClU), which could arise by chlorination and deamination of cytosine residues in DNA and nucleotides or chlorination of uracil bases present in nucleotide precursor pools (6–10) as shown in Fig. 1. Previous studies have demonstrated that significant levels of ClU can be incorporated into the DNA of replicating cells when 5-chloro-2’-deoxyuridine (CldU) is added as a nucleotide precursor (11–14).
Fig. 1. Formation of 5-chlorouracil (ClU).
ClU can be generated by two routes. Initial reaction of cytosine with HOCl can result in formation of the dihydro intermediate, which can then deaminate and dehydrate, generating ClU (upper pathway). Alternatively uracil can react with HOCl forming a dihydro intermediate and then dehydrate forming ClU (lower pathway).
The impact of the replacement of thymine with ClU in duplex DNA is not yet known. Although CldU is incorporated into the DNA of replicating cells, it does induce toxicity, senescence and sister-chromatid exchanges (15–20). In this paper, we have constructed synthetic oligonucleotides containing ClU residues at a defined position in a self-complementary duplex paired opposite adenine. The synthetic oligonucleotides have been characterized by mass spectrometry, thermal melting studies and NMR spectroscopy. In the duplex, the ClU residue pairs with adenine, forming two hydrogen bonds in a geometry approaching that of a normal thymine-adenine Watson-Crick base pair. We observe that the replacement of thymine with ClU paired opposite adenine has only a slight impact on oligonucleotide stability, although the linewidth of the ClU imino proton broadens much faster with increasing temperature than does the corresponding thymine imino proton. The potential for single-proton transfer from within the intact base pair is considered. The ClU-containing oligonucleotide was probed with the repair glycosylase MUG, which is known to exploit reduced duplex stability in its search for target bases. Whereas ClU mispaired with guanine is readily repaired, ClU paired opposite adenine within the same sequence context is not repaired. These data suggest that the replacement of thymine with ClU does not significantly perturb the structure or dynamics of a DNA duplex.
Materials and methods
Oligonucleotide synthesis
Commercially available 2'-deoxyuridine was converted to CldU using the method of Kumar et al. (21). The corresponding phosphoramidite of CldU was prepared by standard methods, as previously described (22). All other phosphoramidites were obtained from Glen Research (Sterling, VA). Oligonucleotide synthesis was conducted with an Expedite synthesizer from Applied Biosystems (Foster City, CA). Oligonucleotides containing CldU were deprotected with concentrated aqueous ammonia at room temperature for 24 h.
The sequence of the 12-mer oligonucleotide examined here is shown in Fig 2. Oligonucleotides were purified by two rounds of HPLC, first with the DMT group on, and the second with the DMT group off. Oligonucleotides were examined by MALDI-ToF MS (23) and the free base composition was verified by GC/MS following acid hydrolysis (24).
Fig. 2. Oligonucleotide sequences.
a 12 mer oligonucleotide sequence used for the study, X= 5-chlorouracil. b Watson-Crick base pairing of ClU-A base pair.
Oligonucleotide UV Melting studies
The melting temperature (Tm) and thermodynamic values of the oligonucleotide were obtained as previously described (23, 30) using a Varian Bio 300 Cary UV Vis spectrophotometer (Palo Alto, CA). The self complementary oligonucleotide at various concentrations (2 µM to 60 µM) was dissolved in a buffer containing 100 mM NaCl, 0.1 mM EDTA, and 10 mM sodium phosphate at pH 7. The sample was then treated with a thermal cycle from 10 °C to 90 °C at an interval of 0.5 °C. The thermodynamic values are obtained from the average of five such temperature cycles. Tm values are reported at 28 μM total strand concentration.
Nuclear Magnetic Resonance Spectroscopy (NMR)
NMR spectra were obtained with a 500 MHz Bruker NMR system (Billerica, MA.). The proton NMR spectra of the oligonucleotide were taken in a solution containing 10% D2O, 100 mM NaCl, 10 mM sodium phosphate and 0.2 mM EDTA at pH 7.0 and compared with results from similar sequences published by other laboratories (25–28). The oligonucleotide (200 A260 OD units, 2.5 mM) was annealed at 86 °C for 2 min and slowly cooled prior to acquisition of NMR spectra. Each spectrum was calibrated using DSS (4,4-dimetyl-4-silapentane-1-sulphonate) as an internal standard reference. Proton NMR spectra of oligonucleotides in 90% H2O were acquired with a water suppression double gradient echo WATERGATE W5 pulse program (29). The temperature of the sample was controlled by a variable temperature monitor (Eurotherm, BVT 3000) from Bruker.
A typical spectrum was acquired with a binomial water suppression delay (d19) of 160 µsec. Since the delay (d19) of 160 µsec affected some of the imino resonances near 11 ppm, it was changed to 40 µsec when high peaks for the imino protons were desired. The null point repetition was every 6.25 ppm from the suppression point when the d19 was 160 µsec. When the delay was 40 µsec, the null point repetition was far from any of the imino proton resonances. The line width studies were performed using the d19 delay of 40 µsec. The 2D NOE spectra were acquired using a mixing time of 300 ms
Mispaired uracil glycosylase (MUG) reaction with ClU oligo
The glycosylase studies were performed using mispaired uracil glycosylase (MUG) from Escherichia Coli (30). A typical reaction was done by reacting 300 pmol oligonucleotide with 14.7 pmol of enzyme in a buffer containing 20 mM NaCl, 20 mM tris-HCl, 1 mM EDTA, 1 mM DTT at pH 8. The reaction mixture was incubated at 37 °C overnight. The sample was then desalted for MALDI-TOF analysis with a P6 column and a cation exchange column containing AG50W-X8 beads on the H+ form (Bio-Rad, Hercules, CA). MALDI-TOF experiments were performed with a Bruker Autoflex time of flight mass spectrometer operated in positive ion and reflectron modes (23).
Results
Oligonucleotides containing ClU at a defined site were prepared by standard phosphoramidite methods. Commercially available dU was converted to CldU using a previously reported method (22). Oligonucleotides containing ClU were synthesized, deprotected and purified using standard conditions (Fig. 2), except that deprotection was conducted in concentrated aqueous ammonia at room temperature for 24 h. The base composition of each oligonucleotide was verified by GC/MS following acid hydrolysis (Fig. 3). Due to the high natural abundance of both 35Cl and 37Cl isotopes, the mass spectrum of chlorine-containing molecules has a significant M+2 line for the parent ion and M-methyl ion. Intact oligonucleotides were also examined by MALDI-TOF-MS as shown in Fig. 3. The isotope envelope of larger molecules can also be simulated, and the presence of one chlorine atom in the oligonucleotide profoundly impacts the shape of the isotope envelope. The simulated ion cluster of an oligonucleotide of composition C116H145N45O70P11Cl is shown above and matches the experimental spectrum in Fig. 3a. The theoretical and observed mass, respectively, for each of the oligonucleotides examined here are: T-A 3644.65, 3644.61; ClU-A 3664.59, 3664.54 and ClU-G 3680.59, 3680.54.
Fig. 3. Oligonucleotide analysis by mass spectrometry.
a Experimental (bottom) and theoretical (top) isotope envelope of the 12mer oligonucleotide 5’-CGClUGAATTCACG-3’ used in this study. The experimental isotope envelope is obtained from MALDI-TOF mass spectrometry and the theoretical envelope is obtained using a computer simulation program (ref 23). b GC-MS spectrum of ClU after acid hydrolysis of ClU-A oligonucleotide. Molecules containing chlorine are easily recognized by the prominent M+2 peaks (277 and 292 amu) in the mass spectrum.
The thermal stability of the oligonucleotide duplex (31–33) containing ClU was examined and compared with that of an oligonucleotide containing T. The melting temperature of the ClU-containing oligonucleotide was observed to be 55.6 °C, while that of the control oligonucleotide containing T was 53.4 °C. Thermodynamic parameters obtained are shown in Table 1. The thermal melting profiles of the T and ClU-containing oligonucleotides are shown in Fig. 4.
Table 1.
Thermal melting and thermodynamic parameters for the helix-coil transition of 5’-d[CGXGAATTCACG]-3’ at 10 mM sodium phosphate, 100 mM sodium chloride, 0.1 mM EDTA, pH 7.0.
| T-A | ClU-A | |
|---|---|---|
| Tm (°C) | 53.4 ± 0.3 | 55.6 ± 0.3 |
| ΔH° (kcal/mol) | −84.7 ± 13 | −80.7 ± 14 |
| ΔS° (cal/mol/k) | −238.0 ± 40 | −233.8 ± 4 |
| ΔG° (kcal/mol) | −11.9 ± 1.0 | −12.2 ± 1.0 |
| Hyperchromicity (%) | 22.0 ± 0.6 | 21.3 ± 0.8 |
Fig. 4. Oligonucleotide melting profiles examined by temperature dependent absorbance at 260 nm.
UV melting profiles (normalized) of 12 mer oligonucleotide 5’-CGXGAATTCACG-3’ at 28 µM concentration in a buffer containing 100 mM sodium chloride, 10 mM sodium phosphate, 0.1 mM EDTA at pH 7.0. T-A (solid line) and ClU-A (dotted line).
Proton NMR spectra of the monomers of thymidine and CldU were obtained to serve as references for oligonucleotide-induced changes in chemical shifts. The H6 proton resonances of dT and CldU in D2O were observed to be 7.64 and 8.17 ppm, respectively. The chemical shifts of the N3 imino proton resonances of dT and CldU in DMSO were observed to be 11.26 and 11.82 ppm respectively. The imino proton of ClU is substantially more acidic than the corresponding imino proton of T. The pK of the CldU imino proton was measured by titration of the chemical shift of the H6 resonances and found to be 8.0 (supplementary data).
Proton NMR spectra of the non-exchangeable protons of each duplex were obtained and shown in Fig. 5. Proton resonances were assigned by standard 2D methods (34) and are shown in Table 2. The corresponding 2D spectra are shown as supplementary figures. The observed proton connectivities indicate that both the T-A and ClU-A duplexes are predominantly in a B-form geometry and all bases are intrahelical. The H6 proton for T3 of the T-A oligonucleotide is observed at 7.12 ppm, whereas the corresponding H6 proton of the ClU residue of the ClU-A duplex was observed at 7.61 ppm.
Fig. 5. NMR spectrum of non-exchangeable protons.
1H NMR spectra of the ClU-A (bottom) and T-A (top) oligonucleotide in the aromatic region. The H6/H8 and H2 assignments are written in the top of the spectrum. H6 of ClU3 and T3 is indicated by the arrow mark. The spectra were taken at 30 °C with 256 scans using a water suppression WATERGATE program with binomial water suppression delay (d19) of 160 µsec. The oligonucleotide was dissolved in a buffer containing 10% D2O, 100 mM sodium chloride, 10 mM sodium phosphate, 0.2 mM EDTA at pH 7.
Table 2.
Chemical shifts of aromatic protons of the oligonucleotide, 5’-d[CGXGAATTCACG]-3’ at pH 7.0 and 30 °C.
| Nucleobase | H6/H8 | H2/H5 | ||
|---|---|---|---|---|
| T-A | ClU-A | T-A | ClU-A | |
| C1 | 7.64 | 7.65 | 5.91 | 5.92 |
| G2 | 7.97 | 7.97 | ||
| T3/ClU3 | 7.12 | 7.61 | ||
| G4 | 7.84 | 7.86 | ||
| A5 | 8.09 | 8.10 | 7.17 | 7.18 |
| A6 | 8.09 | 8.09 | 7.59 | 7.59 |
| T7 | 7.08 | 7.08 | ||
| T8 | 7.35 | 7.35 | ||
| C9 | 7.50 | 7.50 | 5.68 | 5.81 |
| A10 | 8.28 | 8.29 | 7.75 | 7.73 |
| C11 | 7.25 | 7.24 | 5.35 | 5.34 |
| G12 | 7.88 | 7.88 | ||
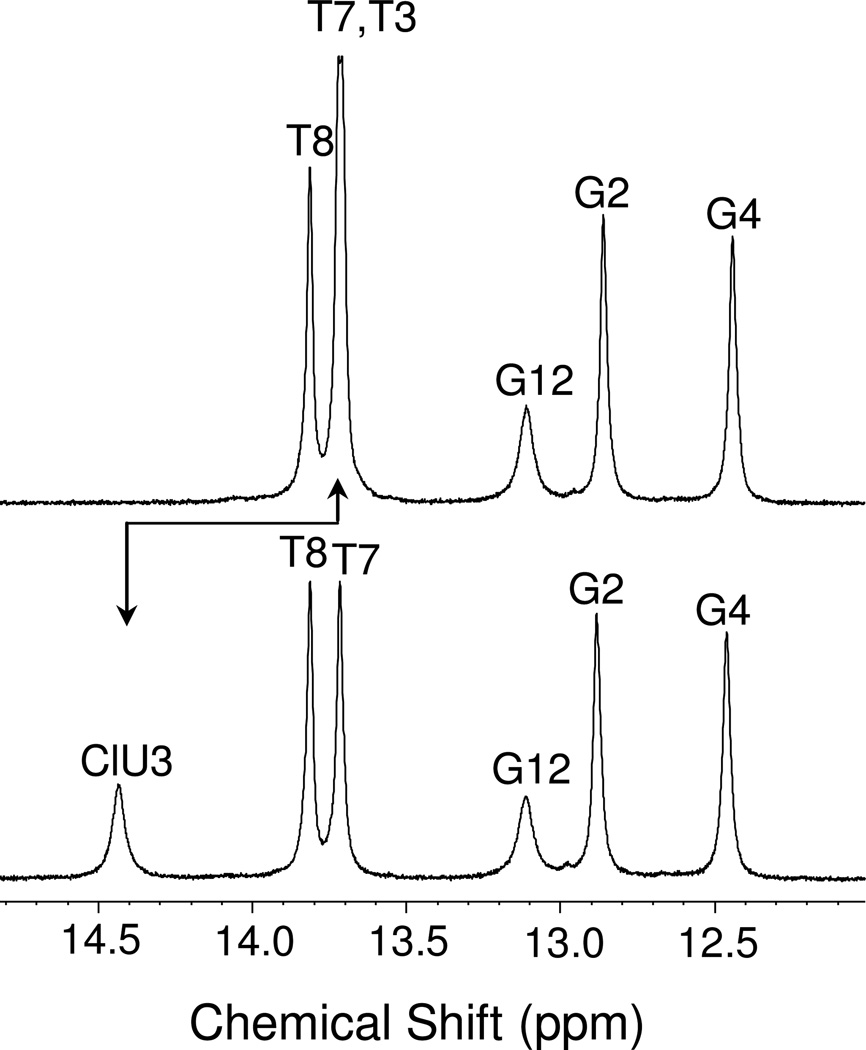
Proton spectra of the exchangeable proton resonances were obtained in 90% H2O, 10% D2O, and the spectra of the T-A and ClU-A duplexes are shown in Fig. 6. Proton resonances were assigned by standard 2D methods, as described previously. The assignments are recorded in Table 3. The N3 imino proton of dT in position 3 of the oligonucleotide was observed at 13.71 ppm and for CldU 14.43 ppm. The linewidth of the proton resonance assigned to the ClU imino proton was greater than most others in the duplex, and significantly more broad than the imino resonance of the corresponding T-A proton. The width of this proton increased rapidly with increasing temperature (Fig. 7). The rapid line broadening of the ClU imino proton appeared to be independent of solution pH (Fig. 7c).
Fig. 6. NMR spectrum of exchangeable protons.
1H NMR spectrum of the ClU-A (bottom) and T-A (top) oligonucleotides in the imino region. The spectra were taken at 5 °C with 256 scans using a water suppression WATERGATE program with binomial water suppression delay (d19) of 40 usec. The oligonucleotide was dissolved in a buffer containing 10% D2O, 100 mM sodium chloride, 10 mM sodium phosphate, 0.2 mM EDTA at pH 7.
Table 3.
Chemical shifts of imino protons of the oligonucleotide, 5’-d[CGXGAATTCACG]-3’ at pH 7.0 and 5 °C.
| Nucleobase | Imino protons | |
|---|---|---|
| T-A oligo |
ClU-A oligo |
|
| G2 | 12.86 | 12.88 |
| T3/ClU3 | 13.71 | 14.43 |
| G4 | 12.44 | 12.46 |
| T7 | 13.72 | 13.72 |
| T8 | 13.81 | 13.81 |
| G12 | 13.11 | 13.11 |
Fig. 7. Linewidths of imino protons as a function of temperature and pH.
a Temperature dependent imino proton line widths of different base pairs of T-A oligonucleotide at pH 7.0. b Temperature dependent imino proton line widths of different base pairs of ClU-A oligo at pH 7.0. c Temperature dependent imino proton line widths of ClU-A (3) base pair of ClU-A oligo at pH, 5.9, 7.0, 8.0, and 8.8. The oligonucelotide was dissolved in a buffer containing 10% D2O, 100 mM sodium chloride, 10 mM sodium phosphate, 0.2 mM EDTA.
The imino proton region of the ClU-A base pair examined here is shown in Fig. 8. The imino proton region of the duplex at 5 °C and pH 7.0 is shown in Fig. 8a. Under these conditions, the ClU and terminal G imino proton resonances are significantly broader than the remaining imino proton resonances. Upon increasing the solution pH to 8.8, the imino proton of the terminal G:C base pair broadens further and is lost from the spectrum, whereas the ClU imino proton has neither broadened nor shifted (Fig. 8b). Increase in sample temperature from 5 °C to 22 °C at pH 7.0 results in broadening and loss of both the terminal G:C and ClU imino protons (Fig. 8c).
Fig. 8.
1H NMR spectrum of the ClU-A oligonucleotide in the imino region at different pH and temperature: a at pH 7.0 and 5 °C b at pH 8.8 and 5 °C and c at pH 7.0 and 22 °C. The spectra were taken with 256 scans using a water suppression WATERGATE program with binomial water suppression delay (d19) of 40 µsec. The oligonucelotide was dissolved in a buffer containing 10% D2O, 100 mM NaCl, 10 mM sodium phosphate, 0.2 mM EDTA.
The enzymatic repair of ClU was investigated by probing the ClU-containing oligonucleotide with the repair glycosylase, MUG. A positive control sequence in which the ClU was paired opposite guanine was probed with MUG and the ClU was rapidly removed (Fig. 9). Enzymatic removal of the ClU residue generates an oligonucleotide containing an abasic site. The theoretical mass of the abasic-site containing oligonucleotide is 3552.59 amu and the observed mass is 3552.50 amu. The observed reduction in the oligonucleotide mass by 128 amu results from the displacement of ClU (−145 amu) by a hydroxyl group (+17 amu). Under identical reaction conditions, neither ClU (Fig. 9) nor thymine (data not shown) paired opposite adenine are repaired.
Fig. 9. Activity of MUG on synthetic oligonucleotides containing ClU-A or ClU-G.
MALDI spectra of oligonucleotides. a Mixture of T-A, ClU-A and ClU-G oligonucleotides without enzyme reaction, b ClU-A oligo reacted with MUG, and c ClU-G oligonucleotide incubated with MUG. The enzyme reaction was carried out by reacting 300 pmole oligonucleotide with 14.7 pmol of enzyme in a buffer containing 20 mM sodium chloride, 20 mM tris-HCl, 1 mM EDTA, 1 mM DTT at pH 8. The incubation was at 37 °C for 20 hrs.
Discussion
Emerging studies show that DNA can be damaged by reactive molecules derived from activated neutrophils and eosinophils at sites of inflammation (1–9). Among the damage products are oxidized and halogenated pyrimidines, including ClU (Fig. 1). It has been suggested that these damage products might provide a mechanistic link between inflammation and human diseases including cancer (5).
It is well established that halogenated pyrimidines can be incorporated into nucleotide precursor pools and ultimately into DNA. FU (5-fluorouracil) is used as a chemotherapy agent as its metabolite (5'-fluoro-2'-deoxyuridine-5'-monophosphate) inhibits thymidylate synthase needed for the conversion of dUMP to TMP (35, 36). Some FU is also incorporated into DNA where it can be removed by several glycosylases (37, 38). BrU is a mutagenic base analog long known to induce transition mutations, presumably by miscoding more frequently than T (39, 40). Replicating cells can tolerate substantial replacement of T with BrU (41, 42). Similarly, ClU can be incorporated into the DNA of replicating cells (6, 10, 11–15). However, ClU has been shown to induce cellular toxicity, cause senescence, and increase the frequency of sister-chromatid exchanges (15–19). It has been proposed that the toxicity might in part be attributed to interference with nucleotide metabolism, which promotes dUTP misincorporation and repair by a mechanism similar to that proposed for FU (18). However, the mechanism of sister chromatid exchange remains to be resolved.
Due to the recent interest in ClU in DNA, we have incorporated ClU into synthetic oligonucleotides to ascertain the base-pairing configuration when paired opposite adenine and to examine the impact of ClU substitution on the stability of an oligonucleotide duplex. Oligonucleotides containing ClU were prepared and purified by standard methods (Fig. 2). The phosphoramidite of ClU is prepared by standard methods, and no special deprotection conditions are required. The composition of oligonucleotides containing ClU can be verified by acid hydrolysis followed by GC/MS analysis (Fig. 3). The presence of chlorine in a molecule profoundly impacts the corresponding mass spectra, allowing unambiguous identification of the chlorinated pyrimidine. Similarly, the presence of chlorine alters the MALDI-TOF-MS of the intact oligonucleotide (Fig. 3).
In order to investigate the impact of the ClU substitution on oligonucleotide stability, oligonucleotide melting temperatures were determined by measuring UV absorbance as a function of temperature. Example results for the T-A and ClU-A duplexes are shown in Fig. 4. In the ClU-A duplex, two ClU-A pairs are substituted for two T-A base pairs. The results show that substitution of dT with CldU increases the oligonucleotide melting temperature by 2.2 °C. We note that the self-complementary sequence used here has two substitutions per duplex. Therefore, the change in melting temperature is approximately 1.1 °C per CldU substitution. Thermodynamic parameters were obtained by measuring Tm's as a function of oligonucleotide concentration. The values obtained are recorded in Table 1. The substitution of T with ClU, therefore, increases slightly the thermal and thermodynamic stability of an oligonucleotide duplex.
Duplex conformation and base-pairing configuration were examined with NMR spectroscopic methods. All of the expected non-exchangeable proton resonances were observed for the T-A and ClU-A duplexes, as shown in Fig. 5. The only difference between the two duplexes is that the protons of T3, and in particular the T3 H6 proton resonance at 7.12 ppm is lost upon substitution of T with ClU, with a corresponding gain of a proton resonance at 7.61 ppm assigned to the H6 proton of the ClU residue. The difference in the chemical shifts of the H6 protons of T and ClU is attributed to the difference in the electronic-inductive property of the 5-methyl versus 5-chloro substituent. The chemical shift of T3 H6 moves upfield, relative to the monomer chemical shift, by 0.52 ppm, whereas that of the H6 of ClU moves upfield by 0.56 ppm. The similar magnitude of the stacking-induced upfield shift indicates both the T and ClU experience similar base stacking and geometry relative to the bases above and below in the helix. However, the greater relative stacking induced shift of the ClU H6 proton is consistent with the observed increase in the thermodynamic stability of the ClU-containing duplex discussed above. The observed chemical shift of the H6 proton of ClU when paired with A indicates it is in the neutral and keto tautomeric form (28). The base-sugar connectivities observed indicate that the ClU-A duplex, as with the T-A duplex, is predominantly B-form.
The impact of the ClU substitution on base pair formation was investigated by examining exchangeable proton resonances. Exchangeable spectra are shown in Fig. 6. The T3 imino proton of the T-A duplex at 13.71 ppm is lost upon substitution with ClU; however, a new resonance is observed at 14.43 ppm, assigned to the ClU imino proton. The downfield shift in the N3 resonance from dT to CldU of 0.72 ppm can be attributed primarily to the opposing electronic inductive properties of the methyl and chloro substituents. The N3 resonances of dT and CldU monomers in DMSO are 11.26 and 11.82 ppm, respectively, a difference of 0.56 ppm. The observed chemical shift for an imino proton in an oligonucleotide is a function of both the intrinsic chemical shift and hydrogen bond formation. Upon substitution of dT by CldU, the observed imino proton chemical shift moves further downfield than can be attributed exclusively to the intrinsic chemical shift difference. The observed data would suggest that the hydrogen bond formed by the N3 proton of CldU is stronger than that of the corresponding dT base pair. Increased strength of this hydrogen bond is consistent with the increase in the observed acidity of the CldU imino proton as well as with results of theoretical studies (43). Consistent with the results reported here for the ClU-A base pair, previous NMR studies with both the FU-A (44, 45) and BrU-A (46) base pairs indicate base pair configurations similar to the T-A base pair.
The structural and thermodynamic properties of the T-A and ClU-A duplexes appear to be very similar in many ways. However, a distinguishing feature of the ClU-A pair is that the proton resonance assigned to the ClU imino proton is substantially more broader than the corresponding T-A proton within the same sequence context (Fig 7 and 8): the linewidth of the T imino proton of the T-A duplex at 30°C is 10.7 Hz whereas the linewidth of the corresponding ClU-imino proton of the ClU-A duplex is 110 Hz. We considered the possibility that base-catalyzed exchange of the more acidic ClU imino proton might destabilize and reduce the lifetime of the ClU-A base pair as shown in Fig. 10B. Increasing the solvent pH from 7.0 to 8.8 at 5 °C results in broadening of the terminal G:C imino protons, attributable to an increased rate of base pair opening (47, 48). However, the ClU-A imino proton is neither shifted nor broadened although the solution pH has been increased to nearly 1 pH unit above the pKa of the monomer, CldU. This result indicates that formation of the ClU-A base pair sequesters the imino proton, suppressing the transfer of the ClU imino proton to solvent water. Indeed, the results of theoretical calculations (43) suggest that the ClU-A imino proton hydrogen bond is both shorter and stronger than the corresponding bond in the T-A base pair, consistent with the hydrogen-bonding induced shifts as discussed above.
Fig. 10. Proposed proton tunneling between ClU and A residues in a DNA duplex.
A) neutral base pair in pseudo Watson-Crick geometry, B) ionized base pair resulting from exchange of ClU N3 proton with solvent, and C) Base pair following proton transfer from ClU to A.
The observed broadening of the ClU-A cannot be attributed to reduced duplex stability as the oligonucleotide containing the ClU-A base pair is more thermally and thermodynamically stable than the corresponding T-A control oligonucleotide (Fig. 4 and Table 1). Previously, Sternglanz and Bugg concluded from examination of the crystal structures of 5-chloro and 5-bromouracil that halogenated uracil analogs may have enhanced base-stacking interactions (49). The linewidths of the imino protons of the base pairs above and below the ClU-A base pair are identical with the linewidths of the corresponding protons observed for the T-A oligonucleotide control, indicating that the ClU-A base pair does not create a denaturation bubble within the helix. The observed broadening of the ClU-A imino proton cannot be attributed to enhanced exchange with solvent as discussed above. We therefore considered the possibility that the broadening of the ClU-A imino proton might result from proton exchange from within the intact base pair.
Previously, Shulman (50) considered single proton transfer of the N3 imino proton of T to the complementary A residue in a Watson-Crick T-A base pair. Single proton transfer renders the T residue anionic and the A residue protonated. The energy cost for this transfer is the sum of the energetic cost of ionizing both the T and A monomers at pH 7.2, and is estimated to be approximately 7.6 kcal/mol so that the Zwitterionic base pair would be present at a frequency of approximately one in 3.8×105 A:T base pairs. Replacement of the electron-donating 5-methyl group of T with the electron-withdrawing chloro substituent of ClU reduces the pKa of the N3 imino proton from 9.8 to 7.9–8.0. The energy cost of ionizing the ClU therefore drops the free energy of proton transfer by 2.6 kcal/mol to approximately 5.0 kcal/mol (51), increasing the frequency of the Zwitterionic base pair shown in Fig. 10C by a factor of approximately 80, relative to the canonical T:A base pair. Previous studies have demonstrated that base damage and ion binding can also facilitate inter-base pair single proton transfer (52, 53).
The pKa of the N3 position of monomeric 2'-deoxyadenosine is approximately 3.8 in solution (28). However, in some base-pairing configurations, the pKa of the N3 position of an adenine residue has been observed to increase to above 7.0 (54–56). Recent studies suggest that adenine protonation in a duplex can further stabilize base stacking interactions and can facilitate the formation of some nucleic acid structural motifs. Possibly, thermal motions that disrupt normal base stacking interactions could be offset by single proton transfer and adenine protonation. The reduced energy of single proton transfer for the ClU-A base pair might partially account for the slight increase in the stability of the ClU-A containing duplex.
The results of the physical studies described above suggest that the ClU substitution does not significantly alter the overall duplex structure or base pairing configuration, and increases slightly duplex stability. The increased broadening of the ClU imino proton suggests possible increased base pair mobility. We therefore sought to determine if these modest structural fluctuations could be detected by DNA repair glycosylases. Previous studies from this lab and others (30, 37, 38) have demonstrated that repair glycosylases can find and distinguish damaged and modified bases in DNA upon the basis of reduced stability and changes in glycosidic bond strength. The electron-withdrawing 5-chloro substituent of ClU enhances glycosylase removal by several glycosylases relative to T, which has an electron-donating methyl substituent. Target bases mispaired with G are more easily repaired than those paired with A due to the reduced stability of the mispair. We probed the ClU-containing duplex examined here with the Escherichia coli mispaired uracil DNA glycosylase, MUG. We observed that ClU mispaired with G is readily removed by MUG, as shown in Fig. 9. In contrast, ClU paired with A is not repaired. The apparent increased mobility of ClU when paired with A is insufficient to render the ClU base susceptible to glycosylase removal.
The similarity of the structures and stabilities of the oligonucleotides containing T-A and ClU-A base pairs demonstrated here is consistent with the results of studies showing that substantial amounts of CldU can be incorporated into the DNA of replicating cells (11–14). Substitution with ClU is not overtly toxic, but is more subtle in causing chromosomal aberrations (15–20). The results of this study suggest that once incorporated, ClU can form a fraudulent base pair with adenine, undetected by DNA repair glycosylases. The adverse effects of ClU substitution might only reveal themselves in biologically important unusual DNA structures required for DNA replication, repair or transcription. It is known that the thymine methyl group is important for the binding of sequence-specific DNA-binding proteins (57). While 5-bromo can substitute for 5-methyl in many DNA-protein interactions (58–60), 5-chloro is smaller and might perturb some DNA-protein interactions (61). On the other hand, one recent study suggests that 5-halogenated uracil residues in DNA could direct the formation of unusual macromolecular conformation (62).
Conclusions
In this paper we have investigated the base pairing configuration, duplex conformation and thermodynamic stability of a model oligonucleotide duplex containing ClU. The ClU-A duplex is equivalent to the corresponding T-A duplex with respect to overall conformation and base pairing. The ClU-A base pair is slightly more stable than the T-A base pair, and appears to adopt a similar geometry. Single proton transfer would be more likely in the ClU-A base pair relative to T-A, and the increased proton transfer from ClU to A could account in part for the apparent increased stability of the ClU-A-containing oligonucleotides. The ClU base, when paired with A is sufficiently stable to escape detection and removal by repair glycosylases. The adverse biological effects of ClU could result from the disruption by ClU of biologically important unusual DNA structures or sequence-specific DNA protein interactions. Studies are currently in progress to investigate these possibilities.
Supplementary Material
Abbreviations
- ClU
5-Chlorouracil
- MUG
Mispaired uracil DNA glycosylase
- DMT
Dimethoxytrityl
- GC/MS
Gas chromatography/Mass spectrometry
- MALDI-TOF-MS
Matrix-Assisted Laser Desorption/Ionization -Time of Flight Mass Spectrometry
- WATERGATE
Water suppression through gradient tailored excitation
- DSS
4,4-dimethyl-4-silapentane-1-sulphonate
- Tm
Melting temperature
- EDTA
Ethylenediaminetetraacetic acid
- NOE
Nuclear Overhauser Effect
- DTT
Dithiothreitol
- DMSO
Dimethyl sulfoxide
- FU
5-Fluorouracil
- BrU
5-Bromouracil
- dUMP
2’-deoxyuridine monophosphate
- TMP
Thymidine monophosphate
Footnotes
This work is supported by grants from the Grants of the National Institute of Health.
References
- 1.Henderson JP, Byun J, Takeshita J, Heineke JW. Phagocytes produce 5-chlorouracil and 5-bromouracil, two mutagenic products of myeloperoxidase, in human inflammatory tissue. J. Biol Chem. 2003;278:23522–23528. doi: 10.1074/jbc.M303928200. [DOI] [PubMed] [Google Scholar]
- 2.Jiang Q, Blount BC, Ames BN. 5-Chlorouracil, a marker of DNA damage from hypochlorous acid during inflammation- A gas chromatography-mass spectrometry assay. J. Biol. Chem. 2003;278:32834–32840. doi: 10.1074/jbc.M304021200. [DOI] [PubMed] [Google Scholar]
- 3.Takeshita J, Byun J, Nhan TQ, Pritchard DK, Pennathur S, Schwartz SM, Chait A, Heineke JW. Myeloperoxidase generates 5-chlorouracil in human atherosclerotic tissue- a potential pathway for somatic mutagenesis by macrophages. J. Biol. Chem. 2006;281:3096–3104. doi: 10.1074/jbc.M509236200. [DOI] [PubMed] [Google Scholar]
- 4.Knaapen AM, Güngör N, Schins RP, Borm PJ, Van Schooten FJ. Neutrophils and respiratory tract DNA damage and mutagenesis: a review. Mutagenesis. 2006;21:225–236. doi: 10.1093/mutage/gel032. [DOI] [PubMed] [Google Scholar]
- 5.Valinluck V, Sowers LC. Inflammation-mediated cytosine damage: A mechanistic link between inflammation and the epigenetic alterations in human cancers. Cancer Res. 2007;67:5583–5586. doi: 10.1158/0008-5472.CAN-07-0846. [DOI] [PubMed] [Google Scholar]
- 6.Hale JT, Bigelow JC, Mathews LA, McCormack JJ. Analytical and pharmacokinetic studies with 5-chloro-2’-deoxycytidine. Biochem. Pharmacol. 2002;64:1493–1502. doi: 10.1016/s0006-2952(02)01413-2. [DOI] [PubMed] [Google Scholar]
- 7.Kang JI, Sowers LC. Examination of hypochlorous acid-induced damage to cytosine residues in a CpG dinucleotide in DNA. Chem. Res. Toxicol. 2008;21:1211–1218. doi: 10.1021/tx800037h. [DOI] [PubMed] [Google Scholar]
- 8.Morris SM. the genetic toxicology of 5-fluoropyrimidines and 5-chlorouracil. Mutation Res. 1993;297:39–51. doi: 10.1016/0165-1110(93)90006-9. [DOI] [PubMed] [Google Scholar]
- 9.Whiteman M, Jenner A, Halliwell B. Hypochlorous acid-induced base modifications in isolated calf thymus DNA. Chem. Res. Toxicol. 1997;10:1240–1246. doi: 10.1021/tx970086i. [DOI] [PubMed] [Google Scholar]
- 10.Visser DW, Frisch DM, Huang B. Synthesis of 5-chlorodeoxyuridine and a comparative study of 5-halodeoxyuridines in E. coli. Biochem. Pharmacol. 1960;5:157–164. doi: 10.1016/0006-2952(60)90017-4. [DOI] [PubMed] [Google Scholar]
- 11.Jaunin F, Visser AE, Cmarko D, Aten JA, Fakan S. A new immunocytochemical technique for ultrastructural analysis of DNA replication in proliferating cells after application of two halogenated deoxyuridine. J. Histochem. Cytochem. 1998;46:1203–1209. doi: 10.1177/002215549804601014. [DOI] [PubMed] [Google Scholar]
- 12.Svetlova M, Solovjeva L, Blasius M, Shevelev I, Hubscher U, Hanawalt P, Tomilin N. Differential incorporation of halogenated deoxyuridines during UV-induced DNA repair synthesis in human cells. DNA Repair. 2005;4:359–366. doi: 10.1016/j.dnarep.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Yamada K, Semba R, Ding X, Ma N, Nagahama M. Discrimination of cell nuclei in early S-phase, mid-to-late S-phase, and G2/M-phase by sequential administration of 5-bromo-2’-deoxyuridine and 5-chloro-2’-deoxyuridine. J. Histochem. Cytochem. 2005;53:1365–1370. doi: 10.1369/jhc.4A6601.2005. [DOI] [PubMed] [Google Scholar]
- 14.Vecchio L, Solimando L, Biggiogera M, Fakan S. Use of halogenated precursors for simultaneous DNA and RNA detection by means of immunoelectron and immunofluorescence microscopy. J. Histochem. Cytochem. 2008;56:45–55. doi: 10.1369/jhc.7A7225.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heartlein MW, O’Neill JP, Pal BC, Preston RJ. The induction of specific-locus mutations and sister-chromatid exchanges by 5-bromo and 5-chloro-deoxyuridine. Mutat. Res. 1982;92:411–416. doi: 10.1016/0027-5107(82)90239-1. [DOI] [PubMed] [Google Scholar]
- 16.DuFrain RJ. Probing sister chromatid exchange formation with halogenated pyrimidines. Basic Life Sci. 1984;29:41–58. doi: 10.1007/978-1-4684-4889-4_3. [DOI] [PubMed] [Google Scholar]
- 17.Zwanenburg TS, Hansson K, Darroudi F, van Zeeland AA, Natarajan AT. Effects of 3-aminobenzamide on Chinese hamster cells treated with thymidine analogues and DNA-damaging agents. Chromosomal aberrations, mutations and cell-cycle progression. Mutat. Res. 1985;151:251–262. doi: 10.1016/0027-5107(85)90077-6. [DOI] [PubMed] [Google Scholar]
- 18.Brandon ML, Mi L-J, Chaung W, Teebor G, Boorstein RJ. 5-chloro-2’-deoxyuridine cytotoxicity results from base excision repair of uracil subsequent to thymidylate synthase inhibition. Mutat. Res. 2000;459:161–169. doi: 10.1016/s0921-8777(99)00061-0. [DOI] [PubMed] [Google Scholar]
- 19.Michishita E, Kurahashi T, Suzuki T, Fukuda M, Fujii M, Hirano H, Ayusawa D. Changes in nuclear matrix proteins during senescence-like phenomenon induced by 5-chlorodeoxyuridine in HeLa cells. Experimental Gerontol. 2002;37:885–890. doi: 10.1016/s0531-5565(02)00033-5. [DOI] [PubMed] [Google Scholar]
- 20.Polak MB, Valamanesh F, Felt O, Torriglia A, Jeanny JC, Bourges JL, Rat P, Thomas-Doyle A, BenEzra D, Gurny R, Behar-Cohen F. Controlled delivery of 5-chlorouracil using poly(ortho esters) in filtering surgery for glaucoma. Invest. Ophthalmol. Vis. Sci. 2008;49:2993–3003. doi: 10.1167/iovs.07-0919. [DOI] [PubMed] [Google Scholar]
- 21.Kumar R, Wiebe LI, Knaus EE. A mild and efficient methodology for the synthesis of 5-halogeno uracil nucleosides that occur via a 5-halogeno-6-azido-5,6-dihydro intermediate. Can. J. Chem. 1994;72:2005–2010. [Google Scholar]
- 22.Van Aerschot A, Everaert D, Balzarini J, Augustyns K, Jie L, Janssen G, Peeters O, Blaton N, De Ranter C, De Clercq E, et al. Synthesis and anti-HIV evaluation of 2’,3’-dideoxyribo-5-chloropyrimidine analogues: reduced toxicity of 5-chlorinated 2’,3’-dideoxynucleosides. J. Med. Chem. 1990;33:1833–1839. doi: 10.1021/jm00168a046. [DOI] [PubMed] [Google Scholar]
- 23.Cui Z, Theruvathu JA, Farrel A, Burdzy A, Sowers LC. Characterization of synthetic oligonucleotides containing biologically important modified bases by matrix-assisted laser desorption time-of-flight mass spectrometry. Anal. Biochem. 2008;379:196–207. doi: 10.1016/j.ab.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang JI, Jr, Burdzy A, Liu P, Sowers LC. Synthesis and characterization of oligonucleotides containing 5-chlorocytosine. Chem. Res. Toxicol. 2004;17:1236–1244. doi: 10.1021/tx0498962. [DOI] [PubMed] [Google Scholar]
- 25.Patel DJ, Kozlowski SA, Marky LA, Broka C, Rice JA, Itakura K, Breslauer KJ. Premelting and melting transitions in the d(CGCGAATTCGCG) self-complementary duplex in solution. Biochemistry. 1982;21:428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]
- 26.Hare DR, Wemmer DE, Chou SH, Drobny G, Reid BR. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-TC-G-C-G) using two-dimensional nuclear magnetic resonance methods. J. Mol. Biol. 1983;171:319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- 27.Klevit RE, Wemmer DE, Reid BR. 1H NMR studies on the interaction between distamycin A and a symmetrical DNA dodecamer. Biochemistry. 1986;25:3296–3303. doi: 10.1021/bi00359a032. [DOI] [PubMed] [Google Scholar]
- 28.Sowers LC, Fazakerley GV, Kim H, Dalton L, Goodman MF. Variation of nonexchangeable proton resonance chemical shifts as a probe of aberrant base pair formation in DNA. Biochemistry. 1986;25:3983–3988. doi: 10.1021/bi00362a002. [DOI] [PubMed] [Google Scholar]
- 29.Liu ML, Mao XA, Ye CH, Huang H, Nicholson JK, Lindon JC. Improved WATERGATE Pulse Sequences for Solvent Suppression in NMR Spectroscopy. J. Magn. Reson. 1998;132:125–129. [Google Scholar]
- 30.Liu P, Theruvathu JA, Darwanto A, Lao VV, Pascal T, Goddard W, 3rd, Sowers LC. Mechanisms of base selection by the Escherichia coli mispaired uracil glycosylase. J. Biol. Chem. 2008;283:8829–8836. doi: 10.1074/jbc.M707174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from base sequence. Proc. Natl. Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SantaLucia J, Jr, Allawi HT, Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 33.Allawi HT, SantaLucia J., Jr Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 34.Weiss MA, Patel DJ, Sauer RT, Karplus M. Two-dimensional 1H NMR study of the lambda operator site OL1: a sequential assignment strategy and its application. Proc. Natl. Acad. Sci. USA. 1984;81:130–134. doi: 10.1073/pnas.81.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol. Ther. 1990;48:381–395. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- 36.Wyatt MD, Wilson DM., 3rd Participation of DNA repair in the response to 5-fluorouracil. Cell Mol. Life Sci. 2009;66:788–799. doi: 10.1007/s00018-008-8557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P, Burdzy A, Sowers LC. Substrate recognition by a family of uracil-DNA glycosylases: UNG MUG and TDG. Chem. Res. Toxicol. 2002;15:1001–1009. doi: 10.1021/tx020030a. [DOI] [PubMed] [Google Scholar]
- 38.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miki K, Shimizu M, Fujii M, Hossain MN, Ayusawa D. 5-Bromouracil disrupts nucleosome positioning by inducing A-form-like DNA conformation in yeast cells. Biochem. Biophys. Res. Commun. 2008;368:662–669. doi: 10.1016/j.bbrc.2008.01.149. [DOI] [PubMed] [Google Scholar]
- 40.Yu H, Eritja RE, Bloom LB, Goodman MF. Ionization of bromouracil and fluorouracil stimulates base mispairing frequencies with guanine. J. Biol. Chem. 1993;268:15935–15943. [PubMed] [Google Scholar]
- 41.Dunn DB, Smith JD. Effects of 5-halogenated uracils on the growth of Escherichia coli and their incorporation into deoxyribonucleic acids. Biochem. J. 1957;67:494–506. doi: 10.1042/bj0670494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stetson PL, Normolle DP, Knol JA, Johnson NJ, Yang ZM, Sakmar E, Prieskorn D, Terrio P, Knutsen CA, Ensminger WD. Biochemical modulation of 5-bromo-2’-deoxyuridine and 5-iodo-2’-deoxyuridine incorporation into DNA in VX2 tumor-bearing rabbits. J. Natl. Cancer Inst. 1991;83:1659–1667. doi: 10.1093/jnci/83.22.1659. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z, Rodgers MT. Influence of halogenation on the properties of uracil and its noncovalent interactions with alkali metal ions. Threshold collision-induced dissociation and theoretical studies. J. Am. Chem. Soc. 2004;126:16217–16226. doi: 10.1021/ja045375p. [DOI] [PubMed] [Google Scholar]
- 44.Sowers LC, Eritja R, Kaplan BE, Goodman MF, Fazakerley GV. Structural and dynamic properties of a fluorouracil-adenine base pair in DNA studied by proton NMR. J. Biol. Chem. 1987;262:15436–15442. [PubMed] [Google Scholar]
- 45.Kremer AB, Mikita T, Beardsley GP. Chemical consequences of the incorporation of 5-fluorouracil into DNA as studied by NMR. Biochemistry. 1987;26:391–397. doi: 10.1021/bi00376a009. [DOI] [PubMed] [Google Scholar]
- 46.Fazakerley GV, Sowers LC, Eritja RE, Kaplan BE, Goodman MF. Structural and dynamic properties of a bromouracil-adenine base pair in DNA studied by proton NMR. J. Biomol. Struct. Dynam. 1987;5:639–650. doi: 10.1080/07391102.1987.10506417. [DOI] [PubMed] [Google Scholar]
- 47.Várnai P, Canalia M, Leroy JL. Opening mechanism of G.T/U pairs in DNA and RNA duplexes: a combined study of imino proton exchange and molecular dynamics simulation. J. Am. Chem. Soc. 2004;126:14659–14667. doi: 10.1021/ja0470721. [DOI] [PubMed] [Google Scholar]
- 48.Guéron M, Leroy JL. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 49.Sternglanz H, Bugg CE. Relationship between the mutagenic and base-stacking properties of halogenated uracil derivatives. The crystal structure of 5-chloro- and 5-bromouracil. Biochim. Biophys. Acta. 1975;378:1–11. doi: 10.1016/0005-2787(75)90130-6. [DOI] [PubMed] [Google Scholar]
- 50.Shulman RG. the double minimum of the adenine-thymine hydrogen bond. Ann. NY Acad. Sci. 1969;158:96–99. doi: 10.1111/j.1749-6632.1969.tb56216.x. [DOI] [PubMed] [Google Scholar]
- 51.Sowers LC, Shaw BR, Veigl ML, Sedwick WD. DNA base modification: Ionized base pairs and mutagenesis. Mutat. Res. 1987;177:201–218. doi: 10.1016/0027-5107(87)90003-0. [DOI] [PubMed] [Google Scholar]
- 52.Zhang JD, Chen Z, Schaefer HF. Electron attachment to the hydrogenated Watson-Crick guanine cytosine base pair (GC+H): conventional and proton-transferred structures. J. Phys. Chem. A. 2008;112:6217–6226. doi: 10.1021/jp711958p. [DOI] [PubMed] [Google Scholar]
- 53.Noguera M, Bertran J, Sodupe M. Cu+2/+ cation coordination to adenine-thymine base pair. Effects on intermolecular proton-transfer processes. J. Phys. Chem. B. 2008;112:4817–4825. doi: 10.1021/jp711982g. [DOI] [PubMed] [Google Scholar]
- 54.Boulard Y, Cognet JA, Gabarro-Arpa J, LeBret M, Sowers LC, Fazakerley GV. The pH dependent configurations of the C.A mispair in DNA. Nucleic Acids Res. 1992;20:1933–1941. doi: 10.1093/nar/20.8.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moody EM, Brown TS, Bevilacqua PC. Simple method for determining nucleobase pK(a) values by indirect labeling and demonstration of a pK(a) of neutrality in dsDNA. J. Am. Chem. Soc. 2004;126:10200–10201. doi: 10.1021/ja047362h. [DOI] [PubMed] [Google Scholar]
- 56.Tang CL, Alexov E, Pyle AM, Honig B. Calculation of pKas in RNA: on the structural origins and functional roles of protonated nucleotides. J. Mol. Biol. 2007;366:1475–1496. doi: 10.1016/j.jmb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Ivarie R. Thymine methyls and DNA-protein interactions. Nucleic Acids Res. 1987;15:9975–9983. doi: 10.1093/nar/15.23.9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brennan CA, Van Cleve MD, Gumport RI. The effects of base analogue substitutions on the cleavage by the EcoR1 restriction endonuclease of octadeoxyribonucleotides containing modified EcoR1 recognition sequences. J. Biol. Chem. 1986;261:7270–7278. [PubMed] [Google Scholar]
- 59.Petruska J, Horn D. Sequence-specific responses of restriction endonucleases to bromodeoxyuridine substitution in mammalian DNA. Nucleic Acids Res. 1983;11:2495–2510. doi: 10.1093/nar/11.8.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jansco A, Botfield MC, Sowers LC, Weiss MA. An altered-specificity mutation in a human POU domain demonstrates functional analogy between the POU-specific subdomain and phage lambda repressor. Proc. Natl. Acad. Sci. USA. 1994;91:3887–3891. doi: 10.1073/pnas.91.9.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valinluck V, Wu W, Liu P, Neidigh JW, Sowers LC. Impact of cytosine 5-halogens on the interaction of DNA with restriction endonucleases and methyltransferase. Chem. Res. Toxicol. 2006;19:556–562. doi: 10.1021/tx050341w. [DOI] [PubMed] [Google Scholar]
- 62.Voth AR, Hays FA, Ho PS. Directing macromolecular conformation through halogen bonds. Proc. Natl. Acad. Sci. USA. 2007;104:6188–6193. doi: 10.1073/pnas.0610531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.