Abstract
Enzymatic methylation of cytosine residues in DNA, in conjunction with covalent histone modifications, establishes an epigenetic code essential for the proper control of gene expression in higher organisms. Once established during cellular differentiation, the epigenetic code must be faithfully transmitted to progeny cells. However, epigenetic perturbations can be found in most if not all cancer cells, and the mechanisms leading to these changes are not well understood. In this paper, we describe a series of experiments aimed at understanding the dynamic process of DNA methylation that follows DNA replication. Cells in culture can be propagated in the presence of 15N-enriched uridine which labels the pyrimidine precursor pool as well as newly replicated DNA. Simultaneous culture in the presence of 2H-enriched methionine results in labeling of newly methylated cytosine residues. An ensemble of 5-methylcytosine residues differing in the degree of isotopic enrichment is generated which can be examined by mass spectrometry. Using this method, we demonstrate that the kinetics of both DNA replication and methylation of newly replicated DNA are indistinguishable. The majority of methylation following DNA replication is shown to occur on the newly synthesized DNA. The method reported here does however suggest an unexpected methylation of parental DNA during DNA replication which might indicate a previously undescribed chromatin remodeling process. The method presented here will be useful in monitoring the dynamic process of DNA methylation and will allow a more detailed understanding of the mechanisms of clinically used methylation inhibitors and environmental toxicants.
Introduction
The proper control of gene expression is essential for all higher organisms that undergo development and differentiation. It has been more than twenty-five years since it was proposed that the presence of 5-methylcytosine (5mC)1 residues in DNA could form the basis of selective gene expression (1–3). In several systems, an inverse correlation between methylation of specific cytosine residues within promoter regions and the expression of downstream genes was demonstrated. Importantly, gene expression could be reestablished in model systems by using a series of small molecules that interfered with enzymatic cytosine methylation.
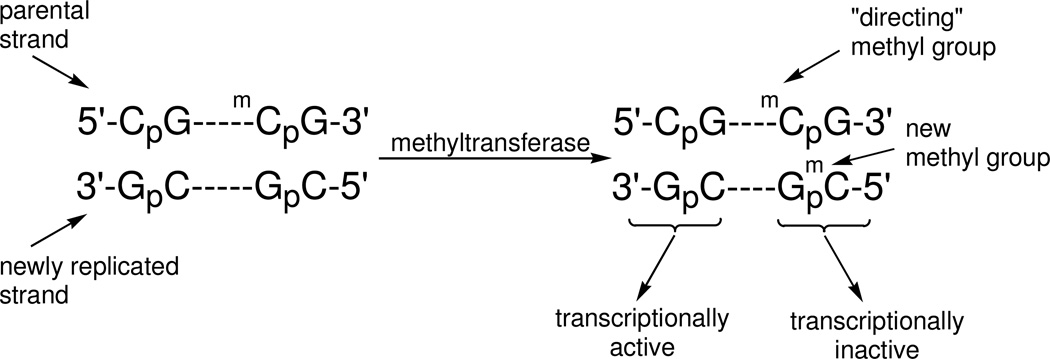
In eukaryotes, cytosine methylation occurs primarily in the CpG dinucleotide following DNA replication. The 5mC residue in a hemimethylated mCpG dinucleotide directs the maintenance methyltransferase (DNMT1) to methylate the newly replicated CpG site (Fig. 1), reestablishing symmetric DNA methylation during each round of DNA replication (4–5). The connection between cytosine methylation and histone modifications has been elucidated more recently. The methyl groups of a symmetrically methylated mCpG dinucleotide are found in the major groove of a B-form DNA helix. A motif common to a series of proteins known to bind with high affinity to methylated DNA is known as the methyl-binding domain, MBD (6). Cytosine methylation increases the affinity of proteins containing the MBD to DNA by a factor of 100 (7). Histone modifying enzymes then bind to the protein containing the methyl binding domain (8). Cytosine methylation therefore initiates a cascade of events that result in the formation of a highly compact chromatin structure in which genes in the vicinity are rendered inaccessible to transcriptional machinery.
While the heritable transmission of cytosine methylation patterns is essential to development and differentiation in higher organisms, defects in the transmission of methylation patterns can result in inappropriate silencing of tumor suppressor genes or the inappropriate derepression of transforming genes (3, 4, 9–12). Such defects are routinely found in human cancer cells, which often display paradoxical global loss of 5mC with site-specific increases in methylation (13). The mechanisms leading to alterations in cytosine methylation are not well understood.
Recently, several nucleoside analogs such as 5-azacytidine (14–16) and 5-aza-2’-deoxycytidine (14, 16–19), as well as non-nucleoside analogs (14) have been introduced into clinical trials for the treatment of several diseases including sickle cell anemia (18, 19), myelodysplastic syndrome (14–16) and leukemia (14). Preclinical studies performed with other demethylating agents including zebularine (14, 20, 21) and 5-fluoro-2’-deoxycytidine (14, 22) have also shown promise in effectively treating cancers. Although these drugs do reduce cytosine methylation, the mechanisms of action and toxicity are as yet unknown (14). We are therefore interested in developing a method to monitor cytosine methylation in cultured cells which would exploit the power of mass spectrometry.
Previously, cytosine methylation in cultured cells has been followed using radiolabeled precursors (23). Separate radiolabels have been used for cytosine and methionine, the methyl precursor for the methyltransferase. However, the use of stable isotopes and mass spectrometry would provide more information because the observed mass of the precursors is additive and the relative amounts of enriched isotopes in a given molecule can be determined. In this paper we demonstrate the use of stable isotope precursors with cells in culture that allows simultaneous measurement of DNA replication and DNA methylation. As expected, most of the methylation following DNA replication occurs on the newly replicated DNA. However, we also observed unexpected methylation of the parental DNA following DNA replication, suggesting a previously undescribed chromatin remodeling process. The method described here should prove useful in exploring the activity of several clinically important demethylating agents.
Materials and methods
Stable Isotope precursor of 5mC
Isotopically enriched analogs of uridine and 2'-deoxy-5-methylcytosine were prepared and characterized in this laboratory as described previously (24). Deuterated methionine (2H3-methionine) was obtained from Cambridge isotope laboratories (Cambridge, MA).
Growth of cells in the presence of isotopically enriched precursors
Human erythroleukemia K-562 cells obtained from ATCC (CLL-243) were grown at 37° C in a humidified incubator at 5% CO2 in RPMI-1640 media (Cellgro). Media was supplemented with 10% fetal bovine serum (Gibco) and 1% L-Glutamine-penicillin-streptomycin solution. Human prostate cancer DU-145 and human melanoma-derived SK-MEL-28 cells were also obtained from ATCC (HTB-81and HTB-72 respectively) and grown under identical conditions.
To start the experiment cells were harvested by centrifugation and washed with cold sterile PBS before resuspension in methionine free media (Caisson Laboratories, Inc) supplemented with 10% dialyzed FBS (Gibco). Cells were initially seeded at 3.0 – 4.0 × 105 cells/ml and treated with 100 µM 15N2-Uridine and 100 µM 2H3-methionine (filter sterilized). We chose to use 100 µM 2H3-methionine because this is the concentration of L-methionine normally present in the tissue culture media. Cells were counted daily by Trypan Blue using a hemacytometer, concentration recorded, aliquots of 1 × 107 cells removed and fresh media added containing 100 µM 15N2-uridine and 100 µM 2H3-methionine to bring the final cell concentration to about 3.0 – 4.0 × 105 cells/mL. Cells were placed back in the incubator until next time point. This was done daily, roughly every 30 h, for the treatment phase of the experiments. The 1 × 107 cells removed at each time point were pelleted by centrifugation (500 × g), washed with 10 mL sterile PBS, pelleted and stored at −20° C until DNA extraction and GC/MS analysis.
DNA Extraction and GC/MS analysis
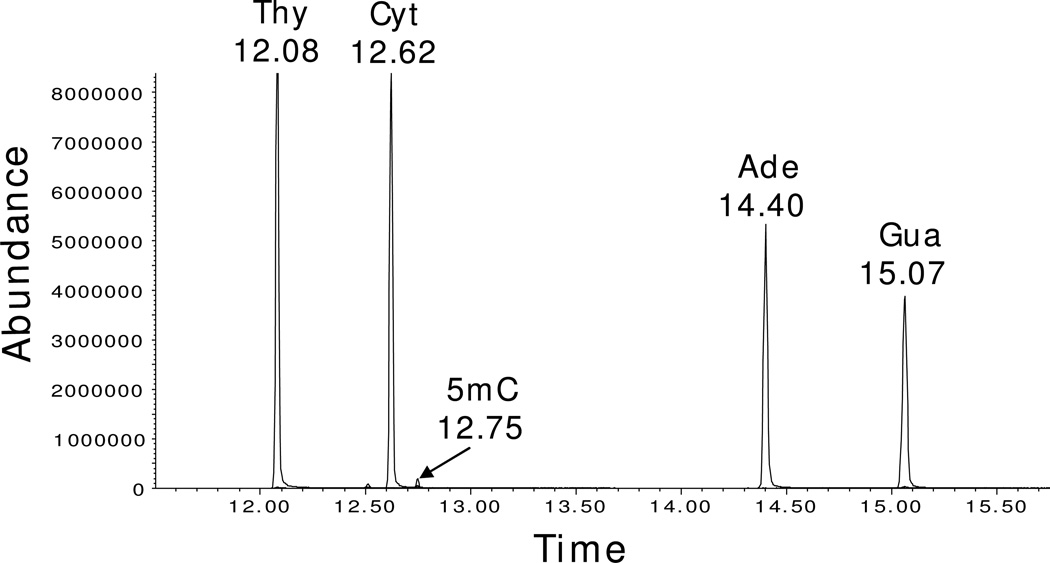
Frozen cell aliquots from samples were thawed and DNA was extracted using the Roche DNA Isolation Kit for Cells and Tissues. The amount of DNA in samples was determined by measuring the UV absorbance at 260 and 280 nm ratio. DNA samples containing approximately 40–50 µg was dried under reduced pressure. DNA was resuspended in 88% formic acid in sealed vials and heated at 140° C for 30 min. Formic acid was removed under reduced pressure and the residue containing purine and pyrimidine free bases was derivatized in 100 µL anhydrous acetonitrile and 100 µL MTBSTFA with 1% TBDMCS (Pierce) in sealed vials at 140° C for 30 min. Samples of approximately 1 µL were injected into the GC/MS using a 20:1 split ratio. Mass spectra were collected in selected ion mode, monitoring a series of masses for the M-57 fragment ion for each pyrimidine examined. The retention times (in min) for the silylated DNA bases were thymine (12.08), cytosine (12.62), 5-methylcytosine (12.75), adenine (14.40), and guanine (15.07).
The data reported in this paper was obtained using the above methods. Since these studies were completed, we have been able to optimize our methods to require significantly less DNA (1 µg). The optimized method is as follows: DNA from frozen cell aliquots was extracted using DNeasy Blood & Tissue Kit (Qiagen). The amount of DNA in samples was determined by measuring the UV absorbance at 260 and 280 nm ratio. DNA samples containing approximately 1 µg were dried under reduced pressure. DNA was resuspended in 88% formic acid in sealed vials and heated at 140° C for 40 min. Formic acid was removed under reduced pressure and the residue containing purine and pyrimidine free bases was derivatized in 20 µL anhydrous acetonitrile and 20 µL MTBSTFA with 1% TBDMCS (Pierce) in sealed vials at 140° C for 40 min. Samples of approximately 1 µL were injected into the GC/MS in splitless mode. Mass spectra were collected in selected ion mode and masses monitored as stated above.
Acid hydrolysis of DNA can potentially result in the deamination of cytosine, 5mC, adenine (25), and guanine (25) to uracil, thymine, hypoxanthine and xanthine, respectively. The hydrolysis method employed here was designed to minimize deamination of cytosine and 5mC. Several studies were conducted with unenriched and enriched 5mC isotopomers to determine the degree of deamination to the corresponding thymine isotopomers. Under the conditions of the experiments described here, using amounts of 5mC up to ten fold higher than found in the DNA samples, only trace amounts of thymine were observed and in all cases examined here, less than 0.1% deamination was observed. In the experiments described here, the relative abundance of the isotopomers is the important observable. In no case did we observe that the degree of deamination differed among the 5mC isotopomers examined.
Deconvolution of mass spectra
The mass spectrum of a given molecule is a composite of mass lines arising from the parent molecule and several fragments generated during ionization in the mass spectrometer. In the studies reported here, the predominant ion in the mass spectrum resulted from the loss of a 57 amu TBDMS fragment (M-57). The lines for this fragment ion are comprised of the monoisotopic peak and additional lines due to the presence of heavier stable isotopes at natural abundance.
If the molecular formula of a parent or fragment ion is known, the expected relative sizes of the mass lines in the mass spectrum of an isotopically unenriched compound can be calculated (26, 27) using the natural abundance of heavier isotopes (27). Enrichment with heavier isotopes alters the mass spectrum in a predictable manner, and the mass spectrum of the enriched molecule can be calculated similarly if the degree of enrichment is known.
In the studies reported here, isotopically-enriched precursors were used to follow the formation of cytosine, thymine and 5mC in replicating cells. The resulting mass spectrum of the M-57 ion for each pyrimidine is a linear combination of the mass spectrum for each of the isotopomers present in the mixture. The experimental mass spectra can be "deconvoluted" to yield the relative contribution of each of the possible isotopomers to the observed mass spectrum as described further in the Results and Discussion section. The relative peak height of the monoisotopic peak was used to obtain an initial value for the percentage of the unenriched isotopomer in the mixture. Initial values were subsequently obtained for each of the possible heavier isotopomers by comparison with the experimental spectrum. Using these initial values of percent contribution for each isotopomer to the experimental spectrum, a theoretical mass spectrum was generated and compared with the experimental spectrum. The relative contributions of the isotopomers was varied over a range of ± 5% from the initial values at 0.05% intervals, and the set of normalized coefficients reflecting the relative percentages of the possible isotopomers providing the best fit between the theoretical mass spectra and the experimental mass spectra was determined.
Determination of DNA replication and methylation kinetics
As cells are grown in the presence of stable isotope precursors, the mass spectrum of a given pyrimidine changes due to the incorporation of the stable isotopes. The mass spectrum of a given pyrimidine can be deconvoluted to reveal the percent contribution of the possible isotopomers present in the mixture as described in the above section. Following the percentage of the various isotopomers as a function of time can then be used to determine DNA replication kinetics as described below.
The contribution of the enriched isotope to the mass spectrum will increase and then plateau as the percentage of the heavier isotopomer approaches the concentration of the isotope-enriched precursor in the cell.
The percentage of enriched cytosine, %C+2, will increase with time (t), and can be determined from the following equation:
| (1) |
where is the fraction of C+2 enriched molecules in the dCTP precursor pool and k is the observed rate constant. The value of is determined by calculating the value approached asymptotically by %C+2 at the longest time points.
The percent unenriched cytosine (%C+0) at time t will decline due to dilution with C+2 enriched dCTP incorporated into the replicated DNA, but this decline is partially offset by incorporation of a lesser amount of unenriched dCTP which is also present in the precursor pool. The %C+0 can be expressed by the following equation where fC+0 is the fraction of unenriched cytosine in the DNA at time equal zero:
| (2) |
Similar equations represent the percent of unenriched thymine, %T+0 and enriched thymine, %T+2 in the DNA as a function of time beginning with the fraction of unenriched thymidine at time zero, fT+0:
| (3) |
| (4) |
The observed rate constants in all four cases above represent the rate of DNA replication, therefore the experimentally determined rates for all four cases measured independently should be identical. The relative enrichment of the dCTP () and dTTP () need not be the same, however. The relative enrichment of the corresponding dCTP and dTTP pools depends upon the activities of the respective de novo and salvage pathways that need not be the same for the cytosine and thymine precursors.
Monitoring the changes in the 5mC mass spectrum as a function of DNA replication is more complicated, as two enriched precursors, 15N2-uridine and 2H3-methionine are involved yielding four possible isotopomers of 5mC:
5mC+0: unenriched 5mC
5mC+2: 5mC derived from enriched cytosine C+2 and unenriched methionine
5mC+3: 5mC derived from unenriched cytosine and enriched methionine methyl+3
5mC+5: 5mC derived from enriched cytosine C+2 and enriched methionine methyl+3
Methylation of cytosine in the newly replicated DNA in the presence of both pyrimidine and methionine enriched precursors can be monitored by measuring the increase in the 5mC+2 and 5mC+5 isotopomers and will be functions of enrichment of both the cytosine pool and the methionine pool. The enrichment of the methionine pool also reaches a limiting value at long replication times, reflecting the degree of enrichment of the methionine pool, :
| (5) |
| (6) |
The unenriched 5mC isotopomer 5mC+0 is from original 5mC residues in parental DNA as well as residual unenriched cytosine from the dCTP precursor pool incorporated into the newly replicated DNA and methylated by residual unenriched methionine in the methionine pool. The 5mC+3 isotopomer results from the methylation of parental strand DNA or methylation of unenriched residual cytosine from the dCTP pool with new methyl groups.
| (7) |
| (8) |
The rate constant determined from the change in the contribution of the various isotopomers to the 5mC mass spectrum need not be the same as the DNA replication rate constant if the rates of replication and methylation differ from one another. However, if methylation follows closely replication, the two rate constants could be indistinguishable.
Results and Discussion
The analysis of DNA bases by GC/MS methods has been demonstrated to be a powerful and sensitive method applicable to several modified bases including 5mC (24, 26). As shown in Fig. 2, the normal DNA bases, including 5mC are well separated by GC. The integrated peak areas can be used to provide accurate quantitative measurement of the relative amounts of the bases. The level of cytosine methylation, as reflected by the 5mC/(5mC + C) ratio is generally between 2 and 5% in human cells (12, 28). We have extended the utility of this method as reported here to make dynamic measurements of DNA synthesis and methylation through the use of stable isotope precursors that are incorporated into DNA through biosynthetic pathways.
Figure 2. Measuring 5mC in DNA using GC/MS.
Chromatogram of DNA from K-562 cells processed for GC/MS analysis, showing separation of silylated Ade, Cyt, Gua, Thy, and 5mC. The mass spectrum of silylated 5mC produces a major fragment (M-57) at 296 m/z with a retention time of 12.75 min.
When cells are propagated in the presence of 15N-enriched uridine, the isotopically enriched pyrimidine is metabolized to both dCTP and dTTP, and ultimately incorporated into the DNA of replicating cells (29) as illustrated in Fig. 3. The incorporation of the stable-isotope enriched metabolites can be monitored by observing changes in the mass spectra of the corresponding bases hydrolyzed from isolated DNA. The propagation of cells in the presence of 2H-enriched methionine results in the incorporation of heavier stable isotopes into the methyl group of 5mC. By monitoring the changes in the mass spectra of cytosine, thymine and 5mC with cell growth, both DNA replication and DNA methylation could be followed simultaneously.
Figure 3. Dual label DNA enrichment scheme.
When grown in the presence of 15N-enriched uridine, cells metabolize the isotopically enriched pyrimidine to dCTP+2 and dTTP+2 which are then incorporated into DNA. The 2H3-methionine is metabolized to SAM+3, the donor in the enzymatic methylation of cytosine residues in DNA. The relative amounts of enriched and unenriched bases in the DNA are a function of the degree of enrichment of the corresponding precursor pool and the period of time in the enriched media. An ensemble of potential isotopomers of cytosine, thymine and 5mC are generated as indicated.
The mass spectrum of a given pyrimidine is a composite of mass lines arising from the parent molecule and several fragments generated during ionization in the mass spectrometer. Additional lines are present in the mass spectrum due to the presence of heavier stable isotopes at natural abundance. If the molecular formula of a parent or fragment ion is known, the relative heights of the mass lines in the mass spectrum can be calculated by use of a polynomial expression (26, 27). For example, the experimental and calculated ion abundance profiles for the predominant M-57 ion of unenriched 5mC are shown in Fig. 4. The molecular formula of the M-57 ion is C13H26N3OSi2 with a monoisotopic mass of 296 amu. Heavier lines at 297, 298, and 299 amu are observed in the experimental spectrum due to the presence of heavier isotopes of C, H, N, O, and Si at natural abundance. The relative abundance of the 296, 297, 298 and 299 lines are calculated to be 72.5%, 19.1%, 7.2% and 1.2%, respectively. The correlation coefficient (r2) between the observed and calculated relative abundance is 0.99. If the molecule of interest is isotopically enriched, an additional term reflecting the enriched ion and degree of enrichment is added to the polynomial describing that ion.
Figure 4. Comparison of the experimental and theoretical ion cluster for the predominant ion of unenriched 5mC.
The predominant ion observed for the TBDMS derivative of 5mC is the M-57 ion resulting from the loss of a tert-butyl group. The molecular formula of the 5mC ion is C13H26N3OSi2 with a monoisotopic mass of 296 amu. Mass spectral lines corresponding to heavier isotopomers of the 5mC M-57 ion of unenriched 5mC are observed due to the presence of heavier isotopes of C, H, N, O, and Si at natural abundance as indicated by the solid line. Upon the basis of the molecular formula and the known natural abundance of heavier isotopes, the contribution of heavier isotopomers can be calculated, and is indicated by the dashed line. The correlation coefficient (r2) between the expected and calculated relative abundance is 0.99.
When several isotopomers are present simultaneously, the observed mass spectrum for that ion is the sum of the individual spectra, each multiplied by their relative abundances. The calculated spectrum is then the algebraic sum of the calculated spectrum of each isotopomer multiplied by its relative abundance. The incorporation of precursor molecules that alter the abundance of heavier stable isotopes changes the experimental mass spectrum. Therefore, the relative abundance of enriched isotopomers in the mass spectrum can be determined by deconvolution of the mass spectrum.
The deconvolution method used here is demonstrated in Fig. 5. A series of isotopomers of 5-methyl-2'-deoxycytidine were prepared by synthetic methods and characterized by mass spectrometry as described previously (24). These isotopomers contained 13C in the methyl group (5mC+1), 15N in the ring-nitrogen positions (5mC+2) or deuterium in the H6 and methyl group (5mC+4). An equimolar mixture of the three heavier isotopomers and 5mC at natural abundance (5mC+0) was prepared, hydrolyzed, converted to the TBDMS derivative and analyzed by GC/MS (Fig. 5). A series of lines is observed in the mass spectrum of the predominant M-57 ion from 296 to 304 amu (Fig. 5).
Figure 5. Deconvolution of a mass spectrum resulting from an equimolar mixture of 5mC isotopomers.
A series of 5mC isotopomers were chemically synthesized and characterized by standard methods. An equimolar mixture of four isotopomers was prepared, hydrolyzed, derivatized and analyzed by GC/MS. The resulting mass spectrum of the M-57 fragment is shown in panel (A). The mass spectral lines from 296 to 304 amu arise from the ensemble of isotopomers. The experimental mass spectrum was deconvoluted to reveal the relative amounts of each of the isotopomers in the mixture. The relative amounts determined from the analysis (panel A) are within 1% of the expected amount (25% for each), and were used to generate a theoretical mass spectrum. The values of the observed and theoretical relative peaks heights are plotted in panel (B), with the line through the points described by the equation y = 1.0012×, and the correlation coefficient, r2 = 0.9988.
The mass spectrum of each of the component isotopomers can be predicted upon the basis of its molecular formula. For each individual isotopomer examined here, the theoretical and experimental spectrum were reproducibly indistinguishable. The composite spectrum of an isotopomer mixture can be represented as a linear combination of each of the component spectra. In order to determine the relative contribution of each of the four isotopomers in the mixture, the heights of the lines in the spectrum from 296 to 304 amu were summed. The height of the monoisotopic line at 296 amu relative to the sum of the lines in the cluster was used as the initial value for the percent contribution of the unenriched (5mC+0) isotopomer. The contribution of the 5mC+0 isotopomer to the heavier mass lines was calculated and subtracted from the remaining lines of the cluster. The relative contribution of the remaining 297 line to the sum of the lines was determined and used as the initial value for the contribution of the 5mC+1 isotopomer. This procedure was then repeated to determine initial values for contributions of the 5mC+2 and 5mC+4 isotopomers as well. Using these initial values as starting points the percent contributions of each of the isotopomers was varied over a range of ± 5% from the initial values, and the "predicted" spectrum for each possible ensemble of isotopomers was compared with the experimental spectrum. The normalized percent contribution of the isotopomers that best fit the experimental spectrum was determined and the values are shown in Fig. 5A. The fit between the experimental spectrum and the predicted spectrum generated from the relative percentage of isotopomers that best fits the experimental spectrum is shown in Fig. 5B. The expected percentages for this equimolar mixture and those determined by examination of the experimental mass spectrum varied by less than one percent. Using this method, the relative contribution of possible isotopomers can be obtained for any experimental mass spectrum.
When cells are propagated in the presence of 15N-enriched uridine, the uridine is metabolized into the dCTP and dTTP pools, and the nucleoside triphosphates in the partially enriched nucleotide pool are incorporated into the DNA. The mass spectra of the corresponding cytosine and thymine peaks are changed by the increasing proportion of the heavier isotopomers. Deconvolution of the experimental mass spectra can then reveal the relative amount of the heavier isotopomers incorporated into the DNA. With increasing incubation time in the presence of the enriched precursor, the percentage of the unenriched isotopomers falls, and the contribution of the heavier isotopomers increases as shown in Fig. 6. Equations describing the growth curves are indicated in Fig. 6 and are presented in Materials and Methods.
Figure 6. Change in isotopomer levels for cytosine and thymine in cells cultured in 15N-enriched uridine.
Cells (K-562) were cultured in the presence of both 100 µM 15N-enriched uridine and 100 µM 2H-enriched methionine for over five days. A fraction of the cells were harvested and DNA was extracted for GC/MS analysis at selected time intervals. Isotopically labeled 15N-enriched uridine was incorporated into DNA as cytosine+2 (C+2) and thymine+2 (T+2) and the relative incorporation of the heavier isotope increased with time. Experimental data were fitted to exponential growth equations, (Eqns found in Materials and Methods). An observed rate constant, kobs, for DNA replication was determined by analyzing both the C+2 and T+2 curves separately, and the values compare well, as is expected. Experimental data points are averages of 3 replicates. In all cases, the standard deviation is smaller than the size of the data points.
The variables needed to compute the theoretical curves in Fig. 6 are the percent enrichment of the precursor pool and the rate constant (k) for cell division. The enrichment of the nucleotide pool is obtained by observing the percent enrichment of the corresponding pyrimidine as the growth curve approaches an asymptotic value. The numerical values for the pool enrichment and rate constant are obtained by iterative least-squares fit to the experimental data. In the experiment illustrated in Fig. 6, the enrichment of the cytosine and thymine pools was determined to be 84 and 85%, respectively. Upon the basis of the observed rate constants determined from examination of the mass spectral data, the cell doubling time for the K-562 cells was 32.1 ± 0.1 h based upon incorporation of heavy uridine into DNA cytosine (kobs = 0.0216 ± 0.0001 h−1), and 30.8 ± 0.5 h, based upon incorporation of heavy uridine into DNA thymine (kobs = 0.0225 ± 0.0004 h−1). The cell doubling obtained from standard trypan blue cell counting was observed to be 35.1 ± 4.9 h.
High levels of isotope incorporation are obtained when cells are grown in media containing 100 µM 15N-enriched uridine. The degree of enrichment of the pyrimidine bases of DNA from the nucleotide pool represents the salvage pathway, which competes with de novo synthesis. The relative enrichment of the cytosine and thymine pools need not be the same. However, the cell doubling times determined from examination of the cytosine and thymine mass spectra should result in the same half-life or doubling time because the method presented here separates isotopically enriched DNA from unenriched precursors. The method reported here could be used to examine the impact of molecules that alter various nucleotide biosynthetic pathways and DNA replication, irrespective of DNA methylation.
It is generally presumed that the methylation of cytosine residues in DNA follows DNA replication (30). In this scheme (Fig. 1), new methyl groups would be added to newly incorporated cytosine residues. In the experiments reported here, new methyl groups would be enriched (2H3-methionine) and added to enriched cytosine residues. There would potentially be four isotopomers of 5mC: 5mC+0 from 5mC residues in DNA prior to the addition of label, 5mC+2 from the addition of unenriched methionine to newly incorporated enriched cytosine residues, 5mC+3 from the addition of enriched methionine methyl groups to unenriched cytosine residues in either parental strand DNA or to newly incorporated but unenriched cytosine residues, and finally, 5mC+5 from the addition of enriched methyl groups to newly incorporated enriched cytosine residues.
Figure 1. Methylation of cytosine in DNA.
Newly replicated, hemimethylated DNA is acted on by a maintenance DNA methyltransferase for heritable transmission of the cytosine methylation pattern. The 5mC (mC) in the parental strand “directs” methylation to the opposing cytosine residue in the newly replicated strand.
The results of a dual-label experiment (enriched uridine and methionine) are shown in Fig. 7 in which the contribution of the 5mC+2 and 5mC+5 isotopomers is observed to increase as a function of cell culture time. The 5mC+2 and 5mC+5 isotopomers result from the addition of unenriched and enriched methyl groups, respectively to 15N-enriched cytosine residues. The +2 and +5 isotopomers must originate from enriched cytosine precursors, and therefore, the data presented in Fig. 7 correspond to the methylation of the newly replicated DNA when cells are grown in 100 µM 15N-enriched uridine.
Figure 7. Change in isotopomer levels for 5mC in cells cultured in 15N-enriched uridine and 2H-enriched methionine. Newly replicated DNA.
Cells (K-562) cultured in the presence of 15N-enriched uridine and 2H-enriched methionine (as mentioned in Fig. 6) were harvested and DNA analyzed as before. Cells were harvested and DNA was extracted for GC/MS analysis. In the presence of stable isotope enriched uridine (Urd+2), newly synthesized DNA will contain C+2 only in the newly synthesized strand. The newly incorporated cytosine residues in appropriate sequences can be methylated with either enriched methionine (methyl+3) or unenriched methionine (methyl+0), resulting in 5mC+5 or 5mC+2, respectively. The relative amounts of these isotopomers of 5mC were determined by deconvolution of the experimental 5mC mass spectrum and calculated as shown (solid lines – described by Eqns found in Materials and Methods). An observed rate constant, kobs, was determined for DNA replication by analyzing the 5mC curves, comparing well to that determined by C+2 and T+2 above, as expected. Experimental data points are averages of 3 replicates. In all cases, the standard deviation is smaller than the size of the data points.
The rate constant that best fits the experimental data corresponds to a doubling time of 33.0 ± 1.1 h (kobs = 0.0210 ± 0.0007 h−1). This independent measurement of the cytosine methylation rate is the same as the DNA replication rate as determined from incorporation of heavy uridine into the cytosine and thymine pools described above. The data presented here is therefore consistent with previous proposals that DNA methylation follows closely DNA replication (30).
Two additional isotopomers of 5mC, +0 and +3, are also observed by this method as shown in Fig. 8. The 5mC+0 isotopomer represents primarily 5mC residues existing in the DNA prior to the addition of labeled precursors. The 5mC+3 isotopomer corresponds to the addition of new, enriched methyl groups to unenriched cytosine residues. Unenriched cytosine residues are found in the parental strand and in the newly replicated DNA with lower frequency because the cytosine pool is only 84% enriched. The contribution of the +0 and +3 isotopomers declines as a function of time as the unenriched cytosine is replaced by enriched cytosine in the newly replicated DNA.
Figure 8. Change in isotopomer levels for 5mC in cells cultured in 15N-enriched uridine and 2H-enriched methionine. Parental DNA.
Cells (K-562) cultured in the presence of 15N-enriched uridine and 2H-enriched methionine (as mentioned in Fig. 6 and 7) were harvested and DNA analyzed as before. The parental DNA in the cultured cells prior to the addition of stable isotope precursors is unenriched. The relative amount of 5mC+0 at a given time can be predicted from the degree of enrichment of the precursor pool and the rate of DNA replication, both determined independently as shown in Fig. 6 and Fig. 7. The decline in 5mC+0 with time can be predicted from these parameters (solid line above). The experimentally determined initial rate of decline of 5mC+0 is greater than expected. Similarly, enriched methionine could serve as a substrate for the methylation of unenriched cytosine residues found predominantly in the parental DNA, and the increase in the percentage of 5mC+3 can also be predicted. The rate of increase of 5mC+3 is also greater than expected. The unexpected loss of 5mC+0 with time approximately equals the increase in 5mC+3, resulting in overall constant levels of 5mC in the DNA. The kobs displayed here was determined by analyzing the 5mC curves in Fig. 7. Experimental data points are averages of 3 replicates. In all cases, the standard deviation is smaller than the size of the data points.
The asymptotic values for the relative abundance of the 5mC isotopomers (+0, +2, +3 and +5) in Figs. 7 and 8 were found to be 1.1%, 6.0%, 14.7% and 78.2% respectively. The enrichment of the cytosine pool (+2) from the incorporation of heavy uridine described above was found to be 84%. The relative abundance for the 5mC+5 isotopomer is the product of the enrichment of the cytosine pool (+2) and the enrichment of the methionine pool (+3) as indicated in Fig. 7 and Eqn 6. The enrichment of the methionine pool can therefore be determined to be 78.2/84.0 = 0.93, or 93%. The enrichment of the methionine pool can be independently determined from the relative abundance of 5mC+5 and 5mC+2 because the 5mC+5 isotopomer arose from the addition of an enriched methionine methyl group to an enriched cytosine residue whereas the 5mC+2 isotopomer arose from the addition of an unenriched methionine methyl group to an enriched cytosine residue. The enrichment of the methionine pool is then 78.2/(78.2 + 6.0) = 93%, the same value as determined above. All of the data thus far examined for methylation of newly incorporated cytosine resides is consistent with cytosine methylation following closely DNA replication.
With the method reported here, all isotopomers of cytosine, thymine and 5mC are measured simultaneously in the DNA sample. The rates for cytosine and thymine incorporation are identical and are the same as the apparent rate of DNA methylation, as expected. The rate of dilution of 5mC from unenriched cytosine residues should decline with the same rate constant. However, as shown in Fig. 8, the original, unenriched 5mC is observed to initially fall at a rate greater than predicted from the rate constants for DNA replication and methylation described above. Based upon the rate of incorporation of uridine into the cytosine and thymine residues in the DNA at approximately one cell doubling (29 h) the relative abundance of unenriched 5mC would be expected to be 54.9 ± 2.6%. The observed value is 48.9 ± 0.3%. Although the effect is subtle, the unenriched 5mC present in the parental DNA appears to be initially lost at a rate faster than can be attributed to dilution through incorporation of heavy uridine into the DNA.
The 5mC+3 isotopomer arises from the addition of new, enriched methyl groups to unenriched cytosine residues in the DNA. In contrast with the 5mC+0 isotopomer, the rate of increase in the relative abundance of 5mC+3 is initially greater than would be expected based upon addition of new methyl groups to the small number of unenriched cytosine residues incorporated into the newly replicated DNA. Upon the basis of the observed cell doubling time determined from the rate of isotope incorporation into DNA cytosine and thymine, the relative abundance of 5mC+3 would be expected to be 6.57 ± 1.85%. The observed value is 13.0 ± 0.1%, roughly twice the expected value. The total amount of 5mC in the K-562 cell line (5mC/C) was measured to be 1.50 % ± 0.31%. The sum of 5mC from all isotopomers does not change during cell replication, and the unexpected increase in 5mC+3 approximately equals the unexpected decline of the 5mC+0.
Similar studies were conducted on two additional human cell lines, DU-145, a prostate cancer derived cell line, and SK-MEL-28, a melanoma-derived cell line. The total amount of 5mC in these two cell lines was measured to be 4.30 ± 0.18% and 2.67 ± 0.13%, respectively. The DU-145 cell line has a doubling time of approximately 35 h, as determined from the incorporation of uridine label into DNA cytosine and thymine. At 36 h, the expected relative abundance of 5mC+0 was calculated to be 50.0 ± 2.7%, and the corresponding observed value was 43.3 ± 0.6%, less than expected. The expected relative abundance of 5mC+3 was calculated to be 8.67%, ± 1.22% whereas the observed value was found to be 16.7% ± 0.6%, again roughly twice the expected value. The SK-MEL cell line has a doubling time of approximately 71 h, also determined by the rate of labeled uridine incorporation into DNA cytosine and thymine. The expected relative abundance of 5mC+0 at 134 h was calculated to be 27.1 ± 0.5%, whereas the observed value was 15.0 ± 0.5%. The expected relative abundance of 5mC+3 was calculated to be 23.8 ± 0.3% whereas the observed value was 36.0 ± 0.5%.
The observed but unanticipated increased rate of 5mC+0 decline with a compensating increased rate of 5mC+3 formation may suggest a previously undescribed pathway for the modification of DNA cytosine methylation patterns as illustrated in Fig. 9. Upon the basis of the observations reported here, we propose a novel chromatin remodeling pathway. In this proposed pathway, some methylation does occur to the parental DNA while some 5mC residues are lost from the DNA. Our data suggest that 5mC+0 residues are possibly lost from the DNA, and not merely diluted through DNA replication. Our data do not indicate whether the loss of 5mC from the parental DNA results from a demethylation of existing 5mC, glycosylase removal of 5mC or some other as yet unknown process (31). The apparent demethylation and remethylation of the parental DNA may have an important role in overall chromatin structure and segregation during DNA replication. Previously, Jones and coworkers described the addition of methionine tritium to light DNA generated in the presence of 5-bromo-2'-deoxyuridine (23). While the authors suggested that presumptive new methylation of parental DNA might have resulted from incomplete separation of the heavy and light DNA strands, it may possibly be attributed to a remodeling process as described here.
Figure 9. DNA methylation coupled with chromatin remodeling during cell replication.
The methylation of newly incorporated cytosine residues in hemimethylated DNA following DNA replication is shown on the right, and is referred to as maintenance methylation. A novel process referred to as chromatin remodeling is shown on the left, in which parental strand DNA could actively lose or gain a methyl group by an unknown mechanism. This model could potentially explain why unenriched 5mC+0 residues are lost at a faster rate than could be attributed to DNA replication alone, and why enriched 5mC+3 residues increase faster than can be accounted for by DNA replication alone.
The method reported here demonstrates that the incorporation of stable isotope precursors into the DNA of replicating cells, followed by GC/MS analysis, can provide important insights into DNA replication and methylation. Although it is possible that enriched methionine could enter the one-carbon cycle (29), potentially confounding the interpretation of the data reported here, no unexpected changes are observed in the mass spectra of thymine, adenine, or guanine, suggesting that under the conditions of the experiments reported here, methyl recycling does not contribute to the observed changes in the mass spectra of cytosine, thymine or 5mC. Inhibitors of DNA methylation proposed as potential chemotherapy agents or other genotoxic chemicals likely impact multiple biochemical pathways in replicating cells. The method reported here may prove valuable in studying these agents and their impact on nucleotide pools, DNA replication, and DNA methylation. Such studies are currently in progress.
Acknowledgment
This work was supported by the National Institutes of Health.
Footnotes
Abbreviations: 5mC, 5-methylcytosine; ATCC, American Type Culture Collection; dCTP, 2’-deoxycytidine-5’-triphosphate; DNMT1, DNA methyltransferase 1; FBS, fetal bovine serum; M-57, Parent mass of compound minus a 57 amu fragment; MBD, Methyl Binding Domain; MTBSTFA, N-(t-butyldimethylsilyl)-N-methyltrifluoroacetamide; SAM, S-adenosyl-l-ethionine; SAH, S-adenosyl-l-homocysteine; TBDMCS, tert-butyldimethylchlorosilane; TBDMS, tert-utyldimethylsilyl
References
- 1.Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 3.Cooper DN, Taggart MH, Bird AP. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983;11:647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singal R, Ginder GD. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 5.Ehrlich M, Wang RY-H. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 6.Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;25:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valinluck V, Tsai H-H, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain MBD of methyl-CpG binding protein 2 MeCP2. Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alteration in human tumours. J. Pathology. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA. Epigenetics in carcinogenesis and cancer prevention. New York Acad. Sci. 2003;983:213–219. doi: 10.1111/j.1749-6632.2003.tb05976.x. [DOI] [PubMed] [Google Scholar]
- 11.Magewu AN, Jones PA. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational Hot spot in human cancer. Mol. Cell. Biol. 1994;14:4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Research. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 13.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa J-P, Markowitz S, Willson JKV, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurkjian C, Kummar S, Murgo AJ. DNA methylation: its role in cancer development and therapy. Curr. Probl. Cancer. 2008;32:187–235. doi: 10.1016/j.currproblcancer.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, Stone RM, Douglas N, Powell BL, DeCastro CM, Ellerton J, Larson RA, Schiffer CA, Holland JF. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J. Clin. Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 16.Leone G, Teofili L, Voso MT, Lubbert M. DNA methylation and demethylating drugs in myelodysplastic syndromes and secondary leukemias. Haematologica. 2002;87:1324–1341. [PubMed] [Google Scholar]
- 17.Yagi S, Oda-Sato E, Uehara I, Asano Y, Nakajima W, Takeshita T, Tanaka N. 5-Aza-2'-deoxycytidine restores proapoptotic function of p53 in cancer cells resistant to p53-induced apoptosis. Cancer Invest. 2008;26:680–688. doi: 10.1080/07357900701840212. [DOI] [PubMed] [Google Scholar]
- 18.Koshy M, Dorn L, Bressler L, Molokie R, Lavelle D, Talischy N, Hoffman R, van Overveld W, DeSimone J. 2’-deoxycytidine and fetal hemoglobin induction in sickle cell anemia. Blood. 2000;96:2379–2384. [PubMed] [Google Scholar]
- 19.Galanello R, Stamatoyannopoulos G, Papayannopoulou T. Mechanism of Hb F stimulation by S-stage compounds. J. Clin. Invest. 1988;81:1209–1216. doi: 10.1172/JCI113437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng JC, Yoo CB, Weisenberger DJ, Chuang J, Wozniak C, Liang G, Marquez VE, Greer S, Orntoft TF, Thyjaer T, Jones PA. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Scott SA, Lakshimikuttysamma A, Sheridan DP, Sanche SE, Geyer CR, DeCoteau JF. Zebularine inhibits human acute myeloid leukemia cell growth in vitro in association with p15INK4B demethylation and reexpression. Exp. Hematol. 2007;35:263–273. doi: 10.1016/j.exphem.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Kaysen J, Spriggs D, Kufe D. Incorporation of 5-fluorodeoxycytidine and metabolites into nucleic acids of human MCF-7 breast carcinoma cells. Cancer Res. 1986;46:4534–4538. [PubMed] [Google Scholar]
- 23.Jones PA, Taylor SM. Hemimethylated duplex DNAs prepared from 5-azacytidine-treated cells. Nucleic Acids Res. 1981;9:2933–2947. doi: 10.1093/nar/9.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaFrancois CJ, Fujimoto J, Sowers LC. Synthesis and characterization of isotopically enriched pyrimidine deoxynucleoside oxidation damage products. Chem. Res. Toxicol. 1998;11:75–83. doi: 10.1021/tx970186o. [DOI] [PubMed] [Google Scholar]
- 25.Lim KS, Huang SH, Jenner A, Wang H, Tang SY, Halliwell B. Potential artifacts in the measurement of DNA deamination. Free Radic. Biol. Med. 2006;40:1939–1948. doi: 10.1016/j.freeradbiomed.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 26.Rusmintratip V, Riggs AD, Sowers LC. Examination of the DNA substrate selectivity of DNA cytosine methyltransferases using mass tagging. Nucleic Acids Res. 2000;28:3594–3599. doi: 10.1093/nar/28.18.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugentobler E, Loliger J. A general approach to calculating isotope abundance ratios in mass spectroscopy. J. Chem. Educ. 1972;49:610–612. [Google Scholar]
- 28.Ehrlich M, Gama-Sosa MA, Huang L-H. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gazziola C, Ferraro P, Moras M, Reichard P, Bianchi V. Cytosolic high Km 5'-nucleotidase and 5'(3')-deoxyribonucleotidase in substrate cycles involved in nucleotide metabolism. J. Biol. Chem. 2001;276:6185–6190. doi: 10.1074/jbc.M007623200. [DOI] [PubMed] [Google Scholar]
- 30.Araujo FD, Knox JD, Szyf M, Price GB, Zannis-Hadjopoulos M. Concurrent replication and methylation at mammalian origins of replication. Mol. Cell. Biol. 1998;18:3475–3482. doi: 10.1128/mcb.18.6.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusmintratip V, Sowers LC. An unexpectedly high excision capacity for mispaired 5-hydroxymethyluracil in human cell extracts. Proc. Natl. Acad. Sci. USA. 2000;97:14183–14187. doi: 10.1073/pnas.97.26.14183. [DOI] [PMC free article] [PubMed] [Google Scholar]