Abstract
Kelch-like ECH-associated protein 1 (Keap1) is an ubiquitin E3 ligase specificity factor that targets transcription factor Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) for ubiquitination and degradation. Disrupting Keap1-Nrf2 interaction stabilizes Nrf2; resulting in Nrf2 nuclear accumulation, binding to antioxidant response elements (AREs) and transcription of cytoprotective genes. Marburg virus (MARV) is a zoonotic pathogen that likely uses bats as reservoir hosts. We demonstrate that MARV protein VP24 (mVP24) binds the Kelch domain of either human or bat Keap1. This binding is of high affinity and 1:1 stoichiometry and activates Nrf2. Modeling based on the Zaire Ebola virus VP24 (eVP24) structure identified in mVP24 an acidic loop (K-loop) critical for Keap1 interaction. Transfer of K-loop to eVP24, which otherwise does not bind Keap1, confers Keap1 binding and Nrf2 activation; and infection by MARV but not EBOV activates ARE gene expression. Therefore, MARV targets Keap1 to activate Nrf2-induced cytoprotective responses during infection.
Introduction
Kelch-like ECH-associated protein 1 (Keap1) is a cellular adaptor protein that links the Cul3/Rbx1(Roc1) ubiquitin E3 ligase to the oxidative stress response through its interaction with the transcription factor Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) (reviewed in (Copple, 2012)). Under homeostatic conditions, Keap1 suppresses the cellular antioxidant transcriptional program by directing the ubiquitin-mediated degradation of Nrf2 (Itoh et al., 1999; McMahon et al., 2003). Keap1 interacts, via its Kelch domain, with two sites located in the Nrf2-ECH homology-2 (Neh2) domain of Nrf2 (Itoh et al., 1999; Tong et al., 2006). Disruption of Nrf2-Keap1 interaction leads to transcription of genes possessing antioxidant response elements (AREs)(Tong et al., 2007). The upregulated ARE genes encode proteins involved in detoxification reactions, cell survival and immune modulation (reviewed in (Baird and Dinkova-Kostova, 2011; Ma, 2013)).
ARE responses impact the outcome of viral infections. For example, the Nrf2 pathway inhibits influenza virus and respiratory syncytial virus replication in cell culture and in vivo (Cho et al., 2009; Kesic et al., 2011). In contrast, for hepatitis B virus, hepatitis C virus and human cytomegalovirus, induction of ARE responses may protect infected cells from oxidative damage and influence immune responses by modulating immunoproteasome function (Burdette et al., 2009; Ivanov et al., 2011; Lee et al., 2013; Schaedler et al., 2010).
Marburg viruses (MARVs) and Ebola viruses (EBOVs), members of the family Filoviridae, are emerging, zoonotic pathogens that likely use bats as reservoir hosts. Filoviruses are of concern because they cause hemorrhagic fever with a high fatality rate in humans (reviewed in (Brauburger et al., 2012)). Filoviruses encode multifunctional VP24 proteins, which play important roles in the formation of viral nucleocapsids, release of infectious virus particles and modulation of viral RNA synthesis (Bamberg et al., 2005; Beniac et al.; Bharat et al., 2012; Bharat et al., 2011; Hoenen et al., 2006; Huang et al., 2002; Mateo et al.; Noda et al., 2006; Watanabe et al., 2007; Wenigenrath et al., 2010). In addition, EBOV VP24 (eVP24) disrupts interferon (IFN) signaling pathways and interacts with select karyopherin alpha proteins (KPNAs), thereby blocking nuclear accumulation of tyrosine phosphorylated STAT1 (Mateo et al., 2009; Reid et al., 2006; Reid et al., 2007). In contrast, MARV VP24 (mVP24) neither interacts with KPNAs nor inhibits IFN signaling, and functionally relevant interactions with host factors have not previously been defined (Valmas et al., 2010). However, a recent mass spectrometry screen identified Keap1 as a potential mVP24 binding partner (Pichlmair et al., 2012).
To date, the described mechanisms by which viruses engage the ARE response do not involve direct interaction with components of the signaling pathways. Rather, viruses are demonstrated to activate other signaling pathways or induce oxidative stress, indirectly activating antioxidant responses. Here, we demonstrate that mVP24 but not eVP24 directly interacts with the human and bat Keap1 proteins. We further define the basis of the interaction and demonstrate that expression of mVP24 but not eVP24 activates Nrf2, triggering cytoprotecitve responses. Correspondingly, MARV but not EBOV infection activates ARE gene expression. Collectively, these data suggest that MARV evolved to specifically target a host cytoprotective gene expression program to facilitate its replication.
Results
mVP24 interacts with Keap1
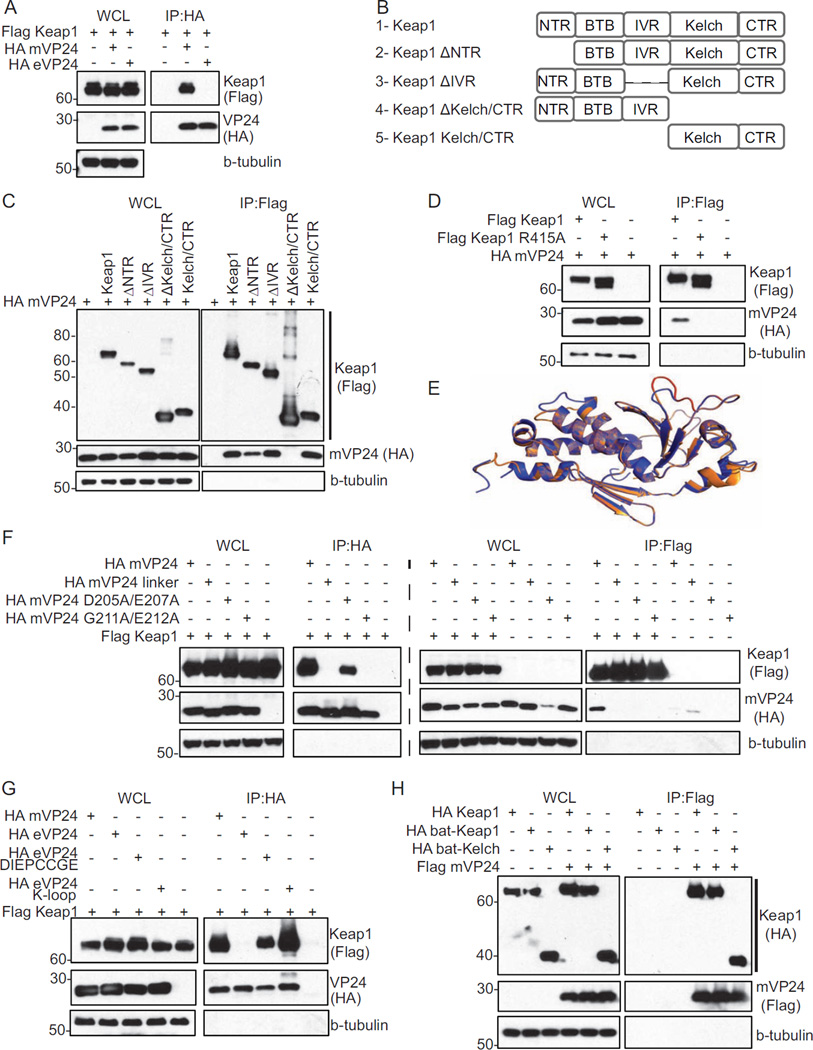
Co-immunoprecipitation (co-IP) assays demonstrated that Flag-tagged Keap1 interacts with HA-mVP24 but not with HA-eVP24 (Fig. 1A). Keap1 contains several previously defined domains: the N-terminal region (NTR); the Bric-a-Brac, Tramtrack, Broad complex (BTB) domain; the intervening region (IVR); and the Kelch domain/C-terminal region (CTR) (Komatsu et al., 2010). Domain deletion mutants of Keap1 and a construct comprising only the Kelch domain/CTR were tested for mVP24 interaction by co-IP (Fig. 1B). The NTR and IVR deletion mutants retained interaction, whereas deletion of the Kelch/CTR resulted in loss of interaction (Fig. 1C). The isolated Kelch/CTR domain also interacted with mVP24 (Fig. 1C). Therefore, the Kelch/CTR domain is necessary and sufficient to interact with mVP24 (Fig. 1C). The mutation to alanine of Keap1 Kelch domain residue R415 disrupts interaction with Nrf2 (Lo et al., 2006). Similarly, Keap1 R415A did not co-precipitate with mVP24 (Fig 1D), suggesting that Nrf2 and mVP24 interact with the Keap1 Kelch region in a similar fashion.
Fig. 1. mVP24 interacts with Keap1 in co-immunoprecipitation assays.
(A) Co-immunoprecipitations (co-IP) with anti-HA antibody were performed on lysates of HEK293T cells co-transfected with plasmids for Flag-Keap1 and HA-mVP24 or HA-eVP24. Western blots were performed for Flag and HA. (B) Schematic diagram of Flag tagged Keap1 domain deletion mutants used in (C). (C) Flag-Keap1 domain deletion mutant constructs were co-expressed in HEK293T cells with HA-mVP24 and analyzed by co-IP with anti-Flag antibody. (D) HA-mVP24 and either Flag-Keap1 or Flag-Keap1 R415A were analyzed by co-IP as in (C). (E) Overlay of the mVP24 structural model (orange) on the determined eVP24 structure (purple). The mVP24 K-loop (amino acids 205–212) is indicated in red. (F) Flag-Keap1 and HA-mVP24 wild-type or mutants were analyzed by co-IP as in (A) and (C). (G) Flag-Keap1 and HA-mVP24, eVP24, eVP24 DIEPCCGE or eVP24 K-loop were co-expressed in HEK293T cells and analyzed by co-IP as in (A). (H) Flag-mVP24 and HA-Keap1, bat-Keap1 and bat-Kelch were co-expressed in HEK293T cells and analyzed by co-IP as in (C). See also Figure S1.
To gain insight into the region(s) of mVP24 required to interact with Keap1, we used our recently solved structure of VP24 from Zaire EBOV, which is very similar to the structures of Sudan and Reston EBOV VP24s (Zhang et al., 2012) (see extended experimental methods and results, and Table S1), and the Phyre2 software package to obtain a molecular model of mVP24 (Kelley and Sternberg, 2009). The resulting structural model identified a loop (the K-loop, amino acids 205–212) that is likely solvent exposed (Fig. 1E). The sequence near the K-loop is not as well-conserved among filoviral VP24 proteins. This loop contains a sequence DIEPCCGE that is reminiscent of the high affinity binding motif of DXXTGE, used by Nrf2 to interact with the Keap1 Kelch domain (Lo et al., 2006). Among the several Keap1 Kelch domain binding determinants, "GE" motifs appear to be the most highly conserved, with nearby upstream acidic residues also playing an important role for several interacting partners (Komatsu et al., 2010; Padmanabhan et al., 2008a). Given this similarity, we made three HA-tagged mVP24 constructs (Fig. 1F). In “mVP24 linker,” the 205-DIEPCCGE-212 sequence was replaced with a serineglycine linker. “mVP24 D205A/E207A” and “mVP24 G211A/E212A” were designed based on analogous loss-of-binding mutants described for cellular Keap1-interactor p62 (Komatsu et al., 2010). By co-IP, wild-type mVP24 strongly interacted with Keap1, mVP24 D205A/E207A interacted weakly, and no interaction was detected with either mVP24 linker or mVP24 G211A/E212A (Fig. 1F). To assess the role of the DIEPCCGE motif for interaction with Keap1, DIEPCCGE was swapped in place of the corresponding residues within eVP24, creating “eVP24 DIEPCCGE.” We also replaced the loop of eVP24 (202-QEPDKSAMDIRHPGPV-217) with the mVP24 K-loop (202-RRIDIEPCCGETVLSESV-219), creating “eVP24 K-loop”. eVP24 DIEPCCGE and eVP24 K-loop interacted with Keap1, with the full K-loop appearing to confer better binding, whereas wild-type eVP24 once again did not interact with Keap1 (Fig. 1G). These results demonstrate that the DIEPCCGE sequence and the K-loop, when placed in the context of the VP24 structural scaffold, play a critical role for mVP24-Keap1 interaction.
MARVs likely use bats as reservoir hosts (Amman et al., 2012; Towner et al., 2009). Therefore, a specific viral interaction with Keap1 likely evolved and should be conserved in bats. Alignment of human Keap1 and two divergent bat species, a microbat, Myotis lucifugus, and a megabat, Pteropus alecto, revealed 97% amino acid identity between human and microbat Keap1 and 98% amino acid identify between human and megabat Keap1 (data not shown). Full-length Keap1 (bat-Keap1) and the Kelch domain (bat-Kelch) constructs were generated from an available microbat (Myotis velifer incautus) cell line. Both co-precipitate with mVP24 with efficiencies similar to that of human Keap1 (Fig. 1H).
Keap1 inhibits ARE gene expression through its interaction with Nrf2 (McMahon et al., 2003). When Keap1 repression is relieved, which can be due to post-translational modification of Keap1 or interaction with select Kelch domain binding partners such as p62, Nrf2 translocates to the nucleus and activates ARE gene expression (Itoh et al., 1999; McMahon et al., 2003). To determine whether the interaction of mVP24 with the Keap1 Kelch domain activates Nrf2, a GFP-Nrf2 fusion protein was expressed alone or in the presence of Flag-Keap1 and HA-tagged wildtype mVP24, mutant mVP24 or wildtype or chimeric eVP24s. Over-expression of Nrf2, which is known to overwhelm the available endogenous Keap1, resulted in nuclear localization of GFP-Nrf2, as expected (Fig. S1). Co-expression of Keap1 retained most of the Nrf2 in the cytoplasm. Additional expression of mVP24 and eVP24-K-loop restored Nrf2-GFP nuclear localization, whereas mVP24 mutants and eVP24-DIEPCCGE, which do not interact efficiently with Keap1, did not (Fig. S1; see Extended Results for details).
mVP24 binds the Keap1 Kelch domain with high affinity and specificity
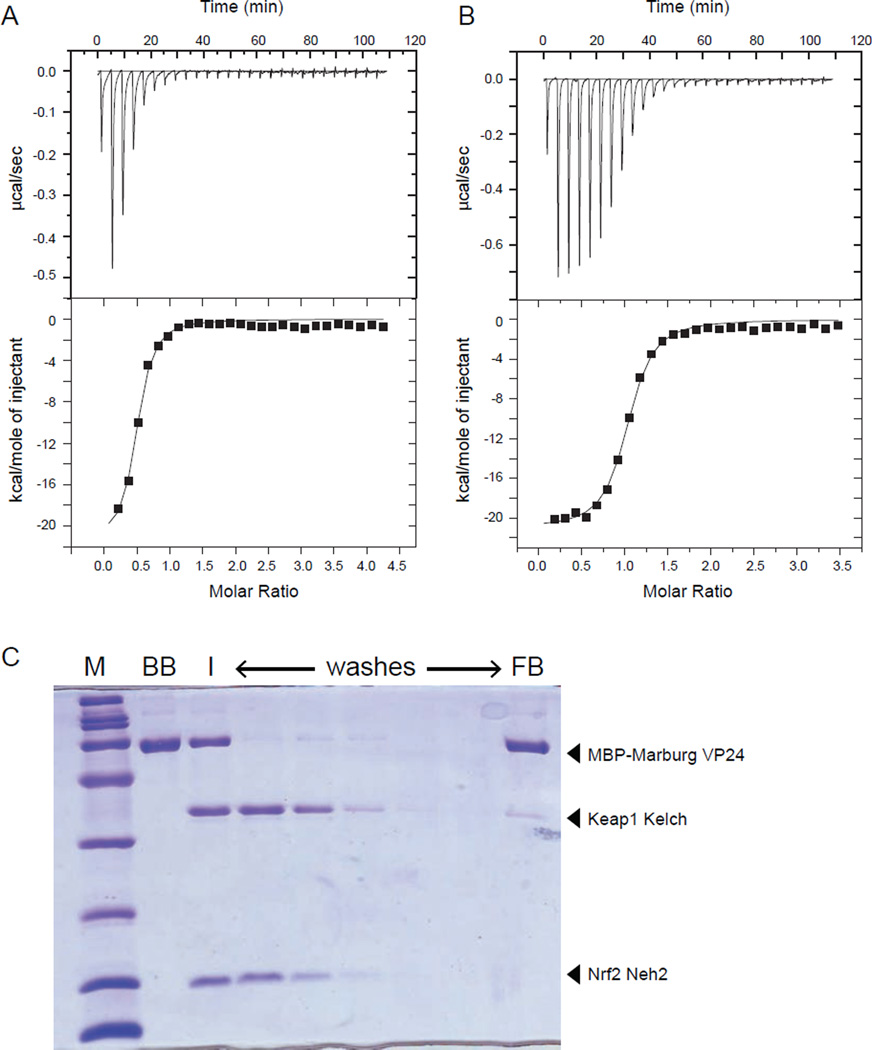
Binding of mVP24 to Keap1 Kelch was further evaluated by isothermal titration calorimetry (ITC), which measures heat generated by these exothermic interactions. ITC results confirmed that Keap1 Kelch binds the Nrf2 Neh2 domain with high affinity (KD = 170 ± 60 nM) and stoichiometry, n, of 0.46 (Fig. 2A) and support a stoichiometry of 2:1 for Kelch binding to Neh2 with thermodynamic parameters similar to those previously reported (Tong et al., 2006). Assays under similar conditions for Kelch-mVP24 resulted in a KD of 158 ± 20 nM (Fig. 2B) with a binding stoichiometry of 1:1.
Fig. 2. Marburg VP24 binds to Keap1 Kelch domain with high affinity and specificity.
mVP24 binds to Keap1 Kelch domain with high affinity and specificity. (A and B) Representative ITC data for KELCH domain of Keap1 binding to (A) Nrf2 Neh2 domain and (B) mVP24. Raw heats of reaction vs. time (top panels) and the integrated heats of reaction vs. molar ratio of ligand to receptor (bottom panels) are shown. Thermodynamic binding parameters of KD = 170 ± 60 nM, ΔH = −1.96 ± 0.1×104 kcal/mol, TΔS = −10.4 kcal/mol, and n (no. of sites) = 0.49 ± 0.02 for (A) and KD = 158 ± 20 nM, ΔH = −2.10 ± 0.03× 104 kcal/mol, TΔS = −11.7 kcal/mol, and n (no. of sites) = 1.00 ± 0.01 for (B) were obtained. (C) mVP24 binding to Kelch prevents Nrf2 Neh2 interaction. Coomassie-blue stained SDS-PAGE of a pulldown assay where MBP-mVP24 was immobilized on amylose resin (BB, bound beads). Keap1 Kelch and Nrf2 Neh2 domain was subsequently added to the resin (I, input), and the resin was washed with buffer (washes). The final bound bead sample (FB, final beads) is indicated. See also Figure S2.
To gain additional mechanistic insight, we performed competition pull-down experiments using wild-type mVP24, eVP24 and eVP24 K-loop which was designed based on the mVP24 structural model (Fig. S2A–C) . We established the basal binding conditions for the Kelch and Neh2 interaction by pulldown (Fig. S2D) as well as Kelch binding to mVP24 (Fig. S2E) and examined the ability of recombinant eVP24 (Fig. S2F) and eVP24 K-loop (Fig. S2G) to bind the Keap1 Kelch domain. Next, we assessed whether mVP24 can out-compete Neh2 binding to the Kelch domain. A complex between the Kelch domain and Neh2 was preformed and the ability of an imobilized mVP24 protein to displace Neh2 from the Kelch/Neh2 complex was assessed. Despite similar affinities of Neh2 and mVP24 for Kelch domain, mVP24 can bind the Kelch domain in the presence of a 2-fold excess of Neh2 (Fig. 2C). Therefore, in the absence of other factors, mVP24 displaces Nrf2 from Keap1. This provides a biochemical explanation as to how the mVP24-Keap1 interaction triggers Nrf2 nuclear localization.
mVP24 expression activates ARE-directed gene expression
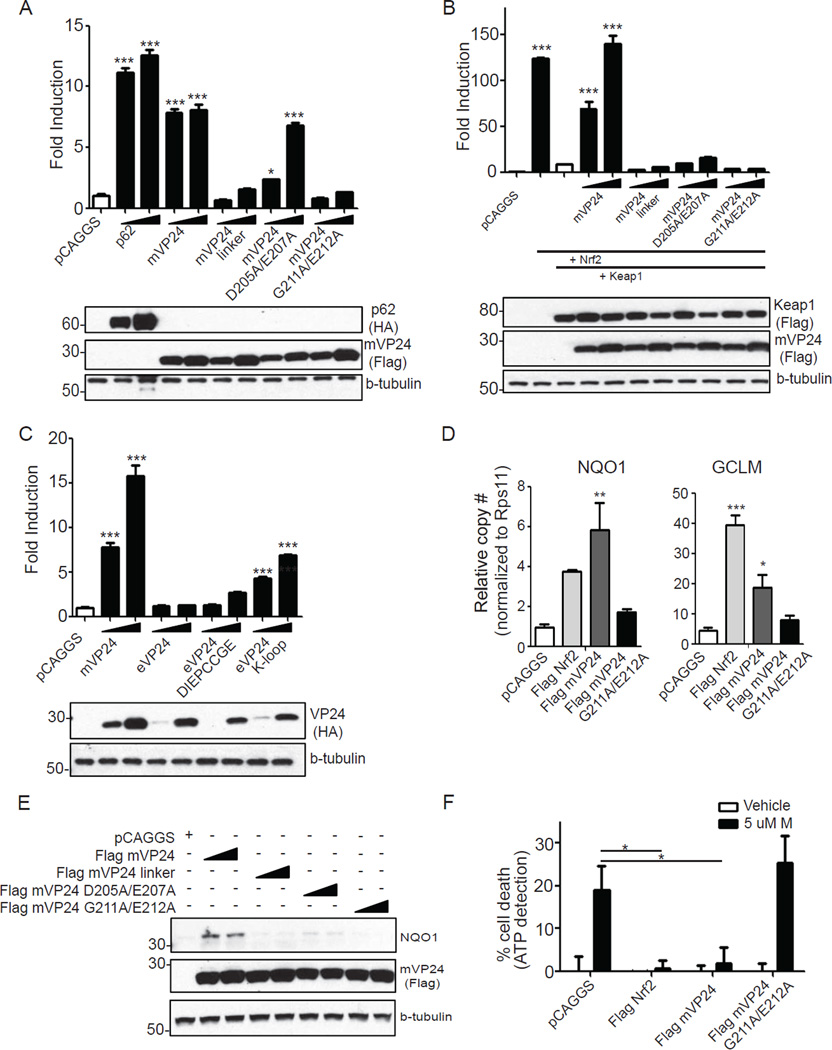
Stimuli that disrupt the Nrf2-Keap1 interaction and promote Nrf2 nuclear localization activate expression of ARE genes (reviewed in (Magesh et al., 2012). We therefore assessed the ability of wild-type or mutant mVP24s to activate an ARE luciferase reporter gene. Cellular Keap1-interacting protein p62, a previously described activator of Nrf2, served as a positive control (Komatsu et al., 2010; Lau et al., 2010). Expression of mVP24 induced the ARE reporter to similar levels as p62 (Fig. 3A). In contrast, mVP24 linker mutant and mVP24 G211A/E212A did not activate the ARE promoter. mVP24 D205A/E207A did activate the ARE promoter but to a lesser extent than wild-type mVP24, reflecting the residual binding activity of this mutant for Keap1 (Fig. 3A). Therefore, Nrf2 activation correlates with Keap1 mVP24 binding activity (Fig. 1F). In a separate experiment, expression of Nrf2 alone resulted in greater than 100-fold ARE reporter activation (Fig. 3B). Keap1 co-expression inhibited the activation. mVP24 expression relieved the repression of Nrf2, resulting in ARE gene expression (Fig. 3B). None of the mutant mVP24s induced significant ARE activation, despite expression comparable to that of wild-type mVP24 (Fig. 3B). This suggests that the residual binding of mVP24 D205A/E207A is not sufficient to disrupt the repressive activity of the overexpressed Keap1 (Fig. 3B). While expression of eVP24 did not activate the ARE reporter, expression of the mutant eVP24-DIEPCCGE resulted in a slight increase in reporter activity, and eVP24 K-loop significantly induced ARE reporter expression (Fig. 3C). Similarly, bat-Keap1 inhibited the activation of the ARE-reporter by over-expressed human Nrf2 (Fig. S3A), and mVP24 expression relieved the repression mediated by bat-Keap1 on the ARE-reporter (Fig S3A). Therefore, mVP24 interaction with Keap1 has functional consequences as it can trigger Nrf2-dependent transcriptional activity in a K-loop-dependent manner.
Fig. 3. mVP24 activates expression of ARE genes.
(A and B) HEK293T cells were transfected with the ARE luciferase reporter plasmid, a constitutively expressed Renilla luciferase plasmid, and pCAGGS (empty vector) or increasing concentrations of HA-p62, Flag-wild-type mVP24 or mVP24 mutants. (B) Same as (A), with the additional over-expression of Flag-Nrf2 and Flag-Keap1. At 18 hours post transfection (hpt) luciferase activity was assayed for (A) and (B). Western blots performed for HA and Flag are indicated. (C) Same assay protocol as (A) but transfected with HA-mVP24, eVP24 or eVP24 mutants. (D) pCAGGS, Flag-Nrf2, mVP24 or mVP24 G211A/E212A were transfected in triplicate in HEK293T cells. At 24 hpt, qRT-PCR was performed to quantify mRNAs for the indicated genes, normalized to the RPS11 mRNA. (E) HEK293T cells were transfected with the indicated plasmids and 18hpt endogenous NQO1 was measured by western blot. (F) Cell viability assay. HEK293T cells were transfected with pCAGGS, Flag-Nrf2, mVP24 or mVP24 G211A/E212A and 24 hpt were treated with vehicle control (ethanol) or 5uM menadione (M) for three hours. (A-D) Represent the mean and SEM of triplicate samples and statistical significance was assessed by a one-way ANOVA comparing columns to the control (white bar), ***p<0.001, **p<0.01 and *p<0.05. Samples in (F) represent the mean and SEM of six samples and significance was assessed by a one-way ANOVA, *p<0.05. See also Figure S3.
mVP24 expression also induced expression of the endogenous ARE genes, NAD(P)H quinone oxidoreductase 1 (NQO1) and glutamate-cysteine ligase, modifier subunit (GCLM) (Lau et al., 2010) as assessed by quantitative RT-PCR (Fig. 3D and Fig. S3B). Neither the mVP24 mutants nor eVP24 induced expression of these genes (Fig. S3). In contrast, eVP24 DIEPCCGE and eVP24 K-loop did induce significant levels of GCLM mRNA (Fig. 3D and Fig. S3B). Correspondingly, NQO1 protein levels increased in the presence of wild-type but not mutated mVP24s, eVP24 or eVP24 mutants (Fig. 3E and Fig. S3C). Interestingly, the eVP24 chimeras did not induce NQO1 and induced GCLM mRNA to a lesser extent than did mVP24. This may reflect in part an as yet uncharacterized inhibitory activity of eVP24 on Nrf2-induced transcription responses which can be seen in ARE reporter gene assays (Fig S3D). Consistent with the ARE induction, cells transfected with Nrf2 (a postive control) or mVP24 were protected from killing by menadione, a compound that induces oxidative damage. In contrast, significant cell death was detected in the pCAGGS and mVP24 G211A/E212A transfected cells (Fig 3F).
MARV infection induces the expression of Nrf2 responsive genes
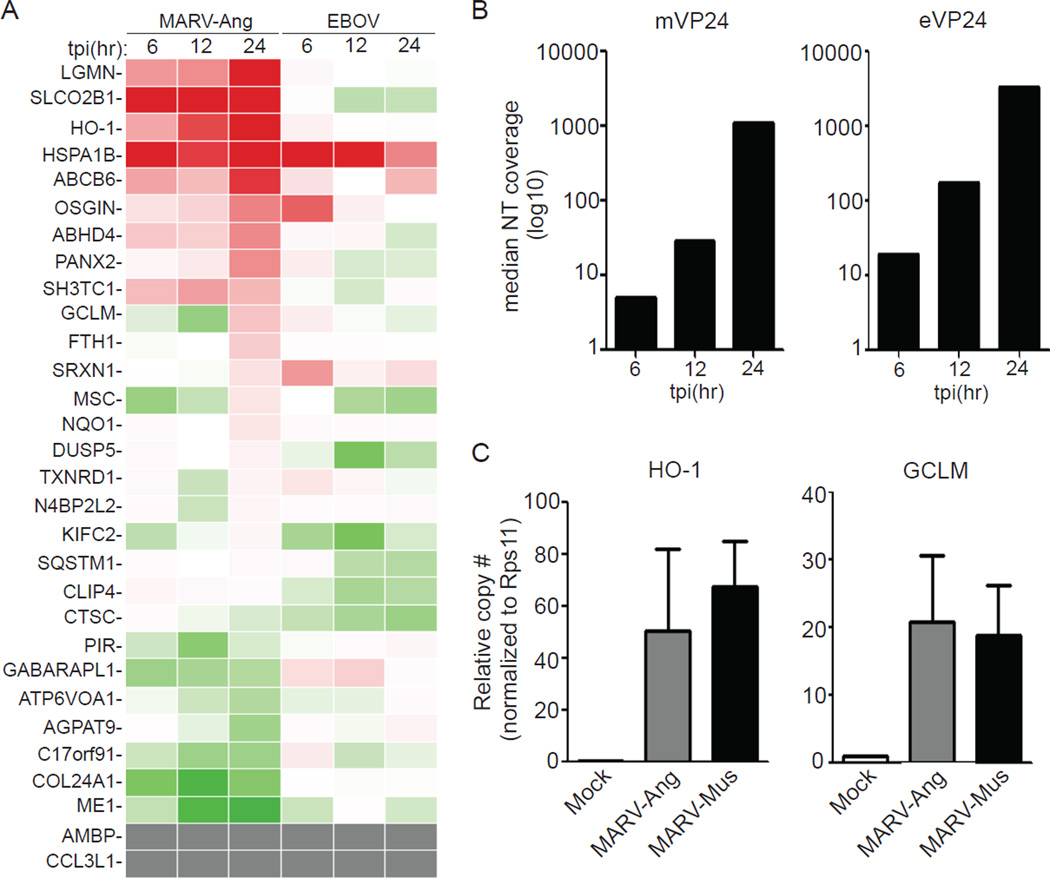
mVP24 activates Nrf2 via interaction with Keap1 but eVP24 does not, suggesting that MARV but not EBOV infection should induce an ARE response. To test this hypothesis, we profiled the expression of select ARE genes in THP-1 cells following MARV Angola strain (MARV-Ang) or Zaire EBOV infection (multiplicity of infection (MOI)=3). A substantial number of ARE genes were upregulated in MARV-infected THP-1 cells as the infection progressed and mVP24 mRNA levels increased (Fig. 4A, B). Although a few ARE genes were upregulated by EBOV infection, the response was not as global as was seen with MARV, and the response did not correlate well with eVP24 expression (Fig. 4A, B). The mVP24 K-loop sequence is conserved among MARV strains, suggesting that ARE activation should also be shared between MARV strains. Indeed, induction of two representative ARE genes, heme oxygenase 1 (HO-1) and GCLM, was demonstrated by qRT-PCR following infection of THP-1 cells with MARV-Ang or Musoke (MARV-Mus) (Fig. 4C). Interestingly, HO-1 is highly upregulated during MARV infection (Fig. 4A) and a recent study has indicated that EBOV replication/transcription is inhibited by HO-1 expression (Hill-Batorski et al., 2013). However, using a MARV minigenome assay we did not detect any inhibition following HO-1 over expression (Fig. S4, see extended results for further detail), suggesting that upregulation of this ARE may not impair MARV replication.
Fig. 4. MARV infection upregulates the Nrf2 antioxidant pathway.
(A and B) THP-1 cells were infected with MARV-Ang or Zaire EBOV (MOI=3) and subjected to expression analysis by mRNA-seq. (A) Heat map displaying the expression profile of 30 Nrf2-activated genes (Chorley et al., 2012)). Red indicates up-regulated genes (max induction = 8.55 fold relative to mock-infected cells). Green indicates down-regulated genes (lowest value=0.2 fold relative to mock-infected cells). Gray indicates genes undetected in the mRNA-seq. (B) mVP24 and eVP24 mRNA expression levels represented as median nucleotide coverage. (C) THP-1 cells were infected with MARV-Ang or MARV-Mus (MOI=1) and subjected to qRT-PCR. Values were normalized to RPS11. Mock sample contains a single replicate; MARV-Ang and MARV-Mus represent the mean and SEM of triplicate samples. See also Figure S4.
Discussion
The host antioxidant response has been increasingly recognized as relevant to virus infections. Here, we demonstrate a direct, high affinity interaction between mVP24 and the Kelch domain of the human and bat Keap1, a major negative regulator of antioxidant responses (see also Supplemental Discussion on bat Keap1). This interaction, for which we define a critical role for the mVP24 K-loop, can disrupt Nrf2-Keap1 interaction and induce a cytoprotective state through transcriptional activation of the ARE promoter. Although other viruses have previously been demonstrated to activate antioxidant responses, the mechanisms of activation appear indirect, with virus infection triggering oxidative stress or other cellular signaling pathways that stimulate Nrf2 nuclear accumulation (Burdette et al., 2009; Cho et al., 2009; Ivanov et al., 2011; Kesic et al., 2011; Lee et al., 2013; Schaedler et al., 2010). In contrast, the direct interaction between mVP24 and Keap1, provides direct evidence that viruses have evolved mechanisms to engage the cellular antioxidant response as part of their replication strategy.
Keap1-Nrf2 interaction is required for negative regulation of the antioxidant response. A number of stimuli, such as oxidative stress, that perturb the Keap1-Nrf2 interaction stabilize Nrf2, allowing it to accumulate in the nucleus where it binds AREs and cooperates with other factors to activate ARE containing promoters (Dinkova-Kostova et al., 2002; Zhang and Hannink, 2003). In addition, the interaction of the Keap1 Kelch domain with p62, an autophagy factor that functions in the clearance of poly-ubiquitinated complexes, activates Nrf2 through the disruption of binding via the lower affinity Keap1 binding site on Nrf2 (Komatsu et al., 2010; Lau et al., 2010). We demonstrated that the mVP24-Keap1 interaction requires the Keap1 Kelch domain, as is true for many other Keap1 interactors (Kim et al., 2010; Komatsu et al., 2010; Lo and Hannink, 2006; Niture and Jaiswal, 2011). Our data further suggest that the interaction of mVP24 with Keap1 can disrupt the high affinity Nrf2-Keap1 binding site, leading to the subsequent nuclear localization of Nrf2 and activation of the antioxidant response.
The structural basis for the Keap1 Kelch interaction with peptides derived from several cellular Keap1 binding partners, including Nrf2, p62 and prothymosin alpha, was previously described (Komatsu et al., 2010; Lo et al., 2006; Padmanabhan et al., 2008b). These peptides bind the bottom of the Keap1 β-sheet propeller, which forms a basic pocket, in part through electrostatic interactions with Keap1 arginine residues. Common features of the binding peptides include acidic residues along with a GE motif (Komatsu et al., 2010; Lo and Hannink, 2006). Data obtained with mutated mVP24 K-loop acidic residues and the GE motif supports a similar mode of binding for mVP24, although we cannot exclude a contribution of other parts of mVP24. Consistent with a model where the mVP24 loop and the acidic residues within the loop make analogous contacts with the Keap1 Kelch domain, substitution of Keap1 R415 to alanine abrogated Keap1-mVP24 interaction.
It is striking that MARVs and EBOVs differ in their interaction with the ARE response (see Extended Discussion for details). While there are no structures of mVP24, several structures of EBOV VP24s, including Sudan and Reston EBOVs (sVP24 and rVP24)(Zhang et al., 2012) as well as Zaire EBOV (eVP24) are available (Fig. 2; PDB 4M0Q). In order to evaluate the mVP24 structure, we used the eVP24 structure, which was most complete as the basis for the Phyre2 threading model of mVP24. In the mVP24 model, the K-loop contains the DIEPCCGE sequence, a sequence that is not conserved between mVP24 and eVP24 but shows similarity to motifs of other Keap1 interacting “GE motifs.” Replacement of the K-loop residues with a heterologous linker sequence or mutation to alanine of the D205 and E207 or of G211 and E212 was sufficient to greatly reduce or abrogate binding, although it should be acknowledged that the nuclear localization confounds interpretation of the G211A/E212A mutant data. That the DIEPCCGE loop is central to binding is confirmed by the fact that transfer of the loop to eVP24, which otherwise does not interact with Keap1, confers binding activity. Further, wild-type mVP24 effectively competes with Nrf2 for binding to Keap1 in vitro and dissociates GFP-Nrf2 from Flag-Keap1 in a K-loop dependent manner. These observations suggest a mechanism by which mVP24 activates an ARE transcriptional response. Interestingly, the mVP24 DIEPCCGE sequence diverges from other Keap1 binding motifs, such as the so-called ETGE motif of Nrf2 (DEETGE), with “PCC” inserted between “GE” and more amino-terminal acidic residues. The presence of the Cys residues is intriguing given that Keap1-Nrf2 interactions are regulated by oxidation. Whether these residues, which are not present in other Keap1 interacting motifs, play an important role in the mVP24-Keap1 interaction will be the subject of future studies.
In addition to the ARE response, Keap1 regulates other stress-induced cell survival pathways through interaction of its Kelch domain with a variety of proteins, including PGAM5, IKKβ and p62 (Kim et al., 2010; Komatsu et al., 2010; Lau et al., 2010; Lee et al., 2009; Lo and Hannink, 2006; Niture and Jaiswal, 2011). mVP24 disruption of these Keap1 interactions could inhibit apoptosis, activate NF-κB-mediated cell survival pathways and influence autophagy (Fan et al., 2010; Kim et al., 2010; Lee et al., 2009; Niture and Jaiswal, 2011). Further, the stable interaction of mVP24 and Keap1, which did not detectably influence mVP24 expression levels, might allow recruitment of Keap1 and binding partners for new functions. Further study is therefore required to fully elucidate the impact of the novel mVP24-Keap1 interaction upon MARV infection.
Experimental Procedures
Co-Immunoprecipitation
24 hours post transfection with the indicated plasmids, HEK293T cells were lysed in NP-40 lysis buffer (50 mMTris (pH 7.5), 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, protease inhibitor (cOmplete; Roche)). Anti-FLAG M2 magnetic beads or anti-HA beads (Sigma-Aldrich) were incubated with lysates for one hour at 4°C, washed five times in NP-40 lysis buffer and eluted using either 3X FLAG peptide (Sigma) or by boiling in sample loading buffer.
Activation of Nrf2
For ARE reporter gene assays, a commercially-available reporter gene, pGL4.37[luc2P/ARE/Hygro] (ARE) (Promega) was co-transfected with a constitutively expressed Renilla luciferase reporter plasmid (pRL-tk (Promega)), and the indicated protein expression plasmids. 18 hours post transfection, a dual luciferase reporter assay (Promega) was performed in triplicate andfirefly luciferase values were normalized to Renilla luciferase values. Statistical significance was assessed with one-way ANOVA using Tukey’s test for comparisons to the control. Protein expression levels were assessed by western blot. Levels of endogenous NQO1, GCLM or HO1 mRNAs were assessed by quantitative RT-PCR, and NQO1 protein levels were assessed by western blot using a commercially available antibody (Santa Cruz).
Virus infections
The following infections were performed under BSL-4 conditions at the Galveston National Laboratory. THP-1 cells were differentiated overnight with 100nM PMA and infected with MARV-Ang (MOI=3 or MOI=1), MARV-Mus (MOI=1) or EBOV (MOI=3). Viral total RNA was extracted with Trizol at the indicated time points for analysis by deep sequencing or qRT-PCR. For deep sequencing, mRNA was purified with oligodT magnetic beads (Invitrogen). cDNA libraries were generated (NEB Next, New England Biolabs) and sequenced on the IlluminaHiSeq 2500 platform and relative expression for each gene of interest was determined. For qRT-PCR, cDNA was generated with oligodT primers and relative expression for each gene of interest was determined by normalizing to the indicated housekeeping gene.
Refer to Extended Experimental Procedures for additional details.
Supplementary Material
Highlights.
Marburg virus (MARV) protein VP24 (mVP24) interacts with both human and bat Keap1
mVP24 interacts with the Keap1 Kelch domain
mVP24 activates transcription factor Nrf2 by blocking its interaction with Keap1
mVP24 expression and MARV infection activate cytoprotective antioxidant responses
Acknowledgements
This work was supported by NIH grants AI059536 to CFB and AI081914 to GKA and DTRA grant HDTRA1-12-1-0051 to CFB and GKA. All microscopy studies were performed with the generous assistance of the Icahn School of Medicine at Mount Sinai Microscopy Shared Resource Facility. Sequencing was performed at the Genomics Sequencing Facility at Mount Sinai. We thank Hardik Shah and the Bioinformatics Group of the Icahn Institute for Genomics and MultiScale Biology for help with sequence analysis. We thank Drs. S. Ginell, N. Duke, and J. Lazarz at the Structural Biology Center (Advanced Photon Source, Argonne, IL) and Dr. J. Nix at Beamline 4.2.2 (Advanced Light Source, Berkeley, CA) for data collection support. Use of Argonne National Laboratory SBC beamlines at APS was supported by the U.S. D.O.E. contract DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, Kemp A, Erickson BR, Comer JA, Campbell S, et al. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 2012;8:e1002877. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird L, Dinkova-Kostova AT. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol. 2011;85:241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- Bamberg S, Kolesnikova L, Moller P, Klenk HD, Becker S. VP24 of Marburg virus influences formation of infectious particles. J Virol. 2005;79:13421–13433. doi: 10.1128/JVI.79.21.13421-13433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac DR, Melito PL, Devarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, Jones SM, Booth TF. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS One. 2012;7:e29608. doi: 10.1371/journal.pone.0029608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Noda T, Riches JD, Kraehling V, Kolesnikova L, Becker S, Kawaoka Y, Briggs JA. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci U S A. 2012;109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Riches JD, Kolesnikova L, Welsch S, Krahling V, Davey N, Parsy ML, Becker S, Briggs JA. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9:e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauburger K, Hume AJ, Muhlberger E, Olejnik J. Forty-five years of Marburg virus research. Viruses. 2012;4:1878–1927. doi: 10.3390/v4101878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette D, Olivarez M, Waris G. Activation of transcription factor Nrf2 by hepatitis C virus induces the cell-survival pathway. J Gen Virol. 2009;91:681–690. doi: 10.1099/vir.0.014340-0. [DOI] [PubMed] [Google Scholar]
- Cho HY, Imani F, Miller-DeGraff L, Walters D, Melendi GA, Yamamoto M, Polack FP, Kleeberger SR. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus disease. Am J Respir Crit Care Med. 2009;179:138–150. doi: 10.1164/rccm.200804-535OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic acids research. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copple IM. The Keap1-Nrf2 cell defense pathway--a promising therapeutic target? Adv Pharmacol. 2012;63:43–79. doi: 10.1016/B978-0-12-398339-8.00002-1. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, Zhu M, Zhong Q. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6:614–621. doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill-Batorski L, Halfmann P, Neumann G, Kawaoka Y. The cytoprotective enzyme heme oxygenase-1 suppresses Ebola virus replication. J Virol. 87:13795–13802. doi: 10.1128/JVI.02422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, Kolesnikova L, Theriault S, Ebihara H, Hartlieb B, Bamberg S, Feldmann H, Stroher U, Becker S. Infection of naive target cells with virus-like particles: implications for the function of ebola virus VP24. J Virol. 2006;80:7260–7264. doi: 10.1128/JVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Smirnova OA, Ivanova ON, Masalova OV, Kochetkov SN, Isaguliants MG. Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells. PLoS One. 2011;6:e24957. doi: 10.1371/journal.pone.0024957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kesic MJ, Simmons SO, Bauer R, Jaspers I. Nrf2 expression modifies influenza A entry and replication in nasal epithelial cells. Free Radic Biol Med. 2011;51:444–453. doi: 10.1016/j.freeradbiomed.2011.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, You DJ, Lee C, Ahn C, Seong JY, Hwang JI. Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation. Cell Signal. 2010;22:1645–1654. doi: 10.1016/j.cellsig.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Koh K, Kim YE, Ahn JH, Kim S. Upregulation of Nrf2 expression by human cytomegalovirus infection protects host cells from oxidative stress. J Gen Virol. 2013;94:1658–1668. doi: 10.1099/vir.0.052142-0. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J Biol Chem. 2006;281:37893–37903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magesh S, Chen Y, Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Medicinal research reviews. 2012;32:687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo M, Carbonnelle C, Martinez MJ, Reynard O, Page A, Volchkova VA, Volchkov VE. Knockdown of Ebola virus VP24 impairs viral nucleocapsid assembly and prevents virus replication. J Infect Dis. 2011;204(Suppl 3):S892–S896. doi: 10.1093/infdis/jir311. [DOI] [PubMed] [Google Scholar]
- Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol. 2009;84:1169–1175. doi: 10.1128/JVI.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- Niture SK, Jaiswal AK. Inhibitor of Nrf2 (INrf2 or Keap1) protein degrades Bcl-xL via phosphoglycerate mutase 5 and controls cellular apoptosis. J Biol Chem. 2011;286:44542–44556. doi: 10.1074/jbc.M111.275073. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Noda T, Ebihara H, Muramoto Y, Fujii K, Takada A, Sagara H, Kim JH, Kida H, Feldmann H, Kawaoka Y. Assembly and budding of Ebolavirus. PLoS Pathog. 2006;2:e99. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan B, Nakamura Y, Yokoyama S. Structural analysis of the complex of Keap1 with a prothymosin alpha peptide. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008a;64:233–238. doi: 10.1107/S1744309108004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan B, Nakamura Y, Yokoyama S. Structural analysis of the complex of Keap1 with a prothymosin alpha peptide. Acta crystallographica Section F, Structural biology and crystallization communications. 2008b;64:233–238. doi: 10.1107/S1744309108004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CF. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol. 2007;81:13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler S, Krause J, Himmelsbach K, Carvajal-Yepes M, Lieder F, Klingel K, Nassal M, Weiss TS, Werner S, Hildt E. Hepatitis B virus induces expression of antioxidant response element-regulated genes by activation of Nrf2. J Biol Chem. 2010;285:41074–41086. doi: 10.1074/jbc.M110.145862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, Swanepoel R, Paddock CD, Balinandi S, Khristova ML, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmas C, Grosch MN, Schumann M, Olejnik J, Martinez O, Best SM, Krahling V, Basler CF, Muhlberger E. Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog. 2010;6:e1000721. doi: 10.1371/journal.ppat.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J Infect Dis. 2007;196(Suppl 2):S284–S290. doi: 10.1086/520582. [DOI] [PubMed] [Google Scholar]
- Wenigenrath J, Kolesnikova L, Hoenen T, Mittler E, Becker S. Establishment and application of an infectious virus-like particle system for Marburg virus. J Gen Virol. 2010;91:1325–1334. doi: 10.1099/vir.0.018226-0. [DOI] [PubMed] [Google Scholar]
- Zhang AP, Bornholdt ZA, Liu T, Abelson DM, Lee DE, Li S, Woods VL, Jr, Saphire EO. The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 2012;8:e1002550. doi: 10.1371/journal.ppat.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.