Abstract
Ras GTPases cycle between active GTP-bound and inactive GDP-bound forms to regulate a multitude of cellular processes, including cell growth, differentiation, and apoptosis. The activation state of Ras is regulated by protein modulatory agents that accelerate the slow intrinsic rates of GDP dissociation and GTP hydrolysis. Similar to the action of guanine-nucleotide exchange factors, the rate of GDP dissociation can be greatly enhanced by the reaction of Ras with small-molecule redox agents, such as nitrogen dioxide, which can promote Ras activation. Nitrogen dioxide is an auto-oxidation product of nitric oxide and can react with an accessible cysteine of Ras to cause oxidation of the bound guanine nucleotide to facilitate Ras guanine nucleotide dissociation. Glutathione has also been reported to modify Ras and alter its activity. To elucidate the mechanism by which glutathione alters Ras guanine nucleotide binding properties, we performed NMR and biochemical analyses of glutathiolated Ras. We determined that treatment of H-Ras, lacking the non-conserved hypervariable region, with oxidized glutathione results in glutathiolation specifically at cysteine 118. However, glutathiolation does not alter Ras structure or biochemical properties. Rather, changes in guanine nucleotide binding properties and Ras activity occur upon exposure of Ras to free radicals, presumably through the generation of a cysteine 118 thiyl radical. Interestingly, Ras glutathiolation protects Ras from further free radical-mediated activation events. Therefore, glutathiolation does not affect Ras activity unless Ras is modified by glutathione through a radical-mediated mechanism. 1
Keywords: Ras GTPases, glutathione, oxidation, reactive oxygen and nitrogen species, oxidative cysteine modifications
Introduction
Ras proteins belong to a large superfamily of GTPases that bind to GDP and GTP with high specificity and affinity. They function as molecular switches that cycle between the inactive GDP-bound and active GTP-bound forms. Once activated, Ras GTPases interact with a variety of effectors to activate signaling pathways that regulate gene expression, cell growth and differentiation, and programmed cell death [1-3]. The structural differences between the GDP- and GTP-bound states of Ras are primarily localized within two regions, Switch I (residues 30-37) and Switch II (residues 60-76) [4]. The intrinsic rates of GDP exchange and GTP hydrolysis are too slow to respond to cell signaling events [5], and consequently, protein factors associate with Ras and accelerate these rates in a regulated manner. Guanine nucleotide exchange factors (GEFs) accelerate exchange of bound GDP for GTP, which leads to Ras activation [6], whereas GTPase activating proteins (GAPs) inactivate the GTPase by facilitating GTP hydrolysis [7, 8]. Ras is considered the most prevalent oncogene in human cancer, and oncogenic mutations have been identified in approximately 30% of human tumors [9, 10]. Oncogenic Ras mutations render Ras constitutively active by impeding down regulation by GAPs [11].
In addition to GEFs, reactive oxygen and nitrogen species (ROS; RNS) have been shown to enhance guanine nucleotide exchange and activate Ras [12]. Two of the most common oxidative modifications identified for Ras in vivo are nitrosation and glutathiolation [13]. Specifically, nitrogen dioxide (•NO2) has been shown to induce Ras guanine nucleotide dissociation by causing guanine base oxidation through reaction with cysteine 118 (Cys118) [14]. As the GTP:GDP ratio in cells is approximately 10:1, oxidation-mediated GDP dissociation can promote GTP loading of Ras, analogous to the action of GEFs [15]. While the reaction of nitric oxide and its auto-oxidation product, •NO2, with Ras have been characterized, it is less clear how other thiol modifications (i.e., glutathiolation) affect Ras activity. As numerous studies have described activity modulation of Ras in the presence of ROS and RNS, we refer the reader to two recent reviews that detail the redox regulation of Ras and Ras-related GTPases [12, 16].
Ras contains a solvent accessible cysteine (X) in the nucleotide-binding NKXD motif. While this cysteine is conserved in H-, K- and N-Ras, it does not form interactions with other residues in Ras or the guanine nucleotide ligand and is poorly conserved in the Ras superfamily. We have previously postulated that Cys118 is conserved in N-, K-, and H-Ras due to its role in the redox regulation of Ras [15]. In fact, Cys118 has been shown to react in vitro and in vivo with a variety of thiol oxidizing agents, including oxidized glutathione, superoxide, and nitric oxide-derived oxidants [13]. We have previously characterized a redox inactive variant of Ras (RasC118S) that retains similar structure and biochemical properties as RasWT [17, 18]. RasC118S has been used in numerous in vitro and cell-based studies to discriminate between direct and indirect mechanisms of Ras activity modulation by Cys118-mediated oxidation [reviewed in 12, 13]. As we have previously shown that S-nitrosation of Ras (RasSNO) at Cys118 does not significantly alter Ras structure, biochemical properties, or binding to the Ras-binding domain of Raf-1 [19], we postulated that NO modulates Ras activity by formation of a thiyl-radical intermediate, which leads to oxidation of the bound guanine base, likely through electron transfer. In support of this premise, we have previously shown that treatment of Ras with •NO2 promotes the oxidation and release of the bound guanine nucleotide [14]. In addition, we have recently detected the presence of a Ras-thiyl radical upon treatment of RasWT with NO using the immuno-spin trapping (IST) reagent 5,5-dimethyl-1-pyrroline N-oxide (DMPO); no radical was detected when RasC118S was used [20]. Thus, we propose that Ras thiyl-radical formation, as opposed to S-nitrosation, induces oxidation and release of the guanine nucleotide base, which can lead to Ras activation.
In addition to S-nitrosation, several studies have observed that Ras can undergo glutathiolation. This oxidative modification was suggested to increase nucleotide exchange and promote Ras activation [21-27]. In particular, detection of Ras glutathiolation coincident with Ras activation was observed after exposure of bovine aortic endothelial cells (BAECs) to peroxynitrite [21, 27], addition of angiotensin II to vascular smooth muscle cells (VSMCs) [26, 28], and exposure of H2O2 to rat ventricular myocytes [23, 25]. However, addition of these agents to cultured cells can produce redox agents capable of protein thiyl radical formation. For example, peroxynitrite can react with cellular CO2 to produce CO3 •− and •NO2 radicals, and under acidic conditions (peroxynitrite pKa is ~6.6), peroxynitrite can decompose to hydroxyl radical and •NO2 [29]. Furthermore, peroxynitrite and angiotensin II can dysregulate several kinase pathways, including the PI3K/Akt pathway, and increase the production of cellular NO by activation of endothelial nitric oxide synthase (eNOS) [30, 31]. The NO produced by eNOS can auto-oxidize to produce •NO2, which is a powerful oxidant capable of thiyl radical formation. Consistent with observations of Liaudet et al. and Ushio-Fukai et al., the PI3K/Akt pathway was activated after exposure of BAECs to peroxynitrite [21]. Intriguingly, in a pancreatic cancer cell line and a severe combined immunodeficiency/Beige mouse model, activation of PI3K has been shown to activate Ras via eNOS stimulation through a redox-mediated mechanism dependent on Cys118 [32]. Furthermore, peroxynitrite and angiotensin II signaling both activate NADPH oxidase, which can produce waves of reactive oxygen species long after peroxynitrite decomposition [33]. Lastly, H2O2 can produce free radicals through the Fenton reaction by reacting with transition metals present in cells. Thus, based on our previous characterization of nitrosated Ras [19], we postulate that Ras activation observed by Clavreul et al. [21, 27] and Adachi et al. [26] results from a radical-mediated mechanism, which occurs prior to the modification by glutathione. However, due to the larger size of glutathione compared to NO, it was hypothesized that Ras glutathiolation at Cys118 causes structural changes in the nucleotide binding pocket, leading to perturbation of guanine nucleotide binding and an increased rate of nucleotide exchange [21]. As Cys118 is critical for the radical-mediated regulation of Ras activity, we sought to circumvent the confounding factors of peroxynitrite and angiotensin II use in cells by directly determining whether glutathiolation at Cys118 alters Ras structure and activity in vitro.
While Ras has been shown to be glutathiolated in cells [21, 23, 26, 27], it is unclear how glutathiolation alters Ras activity. Therefore, we employed NMR and fluorescence-based biochemical assays to assess whether glutathione modification of Ras perturbs Ras structure or activity. We find that treatment of Ras with oxidized glutathione leads to glutathiolation specifically at Cys118, which does not alter Ras tertiary structure or guanine nucleotide binding. These results are consistent with our previous observations that S-nitrosation of Ras Cys118 does not perturb the structure or activity of Ras [19]. Therefore, our data suggests that glutathiolation can only affect Ras activity if modification proceeds through a radical-mediated reaction. Moreover, Ras glutathiolation prevents further redox-mediated activation of Ras by free radical-based mechanisms, which may serve to protect Ras from future radical-mediated oxidation events under conditions of oxidative stress.
Materials and Methods
Ras purification and glutathiolation
Truncated human H-ras (H-Ras1-166) was cloned into the pQlinkH vector (Addgene; Cambridge, MA), which contains an N-terminal 6x-His purification tag followed by a Tobacco Etch Virus protease cleavage site for removal of the affinity tag. The hypervariable region of Ras, including the C-terminal CAAX box, was removed as this region does not undergo post-translational lipid modification in bacteria, is unstructured, and its removal does not affect guanine nucleotide binding or GTP hydrolysis [34]. All proteins were expressed in BL21 (DE3) RIPL cells (Stratagene; La Jolla, CA) and purified following the Qiagen Nickel NTA purification protocol (Germantown, MD). Ras was further purified by size exclusion chromatography (Superdex-75 10/300 GL column; GE Life Sciences; Piscataway, NJ) and judged greater than 95% pure by SDS-PAGE analysis.
Ras was modified with glutathione by the addition of 1000× oxidized glutathione to purified Ras in glutathiolation buffer (50 mM Tricine pH 8.0, 50 mM NaCl, 5 mM MgCl2, and 30 μM GDP) at 37°C for 15 min. Prior the addition of glutathione, Ras was reduced with dithiothreitol (DTT) for 30 min at pH 8.5 before being buffer exchanged into glutathiolation buffer that was flushed with N2 gas to remove dissolved oxygen and prevent auto-oxidation.
Mass Spectrometry of glutathiolated-Ras and Ras
Ras mass measurements were performed on an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific; San Jose, CA). The mass analysis of intact Ras samples was achieved in full-mass spectrometry (MS), selected-ion monitoring (SIM), and higher energy collisional dissociation-(HCD)-MS/MS modes with a resolution of 120,000 at m/z 400 Da. The intact MS spectra were deconvoluted using ProMass, and HCD-MS/MS product ion spectra were processed manually by assigning sequence ions to theoretical masses corresponding to RasSSG. The bottom-up peptide identification of trypsinized Ras samples was performed using reversed phase LC-MS/MS on a nano-LC ultra 2D plus (Eksigent Inc; Dublin, CA) coupled to an LTQ-Orbitrap Velos as described previously [35]. Briefly, data acquisition included a full-MS scan on the Orbitrap (externally calibrated to a mass accuracy of < 1 ppm and instrumental resolving power of 60,000 at m/z 400 Da) followed by intensity-dependent CAD-MS/MS of the top 10 most abundant peptide ions. Mass spectra were processed, and peptide identification was performed using Mascot (Matrix Science Inc.) against a Human Uniprot database. Peptides were confidently identified using a target-decoy approach with a false-discovery-rate (FDR) of 1%. A precursor ion mass tolerance of 200 ppm and product ion mass tolerance 0.5 Da with a maximum of two missed cleavages and variable modifications of cysteine glutathiolation and oxidation were used as a protein database search parameter. All peptides were filtered and reported within a mass accuracy of 5 ppm.
Nucleotide exchange and hydrolysis assays
The rate of GDP dissociation from Ras was measured using 2′-/3′-O-(N’-Methylanthraniloyl)guanosine-5′-O-diphosphate (MANTGDP) as previously reported [36, 37]. One micromolar Ras (RasWT, RasSSG, or RasC118S) loaded with MANTGDP (BioLog; San Diego, CA) was added to 1 mL of degassed assay buffer (50 mM Tris pH 7.4, 50 mM NaCl, 5 mM MgCl2, 2 mM GDP, and 100 μM diethylenetriaminepentaacetic dianhydride; DTPA), and the rate of guanine nucleotide dissociation was measured by monitoring the change in fluorescence (excitation: 365 nm; emission: 435 nm; 25°C) over time using a Perkin Elmer LS50B fluorimeter (Waltham, MA). All experiments were performed in triplicate. Fluorescent nucleotide dissociation curves were fit to a one-phase exponential decay equation using GraphPad Prism version 3.03 (GraphPad Software; San Diego, CA). For GEF-induced Ras-GDP dissociation assays, the minimal catalytic domain of Son of Sevenless (SOScat) was used at a 1:1 ratio to Ras. This concentration of SOScat was selected as the rate enhancement of nucleotide exchange was easy to measure using fluorimetric assays and other labs have published using the GEF at this concentration. The SOScat construct (from John Kuriyan; University of California, Berkeley) was expressed and purified as previously described [38].
Rates of GTP hydrolysis were determined using a Spectramax M5 (Molecular Devices; Sunnyvale, CA) fluorimeter by monitoring phosphate production upon GTP hydrolysis [39]. FLIPPi 5U (Addgene; Cambridge, MA) was used as the phosphate sensor, and the minimal catalytic domain of p120GAP [40] was added to stimulate the rate of Ras GTP hydrolysis. The expression, purification, and use of this sensor have been previously reported [41]. Briefly, all assays used 10 μM FLIPPi with 10 μM GTP-loaded Ras. GTP loading was performed as previously described [39]. Trace phosphate was removed from all buffers using a ‘phosphate mop’ [42]. The rate of GTP hydrolysis was measured by taking the ratio of the 535- and 485-nm emission wavelengths (excitation: 435 nm; 25°C) of kinetic runs performed in triplicate and fit to a one-phase exponential association curve. For these experiments, the ratio of GAP to Ras was 1:200, and the minimal catalytic domain of p120 RasGAP was used.
Nitric oxide-induced nucleotide dissociation
To determine whether addition of nitric oxide to RasWT, RasSSG, and RasC118S alters the rates of GDP dissociation, we used the NO• donor DEANO (diethylammonium (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate; Alexis Chemicals; Farmingdale, NY). Previous studies have shown that NO• in the presence of O2 forms •NO2, which promotes nucleotide dissociation in Ras [14]. Therefore, all buffers used with DEANO were not degassed and were vigorously shaken prior to use. To verify the time-dependent formation of NO• by DEANO in our buffers, diaminofluorescein (DAF; Sigma) was used (data not shown). DAF displays an increased fluorescence upon exposure to NO• radicals with an excitation wavelength of 445 nm and an emission wavelength of 515 nm. All runs were performed in triplicate with 1 μM MANT-loaded Ras in assay buffer. DEANO was added at the indicated concentrations at the start of each run. Fluorescent nucleotide dissociation curves were fit using one-phase exponential decay using GraphPad Prism as described above.
NMR analysis
NMR experiments were collected on a Varian Inova 700 MHz spectrometer at 25°C. The 2D 1H–15N heteronuclear single quantum coherence spectroscopy (HSQC) experiments were performed with pulsed field gradient and water flip-back methods as previously described [43]. Uniformly labeled 15N-enriched Ras was purified as previously described [44]. 2D 1H-15N HSQC experiments were acquired on 0.8 mM 15N-enriched RasWT and RasSSG with 1,024 × 128 complex data points and a spectral width of 8,000 Hz for the 1H dimension and 1,709 Hz for the 15N dimension. Buffer contained 10 mM maleate (pH 6.5), 5 mM MgCl2, 40 mM NaCl, 20 μM GDP, and 10% D2O. NMR data were processed and analyzed using NMR PIPE [45] and NMR ViewJ (One Moon Scientific; Newark, NJ).
Results
Modification of RasC118 by glutathione
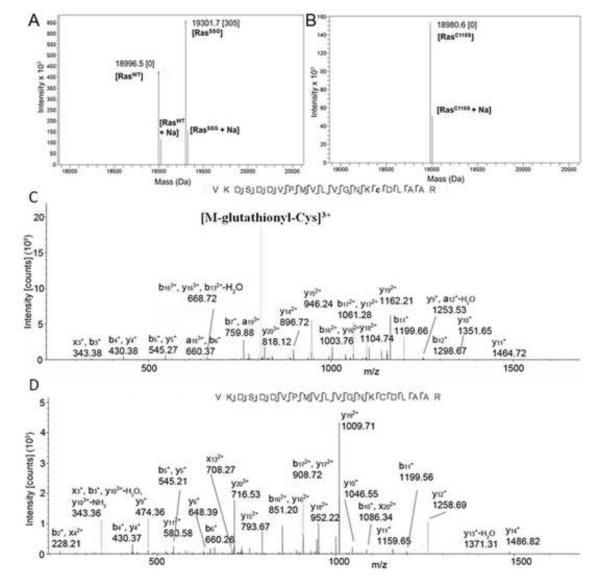
As RasC118 has been previously identified as the site of oxidative modification by nitric oxide [19], we used a combination of top-down and bottom-up MS to confirm that Cys118 is the preferred site of glutathiolation. Whereas partial modification of Ras by glutathione was obtained at a ratio of 50:1 GSSG to Ras upon incubation for 15 min at pH 8.5 (Fig. 1a), increasing the GSSG to Ras ratio to 1000:1 resulted in conversion of hRas1-166 to a predominantly single glutathiolated form. However, Ras glutathiolation was not observed when the redox insensitive RasC118S variant was treated under the same conditions (Fig. 1b). All full-MS and selected-ion monitoring (SIM) scans corresponding to glutathiolated RasC118 and redox-insensitive RasC118S are available in supporting data (Fig. S1). We have also confirmed the site-specificity of hRas1-166 glutathiolation using top-down HCD-MS/MS of the 13+ charge state (Fig. S2) and bottom-up identification of surrogate triply charged tryptic peptides that constitute Cys118 (Fig 1c-d). The top-down data contain diagnostic neutral loss ions corresponding to Cys-glutathionyl and glutathione as well as backbone product ions, which suggests that the glutathione moiety is present exclusively on Cys118. In concordance with hRas1-166, bottom-up peptide mass spectra show diagnostic neutral loss of Cys-glutathionyl and sequence ions that unambiguously localize glutathione on Cys118. Our mass spectrometry data indicate that RasC118, which is solvent exposed in the structure of hRas1-166 (pdb: 1crp), is susceptible to glutathiolation, whereas the two solvent inaccessible cysteine residues (Cys51 and Cys80) were unmodified under the tested conditions.
Figure 1.
(a) Intact deconvoluted mass spectrum resulting from the reaction of RasWT with GSSG. The major peaks correspond to unmodified and singly glutathiolated Ras. (b) Intact deconvoluted MS resulting from the reaction of RASC118S with GSSG. The peak corresponds to unmodified RASC118S. Note: The deconvoluted mass spectra in Fig 1 a-b also consist of gas-phase sodium adducts of Ras. (c) MS/MS-CID spectrum of a triply charged RASWT peptide glutathiolated at Cys118. The glutathione modification is localized at Cys118, and the diagnostic neutral loss of 129 Da corresponds to glutathionyl-cysteine [60]. (d) MS/MS-CID spectrum of the unmodified triply charged peptide counterpart.
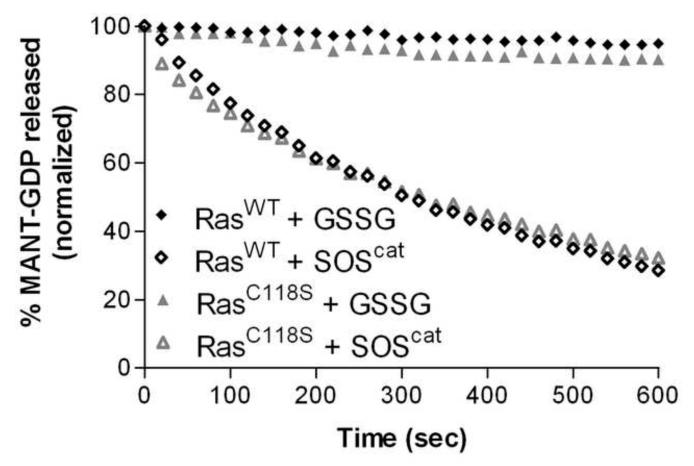
Glutathiolation of RasC118 does not significantly affect Ras-GDP nucleotide dissociation or GTP hydrolysis
It has previously been observed that peroxynitrite addition to BAECs and VSMCs promotes the formation of glutathiolated Ras at Cys118. Moreover, glutathiolation was concluded to be an activating modification as the population of activated Ras was also glutathiolated [21, 22, 26]. As exposure of Ras to peroxynitrite in cells could generate RasC118• and RasSSG, it is difficult to distinguish which modification, the thiyl radical or glutathione, leads to Ras activity changes under the experimental conditions used. To assess whether glutathiolation alters Ras guanine nucleotide binding and/or GTP hydrolysis, we performed nucleotide dissociation and GTP hydrolysis assays on unmodified and glutathiolated hRas1-166 in vitro. As shown in Fig. 2a and b, the intrinsic rates of GDP dissociation (kobs) for RasWT and RasSSG (Fig. 2a-b) were determined to be similar (3.9 ± 0.1×10−5 s−1 vs. 4.7 ± 0.5×10−5 s−1). As the intrinsic rate of nucleotide dissociation for Ras is slow, interactions with GEFs are required to facilitate nucleotide exchange in vivo. Therefore, we measured the rate of GDP dissociation in the presence of SOS, a Ras GEF, as previously described [36]. When the minimal catalytic domain of SOS (SOScat) was added to Ras, the rate of nucleotide dissociation for RasSSG and RasWT was similar (18.5 ± 0.5×10−4 s−1 vs. 20.2 ± 0.7×10−4 s−1, respectively).
Figure 2.
Biochemical characterization of glutathione-modified Ras. (A) Rates of GDP dissociation were determined for RasWT, RasC118S, and RasSSG (1 μM). Nucleotide dissociation kinetic traces in the absence (open symbols) and presence (closed) of the GEF SOScat are shown. (B) The relative rates of GDP dissociation (kobs) are presented. (C) Representative Ras GTP hydrolysis kinetic traces in the absence (open symbols) and presence (closed) of p120RasGAPcat. (D) The relative rates of hydrolysis in the absence and presence of GAP are presented. All reactions were performed in triplicate and were fit to a single exponential dissociation/association curve using GraphPad Prism, and the error is reported as the standard deviation of the replicates. Normalized results are presented for graphical comparison of the experiments. The experimental conditions and data analysis for these assays are described in the Materials and Methods.
To determine the effect of glutathiolation on Ras-GTP hydrolysis rates, we measured the rate of GTP hydrolysis in the presence and absence of the minimal catalytic domain of the Ras-specific GAP, p120GAP [40]. For this assay, the phosphate binding protein FLIPPi 5U was employed as a phosphate sensor. FLIPPi binds to free phosphate in solution and has a measurable change in fluorescence upon phosphate binding [41]. By performing single-turnover hydrolysis assays, the intrinsic rate of GTP hydrolysis for RasWT was determined to be 0.6 ± 0.04×10−4 s−1 at 25°C (Fig. 2c-d), similar to previously reported values of 4.4×10−4 s−1, which were performed at a higher temperature of 30°C [46]. The rates of GTP hydrolysis determined using the FLIPPi sensor compared to previously reported values, indicating that this method is suitable for comparing GTP hydrolysis rates. The intrinsic rate of GTP hydrolysis for RasSSG was determined to be ~4-fold slower (0.14 ± 0.06×10−4 s−1) than for RasWT. However, as the intrinsic rate of GTP hydrolysis is too slow to be biologically relevant and requires catalysis by GAPs in vivo to regulate Ras activity, we measured the rate of hydrolysis in the presence of catalytic amounts of p120GAP. The rate of GAP-mediated GTP hydrolysis for RasWT and RasSSG were observed to be similar (20.6 ± 2.3×10−4 s−1 and 19.4 ± 1.7×10−4 s−1, respectively; Fig. 2c-d). Thus, glutathiolation of RasC118 does not affect p120GAP-mediated GTP hydrolysis.
Glutathiolation at Cys118 does not perturb Ras structure
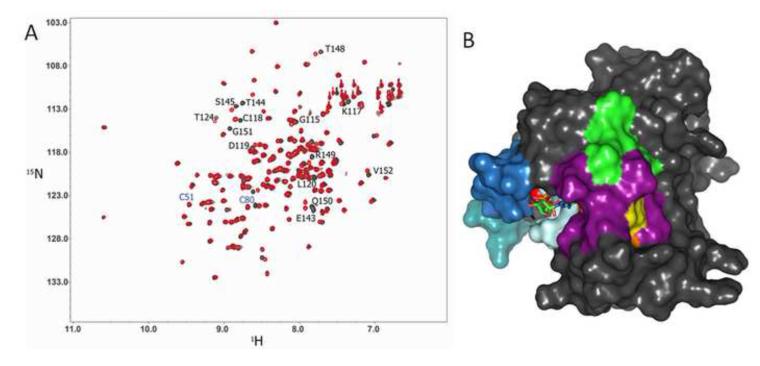
As it was previously suggested that Ras glutathiolation alters the structure of Ras and leads to increased nucleotide exchange [21], we glutathiolated 15N-enriched hRas1-166 and performed 2D NMR HSQC analyses. A 2D 1H-15N NMR HSQC allows for observation of backbone and side chain N-H groups and provides a site-specific probe for every residue in a protein aside from proline. An overlay of the RasWT and RasSSG HSQC spectra is shown in Fig. 3a. Consistent with our MS data that Cys118 is the site of glutathiolation, the amide resonance corresponding to Cys118 is perturbed upon Ras glutathiolation, whereas the chemical shifts associated with Cys51 and Cys80 are unaltered. Glutathione is a tripeptide (Gly-Cys-Glu), and modification of Ras at Cys118 is likely to alter the chemical environment (chemical shift) of resonances proximal to the site of modification. As shown in Fig. 3a, most resonances are unperturbed by glutathiolation of Ras at Cys118. By mapping the residues that show chemical shift perturbations (14 peaks with chemical shift differences greater than one linewidth, 0.05 ppm in the 1H dimension or 0.4 ppm in the 15N dimension) onto the surface of the Ras structure (Fig. 3b), only residues proximal to the modified cysteine show chemical shift changes. Interestingly, the chemical shift perturbations for RasSSG (green in Fig. 3B) compared to RasSNO (orange; overlap is shown in purple, Cys118 in yellow) are similar, and we have previously shown that RasSNO does not alter Ras structure [19]. Thus, the limited chemical shift changes close to the site of glutathiolation and similarity between the HSQC spectra of RasWT, RasSSG, and RasSNO suggest minimal structural perturbation by glutathiolation, consistent with our findings that the biochemical properties of Ras are unaltered. Furthermore, the residues important for Ras recognition by regulatory factors and effectors, including the p-loop (residues 10-17), Switch I, and Switch II, do not show chemical shift changes upon glutathiolation. These data are consistent with our findings that GEF- and GAP-mediated stimulation of GDP dissociation and GTP hydrolysis were unaffected by Ras glutathiolation.
Figure 3.
2D NMR 1H-15N HSQC comparison of RasWT and RasSSG. (A) A 1H-15N 2D HSQC overlay of RasWT (black) with RasSSG (red). The black-labeled residues indicate Ras NH resonances associated with residues that shift upon glutathione modification (peak shifts greater than 1 linewidth, 0.05 ppm in the 1H dimension or 0.4 ppm in the 15N dimension), and the blue-labeled residues correspond to the other two cysteines in Ras (Cys51 and Cys80), which do not show amide peak shifts between RasWT and RasSSG. (B) Residues corresponding to the NH resonances that show chemical shift changes for RasSSG but not RasNO are highlighted in green on a surface representation of hRas1-166, while residues that show chemical shift changes in RasSNO are highlighted in orange. The residues that showed chemical shifts in both RasSSG and RasSNO are highlighted in purple. Cys118, the site of modification, is shown in yellow, GDP is shown as sticks, magnesium is shown as a red sphere, and Switch I (blue), Switch II (teal), and the p-loop (pale blue) are shown for clarity. Surface rendering was generated with PyMol using pdb: 1crp.
Glutathiolation impedes redox-mediated nucleotide dissociation
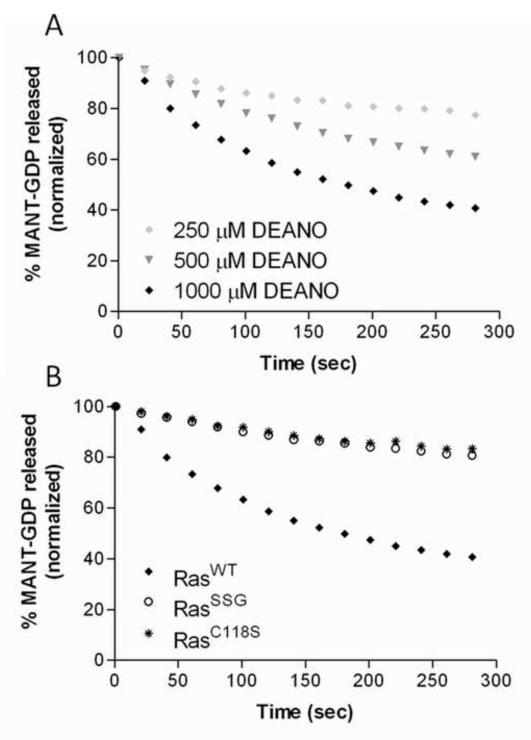
Previous studies have shown that treatment of Ras with •NO2 enhances the rate of nucleotide dissociation, whereas nitrosation of Ras at this site prevents radical-mediated nucleotide dissociation [15]. To determine whether Ras glutathiolation, like nitrosation, impedes free radical-mediated dissociation in Ras, the rate of nucleotide dissociation was determined using a MANTGDP-dissociation assay in the presence of the NO•-releasing agent DEANO [47]. As shown in Fig. 4a, we observe an enhanced rate of RasWT-GDP dissociation in the presence of DEANO (greater than 200-fold faster). In contrast, the rate of GDP dissociation for the redox inactive RasC118S variant was insensitive to the presence of DEANO. Furthermore, RasSSG was resistant towards DEANO-mediated nucleotide dissociation. Although 1 mM DEANO was used in these experiments, only a small fraction generates •NO2. DEANO releases NO• with a half-life of 15 minutes at pH 7.0, and NO•, once generated, reacts with O2 to generate •NO2 [48, 49]. Moreover, the estimated dissolved oxygen content in aqueous solution is only 250 M, which limits the amount of •NO2 that can be formed. Thus, our results show that glutathiolation impedes free radical-dependent Ras GDP dissociation, whereas glutathiolation does not significantly affect Ras structure, activity, or interactions with modulatory proteins.
Figure 4.
MANTGDP nucleotide dissociation assays in the presence of the NO-generating agent DEANO. (A) RasWT was pre-loaded with MANTGDP and exposed to increasing concentrations of DEANO in the absence of unlabeled GDP. All conditions contained 1 μM RasWT and ◆ 250 μM DEANO, ▾ 500 μM DEANO, or ◆ 1 mM DEANO added at the start of the experiment. (B) ◆ RasWT, * RasC118S, and 엯 RasSSG (1 μM) were preloaded with MANTGDP and exposed to 1 mM DEANO. All reactions were performed in triplicate and were fit to a single exponential dissociation curve using GraphPad Prism. Normalized results are presented for graphical comparison of the experiments.
Discussion
A number of studies have reported Ras activity modulation by ROS and RNS [13]. Most of these studies point to RasC118 as the redox-sensitive site because this cysteine has been shown to be oxidized by a number of cysteine-modifying agents and is required for redox-mediated regulation of Ras activity [12]. Two of the most common oxidative modifications identified for Ras in vivo are nitrosation and glutathiolation. We have previously reported that radical-mediated nitrosation of Ras leads to increased guanine nucleotide dissociation [50], whereas non-radical-mediated nitrosation of RasC118 does not affect Ras guanine nucleotide binding [19]. Given these observations, we were intrigued by reports that glutathiolation of Ras alters Ras activity [21, 22, 26, 27].
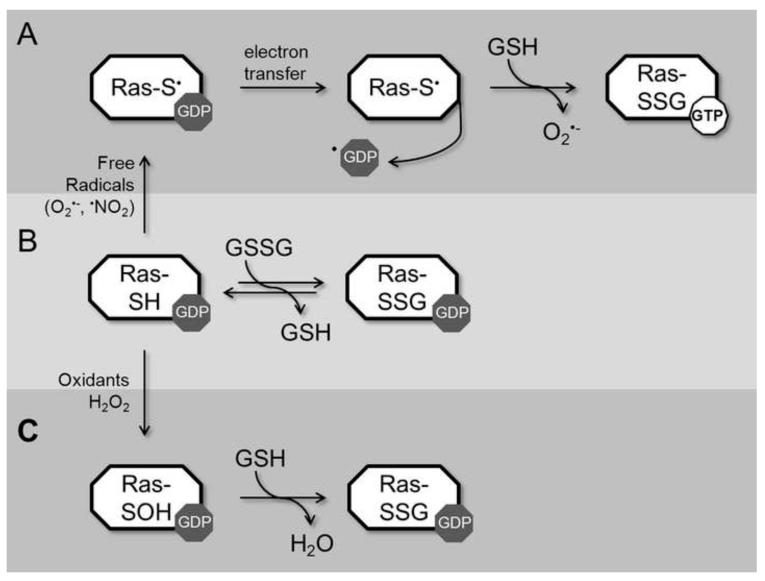
To better understand how Ras activity is modulated by glutathione, we considered radical- and non-radical-mediated reactions that can result in Ras glutathiolation. Three potential pathways of glutathiolation are presented in Fig. 6 (radical-mediated, 6a; and non-radical-mediated, 6b and 6c). Earlier reports have suggested that glutathiolation leads to Ras activation. For example, the fraction of activated GTP-bound Ras was higher in the pool shown to be glutathiolated upon peroxynitrite exposure to BAECs [21] and after administration of angiotensin II to VSMCs [26]. However, angiotensin II and peroxynitrite produce free radicals. Angiotensin II produces ROS through NAD(P)H oxidase activation [31, 51], and peroxynitrite can undergo homolysis to generate ROS and RNS. In fact, under most physiological conditions, CO2 reacts with peroxynitrite to generate CO •−3 and •NO2 byproducts in fairly high yields (>30%) [52, 53]; in addition, under acidic conditions, peroxynitrite can undergo proton catalyzed homolysis to form •OH and •NO2 [54]. An important consideration that must guide in vitro experimentation with peroxynitrite is that in the absence of CO2, peroxynitrite reacts directly with many biochemical targets, particularly thiols. This oxidative reaction produces predominately non-radical oxidative products and is faster than its homolysis to •OH and •NO2. Therefore, peroxynitrite reactions are heavily context dependent. Importantly, given the high glutathione concentration in cells, once peroxynitrite is decomposed and the free radical chain reactions are completed, glutathiolated Ras will predictably be a major end product that is indistinguishable by all means from the end product produced directly by the reaction of oxidized glutathione with RasWT. Therefore, we postulate that the exogenous addition of peroxynitrite and angiotensin II promotes Ras thiyl radical-mediated oxidation of the bound guanine nucleotide, leading to Ras activation. Further, the resulting thiyl radical can react with glutathione to produce RasSSG•, which aerobically decays to RasSSG and O •−2. This route of activation is shown in Fig. 6A, and based on our analysis, is the likely pathway of glutathiolation that would result in Ras activation in vivo. Interestingly, Ras glutathiolation appears to protect Ras from further free radical-mediated events. We evaluated guanine nucleotide binding of RasWT, RasC118S, and RasSSG in the presence of the NO-releasing agent DEANO. In these experiments (Fig. 4), glutathiolated Ras was protected from free radical-mediated nucleotide dissociation. Therefore, glutathione, which likely modifies Ras after an initial activating event, could protect Ras from over-oxidation in cells under conditions of oxidative stress.
Figure 6.
Modification of Ras by glutathione can proceed by three different mechanisms. In pathway A, redox agents capable to generating a Ras thiyl radical can induce radical formation on RasC118; electron transfer results in guanine-radical formation, guanine-base oxidation, and release of the oxidized base from Ras. Given the ratio of GTP/GDP in cells (~30:1), release of GDP promotes GTP binding. Free radical-induced oxidation leaves Ras in the thiyl anion form, which is more reactive to oxidation and can result in glutathione modification. In pathway B, Ras can react with oxidized glutathione through disulfide exchange to form glutathiolated Ras. Given the slow rate of this reaction and concentration of GSSG in cells, this pathway is unlikely to have a large contribution to Ras glutathiolation in vivo. In pathway C, Ras is oxidized by a non-radical oxidant, such as H2O2, which results in sulfenic acid formation at RasC118. Sulfenic acids are an intermediate step in disulfide bond formation; therefore, this form of Ras can interact with reduced glutathione to form glutathiolated Ras. However, RasSSG formed by non-radical means does not affect guanine nucleotide binding, nucleotide hydrolysis, or Ras activation. Thus, Ras activation and glutathiolation likely occurs in cells by reaction of Ras with thiyl radical-generating agents (pathway A), which promotes activation through nucleotide dissociation and GTP binding.
The other potential pathways of Ras glutathiolation shown in Figure 6 are unlikely to lead to Ras activation (Fig. 6b and 6c). To demonstrate this, we generated glutathione-modified Ras through a non-radical-mediated pathway by treating Ras with oxidized glutathione (Fig. 6b), which glutathiolates Ras at Cys118, and find that neither the intrinsic or regulator (GEF/GAP)-mediated rates of guanine nucleotide dissociation and hydrolysis are altered (Fig. 2). However, as the cellular concentration of oxidized glutathione is generally quite low (GSH:GSSG ratio in cells is >100:1) [55] and the reaction rate slow, this is an unlikely pathway of Ras glutathiolation in cells. To experimentally validate the role of glutathione on nucleotide exchange, we exposed RasWT and RasC118S to oxidized glutathione (similar to the pathway presented in Fig. 6b) and found that Ras guanine nucleotide binding was not affected by the presence of oxidized glutathione (Fig. 5). While this data appears contradictory to the results described by Clavreul et al. [21], identical levels of Ras nucleotide dissociation were observed, which was approximately 5% of the bound nucleotide after 300 s. In our experiments, we also tested RasC118S and observed identical levels of nucleotide dissociation, indicating that glutathione does not promote guanine nucleotide release from Ras. We also present GEF-induced dissociation of RasWT and RasC118S within Fig. 5 to highlight the difference in GSSG-mediated dissociation and GEF-mediated dissociation. Furthermore, as the experimental conditions used in Clavreul et al. [21] were not conducive to peroxynitrite-mediated radical formation [7], glutathiolation is unlikely to result in Ras activation. Thus, results from this study do not support Ras activity modulation due to the presence of the glutathione moiety on Ras.
Figure 5.
MANTGDP dissociation measured for RasWT (◆) and RasC118S (▴) in the presence of 100 μM GSSG (closed symbols) and in the presence of SOScat (open symbols). MANTGDP nucleotide dissociation was performed as described in Fig. 2; however, no unlabeled GDP was present and 100 μM oxidized glutathione was added to the reaction cuvette. All reactions were performed in triplicate and were fit to a single exponential dissociation curve using GraphPad Prism. Error bars have been removed for clarity. Ras and RasC118S with 1 μM GEF are shown for comparison. Normalized results are presented for graphical comparison of the experiments.
Seemingly in direct contradiction with our results, Adachi et al. observed that overexpression of glutaredoxin-1 (GRX-1) in VSMCs inhibited Ras glutathiolation and activation upon stimulation with peroxynitrite [26]. However, overexpressed redox proteins can have profound effects on the cellular oxidation state, and it has been previously shown that downregulation of GRX-1 can sensitize lens epithelial cells to oxidative stress-induced apoptosis [56]. Moreover, overexpression of GRX-1 protects against doxorubicin-induced cell death [57, 58], which mediates cell death through increased oxidative stress [59]. Therefore, overexpression of GRX-1, as in Adachi et al., likely decreased the level of oxidative stress, which would result in reduced Ras activation by radical-mediated oxidation.
Another possible reaction pathway to consider is shown in Figure 6c. As disulfide bond formation generally proceeds through a sulfenic acid intermediate, it is possible that oxidation of Cys118 to a sulfenic acid will lead to RasSSG formation in the presence of reduced glutathione, which is present at a high levels in cells (GSH:GSSG ratio in cells is >100:1). As this pathway does not generate radical intermediates, we postulate that it is unlikely to alter Ras activity. While the products of angiotensin II or peroxynitrite exposure to cells can lead to Ras glutathiolation by the pathway presented in Fig. 6c; the relative rate of oxidation is likely too slow to be physiologically relevant. It is more likely that glutathiolation occurs through radical-mediated interactions in cells. However, as Ras glutathiolation can proceed through pathways that do not alter Ras activity, it is difficult to use Ras glutathiolation as a marker of Ras activity regulation unless Ras radical formation can be detected coincident with Ras glutathiolation.
Conclusions
We have previously shown that •NO2 can increase the rate of nucleotide dissociation in Ras and that modification of Ras by NO• renders Ras less sensitive to radical-mediated oxidation [19]. Consistent with these observations, RasSSG does not affect the structure or activity of Ras. When considering the faster reaction rates of radical vs. non-radical-mediated cysteine oxidation, we postulate that activation mediated through oxidation of Ras in vivo occurs through the formation of a Ras thiyl-radical intermediate (Fig. 6). The Ras thiyl radical can promote Ras activation under appropriate redox conditions by facilitating nucleotide exchange and can react with NO• or GS• radicals to form RasSNO or RasSSG (Fig. 6). While these end products do not alter Ras guanine nucleotide binding or hydrolysis, they impede Cys118 from engaging in further free radical-mediated reactions (Fig. 4). Therefore, we postulate that coincident detection of Ras glutathiolation and activation results from Cys118 thiyl radical-mediated oxidation of the guanine base and GDP release.
Supplementary Material
Highlights.
Ras is selectively modified by glutathione at cysteine 118
Ras structure is unaffected by glutathione modification at cysteine 118
Ras glutathiolation is non-activating unless it proceeds through a free radical
Nucleotide exchange and hydrolysis of glutathiolated Ras are similar to RasWT
Free radical-induced modifications at cysteine 118 activate Ras
Acknowledgments
The research efforts described herein, were supported by NIH 5RO1GM075431 to SLC. GAH was partially funded by the Program in Molecular and Cellular Biophysics (NIH T32GM008570). We would also like to thank Min Qi Lu for his help in purifying the proteins and Michael Davis for his assistance with the redox chemistry.
Footnotes
List of Abbreviations: bovine aortic endothelial cells (BAECs); diaminofluorescein (DAF); diethylammonium (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate (DEANO); 5,5-dimethyl-1-pyrroline N-oxide (DMPO); endothelial nitric oxide synthase (eNOS); GTPase activating proteins (GAPs); Guanine nucleotide exchange factors (GEFs); higher energy collisional dissociation (HCD); heteronuclear single quantum coherence spectroscopy (HSQC); 2′-/3′-O-(N’-Methylanthraniloyl)guanosine-5′-O-diphosphate (MANT-GDP); mass spectrometry (MS); nitrogen dioxide (•NO ); S-nitrosation of Ras (RasSNO); glutathiolated-Ras (RasSSG 2 ); reactive oxygen and nitrogen species (ROS; RNS); son of sevenless (SOS); selected ion monitoring (SIM); and vascular smooth muscle cells (VSMCs).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Disclosure Statement All co-authors have seen and agree with the contents of the manuscript and there are no competing financial interests.
References
- [1].Oxford G, Theodorescu D. Ras superfamily monomeric G proteins in carcinoma cell motility. Cancer Lett. 2003;189:117–128. doi: 10.1016/s0304-3835(02)00510-4. [DOI] [PubMed] [Google Scholar]
- [2].Paduch M, Jelen F, Otlewski J. Structure of small G proteins and their regulators. Acta Biochim Pol. 2001;48:829–850. [PubMed] [Google Scholar]
- [3].Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- [4].Kjeldgaard M, Nyborg J, Clark BF. The GTP binding motif: variations on a theme. Faseb J. 1996;10:1347–1368. [PubMed] [Google Scholar]
- [5].John J, Rensland H, Schlichting I, Vetter I, Borasio GD, Goody RS, Wittinghofer A. Kinetic and structural analysis of the Mg(2+)-binding site of the guanine nucleotide-binding protein p21H-ras. J Biol Chem. 1993;268:923–929. [PubMed] [Google Scholar]
- [6].Sprang S. GEFs: master regulators of G-protein activation. Trends Biochem Sci. 2001;26:266–267. doi: 10.1016/s0968-0004(01)01818-7. [DOI] [PubMed] [Google Scholar]
- [7].Geyer M, Wittinghofer A. GEFs, GAPs, GDIs and effectors: taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins. Curr Opin Struct Biol. 1997;7:786–792. doi: 10.1016/s0959-440x(97)80147-9. [DOI] [PubMed] [Google Scholar]
- [8].Sprang SR. G proteins, effectors and GAPs: structure and mechanism. Curr Opin Struct Biol. 1997;7:849–856. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- [9].Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- [10].Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- [11].Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- [12].Davis MF, Vigil D, Campbell SL. Regulation of Ras proteins by reactive nitrogen species. Free radical biology & medicine. 2011;51:565–575. doi: 10.1016/j.freeradbiomed.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lancaster JR., Jr. Protein cysteine thiol nitrosation: maker or marker of reactive nitrogen species-induced nonerythroid cellular signaling? Nitric Oxide. 2008;19:68–72. doi: 10.1016/j.niox.2008.04.028. [DOI] [PubMed] [Google Scholar]
- [14].Heo J, Campbell SL. Mechanism of p21Ras S-nitrosylation and kinetics of nitric oxide-mediated guanine nucleotide exchange. Biochemistry. 2004;43:2314–2322. doi: 10.1021/bi035275g. [DOI] [PubMed] [Google Scholar]
- [15].Heo J, Campbell SL. Ras regulation by reactive oxygen and nitrogen species. Biochemistry. 2006;45:2200–2210. doi: 10.1021/bi051872m. [DOI] [PubMed] [Google Scholar]
- [16].Mitchell L, Hobbs GA, Aghajanian A, Campbell SL. Redox Regulation of Ras and Rho GTPases: Mechanism and Function. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lander HM, Hajjar DP, Hempstead BL, Mirza UA, Chait BT, Campbell S, Quilliam LA. A molecular redox switch on p21(ras). Structural basis for the nitric oxide-p21(ras) interaction. The Journal of biological chemistry. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- [18].Lander HM, Milbank AJ, Tauras JM, Hajjar DP, Hempstead BL, Schwartz GD, Kraemer RT, Mirza UA, Chait BT, Burk SC, Quilliam LA. Redox regulation of cell signalling. Nature. 1996;381:380–381. doi: 10.1038/381380a0. [DOI] [PubMed] [Google Scholar]
- [19].Williams JG, Pappu K, Campbell SL. Structural and biochemical studies of p21Ras S-nitrosylation and nitric oxide-mediated guanine nucleotide exchange. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6376–6381. doi: 10.1073/pnas.1037299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Davis MF, Zhou L, Ehrenshaft M, Ranguelova K, Gunawardena HP, Chen X, Bonini M, Mason RP, Campbell SL. Detection of Ras GTPase protein radicals through immuno-spin trapping. Free radical biology & medicine. 2012 doi: 10.1016/j.freeradbiomed.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Clavreul N, Adachi T, Pimental DR, Ido Y, Schoneich C, Cohen RA. S-glutathiolation by peroxynitrite of p21ras at cysteine-118 mediates its direct activation and downstream signaling in endothelial cells. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:518–520. doi: 10.1096/fj.05-4875fje. [DOI] [PubMed] [Google Scholar]
- [22].Sethuraman M, Clavreul N, Huang H, McComb ME, Costello CE, Cohen RA. Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free radical biology & medicine. 2007;42:823–829. doi: 10.1016/j.freeradbiomed.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kuster GM, Pimentel DR, Adachi T, Ido Y, Brenner DA, Cohen RA, Liao R, Siwik DA, Colucci WS. Alpha-adrenergic receptor-stimulated hypertrophy in adult rat ventricular myocytes is mediated via thioredoxin-1-sensitive oxidative modification of thiols on Ras. Circulation. 2005;111:1192–1198. doi: 10.1161/01.CIR.0000157148.59308.F5. [DOI] [PubMed] [Google Scholar]
- [24].Kuster GM, Siwik DA, Pimentel DR, Colucci WS. Role of reversible, thioredoxin-sensitive oxidative protein modifications in cardiac myocytes. Antioxid Redox Signal. 2006;8:2153–2159. doi: 10.1089/ars.2006.8.2153. [DOI] [PubMed] [Google Scholar]
- [25].Pimentel DR, Adachi T, Ido Y, Heibeck T, Jiang B, Lee Y, Melendez JA, Cohen RA, Colucci WS. Strain-stimulated hypertrophy in cardiac myocytes is mediated by reactive oxygen species-dependent Ras S-glutathiolation. J Mol Cell Cardiol. 2006;41:613–622. doi: 10.1016/j.yjmcc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- [26].Adachi T, Pimentel DR, Heibeck T, Hou X, Lee YJ, Jiang B, Ido Y, Cohen RA. S-glutathiolation of Ras mediates redox-sensitive signaling by angiotensin II in vascular smooth muscle cells. The Journal of biological chemistry. 2004;279:29857–29862. doi: 10.1074/jbc.M313320200. [DOI] [PubMed] [Google Scholar]
- [27].Clavreul N, Bachschmid MM, Hou X, Shi C, Idrizovic A, Ido Y, Pimentel D, Cohen RA. S-glutathiolation of p21ras by peroxynitrite mediates endothelial insulin resistance caused by oxidized low-density lipoprotein. Arterioscler Thromb Vasc Biol. 2006;26:2454–2461. doi: 10.1161/01.ATV.0000242791.28953.4c. [DOI] [PubMed] [Google Scholar]
- [28].Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, Cohen RA. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med. 2004;10:1200–1207. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- [29].Bonini MG, Augusto O. Carbon dioxide stimulates the production of thiyl, sulfinyl, and disulfide radical anion from thiol oxidation by peroxynitrite. The Journal of biological chemistry. 2001;276:9749–9754. doi: 10.1074/jbc.M008456200. [DOI] [PubMed] [Google Scholar]
- [30].Liaudet L, Vassalli G, Pacher P. Role of peroxynitrite in the redox regulation of cell signal transduction pathways. Front Biosci. 2009;14:4809–4814. doi: 10.2741/3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ushio-Fukai M, Alexander RW, Akers M, Yin Q, Fujio Y, Walsh K, Griendling KK. Reactive oxygen species mediate the activation of Akt/protein kinase B by angiotensin II in vascular smooth muscle cells. The Journal of biological chemistry. 1999;274:22699–22704. doi: 10.1074/jbc.274.32.22699. [DOI] [PubMed] [Google Scholar]
- [32].Lim KH, Ancrile BB, Kashatus DF, Counter CM. Tumour maintenance is mediated by eNOS. Nature. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee C, Miura K, Liu X, Zweier JL. Biphasic regulation of leukocyte superoxide generation by nitric oxide and peroxynitrite. The Journal of biological chemistry. 2000;275:38965–38972. doi: 10.1074/jbc.M006341200. [DOI] [PubMed] [Google Scholar]
- [34].John J, Schlichting I, Schiltz E, Rosch P, Wittinghofer A. C-terminal truncation of p21H preserves crucial kinetic and structural properties. J Biol Chem. 1989;264:13086–13092. [PubMed] [Google Scholar]
- [35].Gunawardena HP, Huang Y, Kenjale R, Wang H, Xie L, Chen X. Unambiguous characterization of site-specific phosphorylation of leucine-rich repeat Fli-I-interacting protein 2 (LRRFIP2) in Toll-like receptor 4 (TLR4)-mediated signaling. The Journal of biological chemistry. 2011;286:10897–10910. doi: 10.1074/jbc.M110.168179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lenzen C, Cool RH, Prinz H, Kuhlmann J, Wittinghofer A. Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry. 1998;37:7420–7430. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- [37].Lenzen C, Cool RH, Wittinghofer A. Analysis of intrinsic and CDC25-stimulated guanine nucleotide exchange of p21ras-nucleotide complexes by fluorescence measurements. Methods Enzymol. 1995;255:95–109. doi: 10.1016/s0076-6879(95)55012-7. [DOI] [PubMed] [Google Scholar]
- [38].Sondermann H, Soisson SM, Boykevisch S, Yang SS, Bar-Sagi D, Kuriyan J. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- [39].Shutes A, Der CJ. Real-time in vitro measurement of intrinsic and Ras GAP-mediated GTP hydrolysis. Methods Enzymol. 2006;407:9–22. doi: 10.1016/S0076-6879(05)07002-3. [DOI] [PubMed] [Google Scholar]
- [40].Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- [41].Gu H, Lalonde S, Okumoto S, Looger LL, Scharff-Poulsen AM, Grossman AR, Kossmann J, Jakobsen I, Frommer WB. A novel analytical method for in vivo phosphate tracking. FEBS Lett. 2006;580:5885–5893. doi: 10.1016/j.febslet.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- [43].Grzesiek S, Bax A. Measurement of amide proton exchange rates and NOEs with water in 13C/15N-enriched calcineurin B. J Biomol NMR. 1993;3:627–638. doi: 10.1007/BF00198368. [DOI] [PubMed] [Google Scholar]
- [44].Bax A, Ikura M, Kay LE, Barbato G, Spera S. Multidimensional triple resonance NMR spectroscopy of isotopically uniformly enriched proteins: a powerful new strategy for structure determination. Ciba Found Symp. 1991;161:108–119. doi: 10.1002/9780470514146.ch8. discussion 119-135. [DOI] [PubMed] [Google Scholar]
- [45].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- [46].John J, Rensland H, Schlichting I, Vetter I, Borasio GD, Goody RS, Wittinghofer A. Kinetic and structural analysis of the Mg(2+)-binding site of the guanine nucleotide-binding protein p21H-ras. The Journal of biological chemistry. 1993;268:923–929. [PubMed] [Google Scholar]
- [47].Keefer LK, Nims RW, Davies KM, Wink DA. “NONOates” (1-substituted diazen-1-ium-1,2-diolates) as nitric oxide donors: convenient nitric oxide dosage forms. Methods Enzymol. 1996;268:281–293. doi: 10.1016/s0076-6879(96)68030-6. [DOI] [PubMed] [Google Scholar]
- [48].Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol. 1996;271:C1424–1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- [49].Jourd’heuil D, Jourd’heuil FL, Feelisch M. Oxidation and nitrosation of thiols at low micromolar exposure to nitric oxide. Evidence for a free radical mechanism. The Journal of biological chemistry. 2003;278:15720–15726. doi: 10.1074/jbc.M300203200. [DOI] [PubMed] [Google Scholar]
- [50].Heo J, Prutzman KC, Mocanu V, Campbell SL. Mechanism of free radical nitric oxide-mediated Ras guanine nucleotide dissociation. J Mol Biol. 2005;346:1423–1440. doi: 10.1016/j.jmb.2004.12.050. [DOI] [PubMed] [Google Scholar]
- [51].Ushio-Fukai M, Alexander RW, Akers M, Griendling KK. p38 Mitogen-activated protein kinase is a critical component of the redox-sensitive signaling pathways activated by angiotensin II. Role in vascular smooth muscle cell hypertrophy. The Journal of biological chemistry. 1998;273:15022–15029. doi: 10.1074/jbc.273.24.15022. [DOI] [PubMed] [Google Scholar]
- [52].Kissner R, Koppenol WH. Product distribution of peroxynitrite decay as a function of pH, temperature, and concentration. J Am Chem Soc. 2002;124:234–239. doi: 10.1021/ja010497s. [DOI] [PubMed] [Google Scholar]
- [53].Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- [54].Merenyi G, Lind J, Goldstein S, Czapski G. Peroxynitrous acid homolyzes into *OH and *NO2 radicals. Chem Res Toxicol. 1998;11:712–713. doi: 10.1021/tx980043h. [DOI] [PubMed] [Google Scholar]
- [55].Han D, Hanawa N, Saberi B, Kaplowitz N. Mechanisms of liver injury. III. Role of glutathione redox status in liver injury. Am J Physiol Gastrointest Liver Physiol. 2006;291:G1–7. doi: 10.1152/ajpgi.00001.2006. [DOI] [PubMed] [Google Scholar]
- [56].Lofgren S, Fernando MR, Xing KY, Wang Y, Kuszynski CA, Ho YS, Lou MF. Effect of thioltransferase (glutaredoxin) deletion on cellular sensitivity to oxidative stress and cell proliferation in lens epithelial cells of thioltransferase knockout mouse. Invest Ophthalmol Vis Sci. 2008;49:4497–4505. doi: 10.1167/iovs.07-1404. [DOI] [PubMed] [Google Scholar]
- [57].Franco R, Cidlowski JA. Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ. 2009;16:1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]
- [58].Meyer EB, Wells WW. Thioltransferase overexpression increases resistance of MCF-7 cells to adriamycin. Free radical biology & medicine. 1999;26:770–776. doi: 10.1016/s0891-5849(98)00247-0. [DOI] [PubMed] [Google Scholar]
- [59].Eom YW, Kim MA, Park SS, Goo MJ, Kwon HJ, Sohn S, Kim WH, Yoon G, Choi KS. Two distinct modes of cell death induced by doxorubicin: apoptosis and cell death through mitotic catastrophe accompanied by senescence-like phenotype. Oncogene. 2005;24:4765–4777. doi: 10.1038/sj.onc.1208627. [DOI] [PubMed] [Google Scholar]
- [60].Hung CW, Schlosser A, Wei J, Lehmann WD. Collision-induced reporter fragmentations for identification of covalently modified peptides. Anal Bioanal Chem. 2007;389:1003–1016. doi: 10.1007/s00216-007-1449-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.