Summary
Antibodies of the VRC01 class neutralize HIV-1, arise in diverse HIV-1-infected donors, and are potential templates for an effective HIV-1 vaccine. However, the stochastic processes that generate repertoires in each individual of >1012 antibodies make elicitation of specific antibodies uncertain. Here we determine the ontogeny of the VRC01 class by crystallography and next-generation sequencing. Despite antibody-sequence differences exceeding 50%, antibody-gp120 cocrystal structures reveal VRC01-class recognition to be remarkably similar. B cell transcripts indicate that VRC01-class antibodies require few specific genetic elements, suggesting that naive-B cells with VRC01-class features are generated regularly by recombination. Virtually all of these fail to mature, however, with only a few—likely one—ancestor B cell expanding to form a VRC01-class lineage in each donor. Developmental similarities in multiple donors thus reveal the generation of VRC01-class antibodies to be reproducible in principle, thereby providing a framework for attempts to elicit similar antibodies in the general population.
INTRODUCTION
Despite extraordinary global diversity and immune-evading capabilities of HIV-1, the human immune system can develop antibodies that neutralize HIV-1 effectively. After 2 years, roughly 20% of HIV-1-infected individuals develop antibodies capable of neutralizing diverse strains of HIV-1 (Binley et al., 2008; Dhillon et al., 2007; Li et al., 2007). From these HIV-1-infected donors, a number of human monoclonal antibodies have been identified that individually neutralize over 50% of circulating HIV-1 strains (reviewed in Kwong and Mascola, 2012). Although such antibodies might provide little benefit to the infected individuals in whom they arise because of rapid escape by evolving virus (Richman et al., 2003; Wei et al., 2003; Wu et al., 2012), they nonetheless can prevent infection upon passive infusion or genetic delivery in simian and murine transmission models (Balazs et al., 2012; Hessell et al., 2009; Mascola et al., 2000). Such antibodies have thus been the focus of intense interest because each provides a potential means to prevent HIV-1 infection.
Interest in effective HIV-1-neutralizing antibodies has focused on mechanisms of recognition (epitope specificities and structural features that allow interaction with the envelope trimer) and on B cell ontogeny (the origin and development of B cells through processes by which their antibody genes recombine and antigen-specific B cells mature to produce antibodies with high affinity immunologic recognition). Antibody specificity is gained through V(D)J gene recombination to encode the naive B cell receptor and through antigen-driven somatic mutation of antibody genes to produce a clonal lineage of antibody-producing and memory B cells (Paul, 1999). These processes are highly stochastic. The naive B cell repertoire in each individual is estimated at 1012, and this repertoire is diversified further by somatic mutation (Glanville et al., 2009; Paul, 1999). Highly effective HIV-1-neutralizing antibodies, however, target just a few regions of the viral spike and can employ similar structural modes. Antibodies that recognize the same region, employ the same structural mode of recognition, and develop through similar B cell ontogeny can be considered a “class” (Kwong and Mascola, 2012). Classes that are observed in multiple individuals potentially represent immunological solutions to the challenge of HIV-1 neutralization and might be available to the general human population.
Of the approximately 20 selected donors from whom highly effective HIV-1-neutralizing antibodies have been identified thus far, five have antibodies attributed to a single class (Scheid et al., 2011; Wu et al., 2010, 2011). This class—the VRC01 class named after the first identified class member (Wu et al., 2010)—is among the most effective thus far identified, with individual members capable of neutralizing up to 90% of HIV-1 strains at mean inhibitory (IC50) concentrations of approximately 0.1 µg/ml. Substantial characterization, both crystallographic and with next-generation sequencing of antibody heavy-chain transcripts, has been performed on two donors with VRC01-class antibodies (NIAID 45 and IAVI 74), and additional characterization, involving probe-based isolation of monoclonal antibodies, has occurred with three other donors (RU 3, IAVI 57, and CHAVI 0219) (Scheid et al., 2011; West et al., 2012; Wu et al., 2011; Zhou et al., 2010). Antibodies of the VRC01 class are characterized by a number of specific similarities including heavy-chain mimicry of the CD4 receptor, a heavy chain derived from the IGHV1-2 germline gene, and a light chain with a 5-amino acid third complementary-determining region (CDR L3). Despite this characterization, questions remain concerning the VRC01 class and its appropriateness as a vaccine template.
Antibodies of the VRC01 class show extraordinary degrees of somatic mutation (Scheid et al., 2011; Wu et al., 2010, 2011), sometimes reaching 30% of heavy-chain variable domain nucleotides. Even within the same donor, they are enormously diverse, with variable domain amino-acid sequence differences that exceed 50% (the approximate mean divergence between unrelated antibodies) (Scheid et al., 2011; West et al., 2011, 2012; Zhou et al., 2010). How many B cell lineages generate this diversity in each donor? How similar is VRC01-class recognition and B cell ontogeny in different donors? And more generally, is elicitation sufficiently similar to allow for reelicitation of VRC01-class antibodies in the general population? We sought to answer these questions through a combination of antibody identification, X-ray crystallography (to define recognition), and next-generation sequencing (to provide genetic records). By defining critical structural features and their corresponding genetic signatures, we could delineate VRC01-class transcripts in donor antibodyomes. Phylogeny-based analyses could then be employed to define development pathways, and probabilistic analyses—both frequentist and Bayesian—could be used to delineate frequencies of recombination and to generalize, from a collection of donors, to characteristics of the overall class. Altogether, our findings indicate VRC01-class antibodies in diverse donors require relatively few genetic elements, evolve remarkably similar recognition, and follow similar maturation pathways to related structural solutions, suggesting that the elicitation of these important vaccine templates can in principle be reproduced.
RESULTS
VRC01-Class Antibody with Vλ-Derived Light Chain
One means of defining the ontogeny of an antibody class is to identify antibodies of the class in many donors and then to use these to enumerate class characteristics and pathways of development. We and others have used probe-based methods to identify VRC01-class antibodies, and the resurfaced stabilized core 3 (RSC3) (Wu et al., 2010), a gp120 probe that selectively retains the initial site of HIV-1 engagement with the CD4 receptor, is a particularly discriminating probe. We used RSC3 to select memory B cells from donor IAVI 23 of the Protocol G cohort (Simek et al., 2009). Sequencing of RSC3-binding memory B cells identified four antibodies, VRC-PG19, VRC-PG19b, VRCPG20, and VRC-PG20b, each of which potently neutralized diverse strains of HIV-1 (see Figures S1A–S1E and Tables S1A and S1B available online). To clarify the donor origin of each antibody, from this point forward we explicitly define the donor as a subscript on each antibody name. All four of these new IAVI 23 antibodies appeared to be somatically related, with VRC-PG19d23 and 19bd23 and VRC-PG20d23 and 20bd23 being most closely related to each other. The heavy chains of these antibodies derived from IGHV1-2, and their light chains contained the 5-residue CDR L3 characteristic of the class (West et al., 2012) (Figures S1F and S1G), suggesting that they were VRC01-class antibodies. Notably, the light chains of VRC-PG19d23, 19bd23, 20d23, and 20bd23 derived from a lambda precursor (IGLV2-14), in contrast to the kappa precursors reported for all other previously identified VRC01-class antibodies including VRC01d45 and VRC-PG04d74 (derived from IGKV3-20) or VRC-CH31d0219, 3BNC117d3, and 12A21d57 (derived from IGKV1-33) (Table S1C)(Scheid et al., 2011; West et al., 2012; Wu et al., 2010, 2011). Thus whereas antibody identification is a prerequisite to definition of the ontogeny of a class, additional members of the class with unique properties continue to be found. Nonetheless, once a substantial subset of a class has been identified, these should allow essential properties that allow effective HIV-1 neutralization by the class to be defined.
Structures of Diverse HIV-1 gp120 with VRC01-Class Antibodies from Five Donors
Because similarity in recognition chemistry provides a fundamental means by which to define a class, we sought to answer basic class questions by understanding how VRC01-class antibodies recognize their viral target, the HIV-1 gp120 envelope glycoprotein. All prior cocrystal structures of VRC01-class antibodies were determined with the same core gp120 construct (clade AE strain 93TH057) and from only two donors. We thus sought to define the structures of antibodies from all known donors with VRC01-class antibodies and with HIV-1 gp120s from diverse genetic backgrounds (Table 1, Figure 1).
Table 1.
Crystallographic Data Collection and Refinement Statistics
| Complex Antibody HIV-1 gp120 |
VRC-CH31d0219 93TH057 |
VRC-CH31d0219 93TH0573415v1 |
VRC-CH31d0219 93TH057KER2018_11 |
VRC01d45 KER2018_11 |
VRC01d45 ZM176.66 |
3BNC117d3 C1086 |
3BNC117d3 93TH0573415v1 |
VRC-PG20d23 93TH057 4LSU |
12A21d57 93TH057 |
|---|---|---|---|---|---|---|---|---|---|
| PDB accession code | 4LSP | 4LSQ | 4LSR | 4LSS | 4LST | 4LSV | 4JPV | 4LSU | 4JPW |
| Data collection | |||||||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | I422 | P212121 | C121 | P212121 |
| Cell constants | |||||||||
| a, b, c (Å) α, β, γ (°) | 67.4, 84.0, 175.7 90.0, 90.0, 90.0 | 66.6, 67.7, 221.1 90.0, 90.0, 90.0 | 64.9, 67.7, 220.1 90.0, 90.0, 90.0 | 54.6, 65.3, 259.1 90.0, 90.0, 90.0 | 68.11, 77.98, 200.35 90.0, 90.0, 90.0 | 191.6 191.6, 104.0 90.0, 90.0, 90.0 | 68.6, 66.9, 231.6 90.0, 90.0, 90.0 | 164.3, 66.3, 95.1 90.0, 108.3, 90.0 | 62.0, 66.0, 213.0 90.0, 90.0, 90.0 |
| Wavelength (Å) | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Resolution (Å) | 50.00-2.15 (2.23-2.15)* | 50.00-2.25 (2.29-2.25) | 50.00-2.28 (2.32-2.28) | 50.00-2.73 (2.79-2.73) | 50.00-2.54 (2.54-2.50) | 50.00-3.00 (3.05-3.00) | 50.00-2.80 (2.85-2.80) | 50.00-2.30 (2.34-2.30) | 50.00-2.88 (2.93-2.88) |
| Rmerge | 8.4 (51.9) | 11.6 (52.2) | 13.4 (60.2) | 12.7 (62.8) | 8.4 (33.7) | 12.0 (33.3) | 10.5 (62.1) | 9.5 (39.3) | 10.2 (48.4) |
| I / σI | 18.1 (1.9) | 17.0 (2.0) | 10.4 (2.1) | 13.8 (2.3) | 13.3 (2.1) | 24.8 (6.2) | 14.1 (1.8) | 20.0 (2.0) | 11.0/2.0 |
| Completeness (%) | 95.8 (71.6) | 98.6 (87.9) | 90.7 (75.0) | 90.6 (88.7) | 89.5 (46.4) | 99.1 (98.9) | 98.4 (89.9) | 90.7 (44.6) | 97.8 (86.3) |
| Redundancy | 5.7 (2.8) | 5.5 (4.4) | 5.1 (3.7) | 6.8 (3.6) | 3.7 (3.1) | 7.3 (7.5) | 4.1 (3.2) | 3.2 (1.7) | 3.6 (3.2) |
| Refinement | |||||||||
| Resolution (Å) | 2.15 | 2.25 | 2.28 | 2.73 | 2.50 | 3.00 | 2.80 | 2.30 | 2.88 |
| Unique reflections | 52,604 | 47,596 | 41,144 | 27,290 | 33,762 | 18,566 | 27,017 | 39,655 | 71,104 |
| Rwork / Rfree (%) | 18.6/22.1 | 19.2/22.9 | 19.4/25.8 | 19.2/26.3 | 19.0/24.0 | 26.5/29.8 | 19.9/25.9 | 17.9/23.4 | 20.6/26.4 |
| No. atoms | |||||||||
| Protein | 6175 | 6119 | 6082 | 6134 | 6175 | 6161 | 6120 | 6128 | 6144 |
| Ligand/ion | 76 | 18 | 15 | 37 | 0 | 5 | 27 | 36 | 15 |
| Water | 266 | 382 | 224 | 145 | 76 | 78 | 81 | 234 | 62 |
| B-factors (Å2) | |||||||||
| Protein | 63.8 | 57.9 | 76.5 | 46.5 | 140.4 | 86.6 | 46.5 | 78.2 | 64.0 |
| Ligand/ion | 75.4 | 75.5 | 94.6 | 57.0 | - | 64.7 | 75.5 | 70.3 | 57.2 |
| Water | 53.9 | 49.6 | 57.3 | 34.6 | 89.3 | 34.3 | 49.6 | 55.7 | 46.0 |
| rmsds | |||||||||
| Bond lengths (Å) | 0.004 | 0.008 | 0.005 | 0.005 | 0.004 | 0.002 | 0.004 | 0.008 | 0.002 |
| Bond angles (Å) | 0.858 | 0.863 | 0.854 | 0.891 | 0.896 | 0.567 | 0.876 | 1.147 | 0.619 |
| Ramachandran | |||||||||
| Most favored regions (%) | 96.9 | 96.9 | 96.0 | 94.4 | 97.3 | 93.4 | 93.8 | 93.9 | 96.2 |
| Additional allowed regions (%) | 2.7 | 3.0 | 3.3 | 4.7 | 2.4 | 5.8 | 4.9 | 5.8 | 3.6 |
| Disallowed regions (%) | 0.4 | 0.1 | 0.7 | 0.9 | 0.3 | 0.8 | 1.4 | 0.3 | 0.3 |
Values in parentheses are for highest-resolution shell.
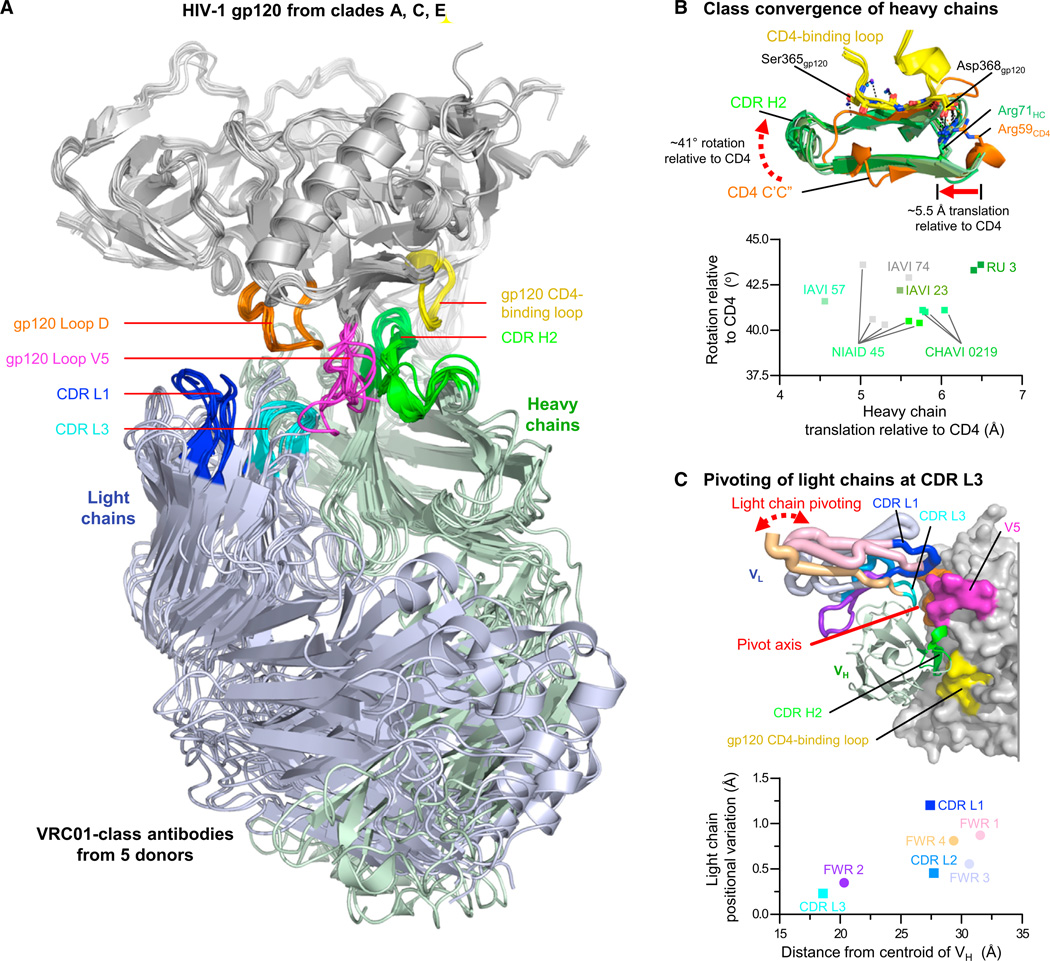
Figure 1. VRC01-Class Recognition of HIV-1 gp120.
(A) Nine cocrystal structures of five VRC01-class antibodies from five donors in complex with HIV-1 gp120s from clades A, C, and AE. Macromolecules are shown in ribbon representation after alignment of gp120, which is shown in gray with CD4-binding loop (yellow), loop D (orange), and V5 (magenta) highlighted. Antibody light chains are colored light blue, and heavy chains are shown in pale green with CDR H2 (green), CDR L1 (blue), and CDR L3 (cyan) highlighted.
(B) Class convergence of heavy chains. Upper panel shows interaction between CD4-binding loop of gp120 (yellow) and CDR H2s of VRC01 class (green) or C’C” ridge of CD4 (orange) with key interacting hydrogen bonds and salt bridges displayed as dotted lines. Lower panel shows mimicry of CD4 by VRC01 class-heavy chains. Rotation (vertical axis) and translation (horizontal axis) relative to CD4 are displayed for each antibody, with the nine new structures colored different hues of green according to the donor from which they were derived (and gray for previously published structures).
(C) VRC01-class variation of light chain relative to heavy chain. Light-chain-variable domain (VL) pivots around an axis that is close to both the CDR L3 and the interface between heavy and light chains. Other regions such as CDR L1 have larger degrees of structural variance. Upper panel shows light-chain positional variation displayed structurally. VL in worm representation is colored to distinguish CDR and framework regions, with worm thickness corresponding to the per-residue variance in Cα position. Heavy-chain variable domain (VH, cartoon) and gp120 (surface) are shown for reference, colored the same as in (A). Lower panel shows the average distance of each light-chain CDR (square) and framework (round) region relative to the centroid of the VH (horizontal axis) is plotted related to the rmsd of the nine VRC01-class antibodies (vertical axis). The symbols and labels of CDR and framework regions are colored to match the worm colors in the upper panel. See also Figure S1 and Table S1.
To define a collection of diverse HIV-1 gp120s for cocrystallization, we selected 15 gp120s with diverse Loop D and V5 regions from five different clades (Table S1D). To assist crystallization, we made chimeras of these 15 gp120s— swapping Loop D and V5 regions—with the clade AE strain 93TH057 gp120 that previously demonstrated robust crystallization (Wu et al., 2011; Zhou et al., 2010). Crystallization trials of VRC01-class antibodies in complexes with diverse gp120s or with chimeric-93TH057 gp120s yielded a total of nine crystals suitable for structural analysis (Table S1E). Crystallized gp120s were from clades A, C, and AE and totaled six different gp120s, including two chimeras. Crystallized antibodies were from five donors, including 3BNC117d3 from donor Rockefeller University 3 (RU3), VRC-PG20d23 from donor IAVI 23, VRC01d45 from donor NIAID 45, 12A21d57 from donor IAVI 57 (also called Rockefeller University Patient 12), and VRC-CH31d0219 from donor CHAVI 0219. Diffraction data were collected, and structures solved by molecular replacement and refined at resolutions limits of 2.15 Å to 3.0 Å (Table 1).
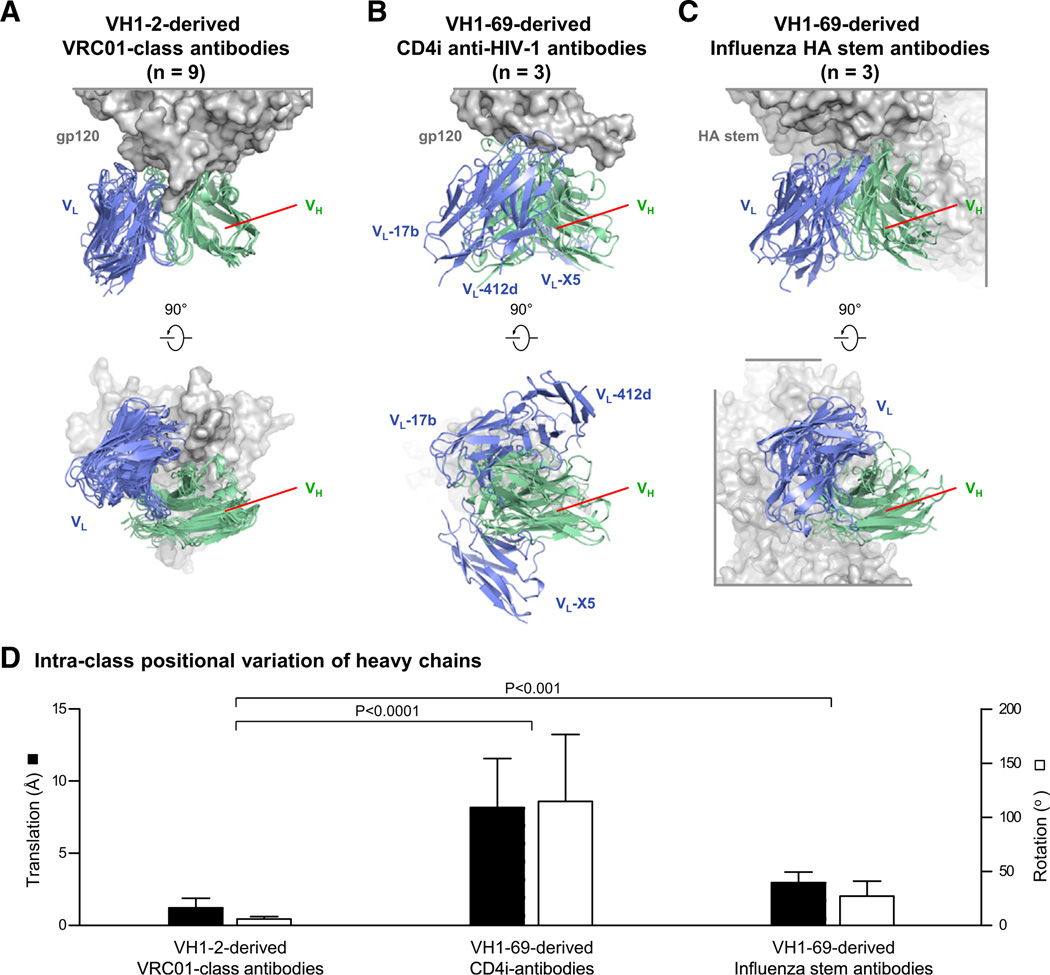
Structures of HIV-1 gp120 from clades A, C, and AE in complexes with VRC01-class antibodies from five different donors, each infected with a different strain of HIV-1 (Table S1F), showed remarkable similarity (Figure 1; Figure S1H–S1L). When gp120s were superimposed, the antibody-variable regions of the heavy chains aligned closely (Cα-rmsd of 1.4 Å), especially in the CDR H2 region (Cα-rmsd of 0.7 Å), which interacts with the CD4-binding loop of gp120. Relative to CD4, the heavy chains in the nine structures converged to within 2 Å, likely reflecting conserved hydrogen-bonding requirements (Figure 1B); salt bridges between heavy chain residue Arg71HC and HIV-1 residue Asp368gp120 were also preserved (Figure S1M). (Kabat numbering [Kabat et al., 1991] is used to define CDR and framework regions and numbering of antibody residues, with subscripts of “LC” or “HC” denoting light chain or heavy chain, respectively.) The antibody-variable regions of the light chains were less well aligned (Cα-rmsd of 1.7 Å) by the gp120 superposition, except for the CDR L3 (Cα-rmsd of 0.18 Å). Examination of light chains positioned by superposition of the heavy chains indicated radial divergence from the heavy-light interface, suggesting that the interface interaction might constrain structural variation of the antibody-variable region (Figure 1C). Overall, the cocrystal structures allow for a precise definition of the epitope boundaries (Figure S1N) and molecular variance for the VRC01 class of HIV-1 neutralizing antibodies. When compared with other classes of neutralizing antibodies such as the VH1-69-derived stem antibodies against influenza virus (Lingwood et al., 2012) or the VH1-69-derived CD4-induced antibodies against HIV-1 (Huang et al., 2004), antibodies of the VRC01 class showed substantially greater structural similarity (Figure 2) (p < 0.001). Thus, although class recognition is observed by several antibodies, antibodies of the VRC01 class are unusual in their degree of similarity.
Figure 2. VRC01 Class Exhibits Greater Structural Convergence than Other Antibody Classes.
(A) Recognition by VH1-2-derived VRC01 class antibodies. Upper panel is shown in the same orientation as in Figure 1A with antigen gp120 depicted in gray surface and variable domains of nine VRC01 class antibodies from five donors displayed in cartoon (VH in pale green and VL in slate blue).
(B) Recognition by VH1-69-derived anti-HIV-1 CD4i antibodies 17b (PDB ID 1RZK), 412d (PDB ID 2QAD), and X5 (PDB ID 2B4C).
(C) Recognition by VH1-69-derived influenza stem antibodies CR6261 (PDB ID 3GBM), CR9114 (PDB ID 4FQI), and F10 (PDB ID 3FKU). In both (B) and (C), to allow comparisons between class members, the VH of one member of each class was first aligned with the VH region of VRC01 in (A) to provide a standard frame of reference. VRC01, 17b, and CR6261 were used to represent their respective classes. Other members of each class were then aligned by superposition of their bound antigen. Antigens are shown in gray, and antibodies are shown in pale green (VH) and blue (VL). Lower panels of (A), (B), and (C) are 90-degree views from upper panels, looking down onto the epitope from the antibody.
(D) Intraclass positional variation of VH domains are shown with average rotation angles and translation distances between antigen-superposed class members. VRC01 class show significantly lower positional variation compared to the other antibody classes. Data are represented as mean ± SEM.
Light-Chain Recognition of HIV-1 gp120 by the VRC01 Class of Antibodies
By quantifying structural similarities, we reasoned that we could identify critical recognition features, and—based on these—derive genetic signatures that could be used to interrogate VRC01-class transcripts in donor antibodyomes. Heavy chains recognize the highly conserved initial site of CD4-attachment, and it was thus possible to identify critical features and a genetic signature (evolutionary similarity as revealed through phylogenetic analysis), from analyses with a single variant of HIV-1 gp120 (West et al., 2012; Wu et al., 2011; Zhou et al., 2010). Light chains, however, protrude beyond the initial site of CD4 engagement and contributed one third of the buried interface between antibody and HIV-1 gp120 (Figure S2A, Table S1F). Roughly half of this recognition occurs in the Loop D region and the other half occurs in the V5 region (Figure S2A). To determine the structural basis for light-chain recognition of diverse gp120s, we analyzed the sites of light-chain interaction in the nine new structures (Figure S2). In these nine structures, diverse surface shapes and chemistries were observed for the light-chain-recognized region of gp120 (Figures S2B and S2C). Light chains of the VRC01 class accommodated this diversity with distinct CDR L1s and conserved CDR L3s (Figure 3).
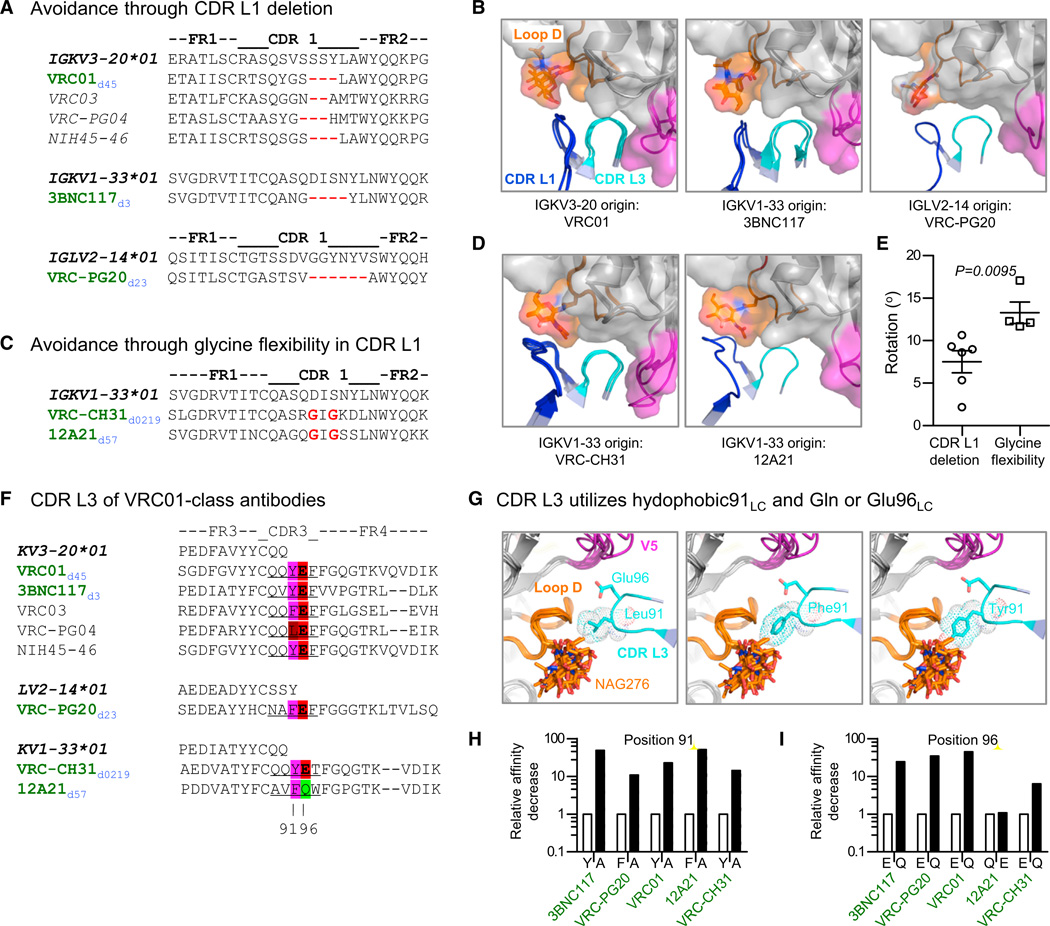
Figure 3. Structural Signatures in CDR L1 and CDR L3.
(A) CDR L1 sequences for VRC01-class antibodies that avoid loop D through deletion, with germline genes italicized and bold, current structures in green text (donor in blue subscript), and structures published elsewhere italicized.
(B) Complex structures for VRC01-class antibodies with CDR L1 deletions. Antibodies are shown in ribbon representation and gp120 with a semitransparent surface, both colored the same as in Figure 1A.
(C) CDR L1 sequences for VRC01-class antibodies that avoid loop D through mutation to glycine. Sequence highlighting scheme is the same as in (A).
(D) Complex structures for VRC01-class antibodies with CDR L1 mutations to glycine. Antibodies are shown in ribbon representation and gp120 with a semitransparent surface, both colored the same as in (B).
(E) Rotation of light chains (vertical axis) for VRC01-class antibodies that use deletion (left) and glycine (right) mechanisms of CDR L1 avoidance. Rotations calculated after gp120 superposition relative to the light chain of the 2010 VRC01-93TH057 gp120 complex structure. Data are represented as mean ± SEM.
(F) CDR L3 sequences. In Kabat numbering, the third position of the CDR L3 is defined as residue 91 and the fourth position is defined as residue 96. Sequence highlighting scheme is the same as in (A).
(G) CDR L3 (cyan) interaction with loop D (orange) of superimposed gp120s shown for VRC01-class antibodies with Leu91LC (VRC-PG04d74; left), Phe91LC (VRC-PG20d23 and 12A21d57; middle), and Tyr91LC (VRC01d45, VRC-CH31d0219 and 3BNC117d3; right).
(H) Relative decrease in affinity (vertical axis) of antibodies of the VRC01 class for gp120 (arranged horizontally) for wild-type (WT, white bars) and 91LC to Ala substitutions (black bars).
(I) Relative decrease in affinity (vertical axis) of antibodies of the VRC01 class for gp120 (arranged horizontally) for WT (white bars) and Gln or Glu 96LC substitutions (black bars). See also Figure S2.
The CDR L1 varies considerably in sequence, length, and position (Figure 3A). IGKV3-, IGKV1-, and IGLV2-derived antibodies, such as VRC01d45, 3BNC117d3, and VRC-PG20d23, have CDR L1s of only six or seven residues in length, the result of deletion during somatic maturation (Figure 3A). These deletions (of two to six amino acids) shorten CDR L1, avoiding a potential clash with Loop D (Figure 3B). By contrast, other IGKV1-derived antibodies, such as VRC-CH31d0219 and 12A21d57, do not have deletions in CDR L1 (Figure 3C). These two antibodies display identical somatic alterations to glycine and adopt similar noncanonical conformations that swing their CDR L1 regions away from Loop D (Figure 3D). Overall, despite providing a substantial portion of the binding interface between light chain and gp120, CDR L1 does not recognize Loop D as much as avoid it, either through deletion or flexibility, both of which exhibit precise structural solutions (Figure 3E) and both of which are acquired through processes of somatic mutation.
In contrast to CDR L1 variation, the CDR L3 of VRC01-class antibodies is constrained to 5-amino acids (corresponding to Kabat numbered [Kabat et al., 1991] residues 89, 90, 91, 96, and 97) with preference for a hydrophobic residue 91LC and a Glu at residue 96LC (Figure 3F) (West et al., 2012). Analysis of the structures of CDR L3 with diverse gp120s (Figure 3G) indicated the packing of hydrophobic 91LC against the backbone of Loop D and the interaction of Glu96LC with an electropositive patch at the conserved base of V5. Mutational substitution of residue 91 to Ala indicated that the packing of Loop D with the signature “hydrophobic91LC” substantially contributes to antibody affinity (Figure 3H). In eight of the nine structures that we determined, Glu96LC made a conserved hydrogen bond to the backbone amide of Gly459gp120 at the N terminus of the V5 region (Figure S2D). With antibody 12A21d57, in which residue 96LC is a Gln, the distance between side chain and amide backbone was too great for hydrogen bonding (4.2 Å) (Figure S2D), and the interacting V5 region was not uniformly electropositive (Figure S2E, top row). To quantify the impact of recognition by the Glu96LC signature residue, we measured changes in affinity with Glu or Gln substitutions (Figure 3I; Figure S2F). When Gln was substituted for Glu, the affinity decreased by ~10-fold, but when Glu was substituted for Gln, the affinity was essentially unchanged. Perhaps relevant to this, a somatic variant of 12A21d57 named 12A12d57 contains a Glu at 96LC (Scheid et al., 2011). Altogether, structural analysis identifies two signatures: a CDR L1 signature with deletion, or mutation to glycine, of two residues; a CDR L3 signature of 5-amino acids with hydrophobic91LC at position 3 and Glu or Gln96LC at position 4.
Genetic Record of Recombination and Maturation for the VRC01 Class
To study the processes of recombination and maturation that generate critical features of VRC01-class antibody light chains, we performed next-generation sequencing and bioinformatics analysis of light-chain-antibody transcripts from peripheral blood mononuclear cells (PBMCs) of donors RU3, IAVI 23, NIAID 45, IAVI 57, and IAVI 74 (Table S2A). We interrogated time points that were previously used to isolate antibodies 3BNC117dRU3, VRC-PG20d23, VRC01d45, VRC03d45, 12A12d57, 12A21d57, and VRC-PG04d74, respectively, from these donors. On identity-diversity plots (Figure 4A, top row), which resolves antibodyome transcripts in two dimensions: identity to a known antibody and divergence from germline V-gene, distinct islands of high diversity and >90% identity to the template antibodies were observed (Figure 4A, top row). None of the reads, however, showed great than 85% identity to isolated antibodies from other donors (Figures S3A–S3E), illustrating a high level of sequence diversity for light chains of the VRC01 class.
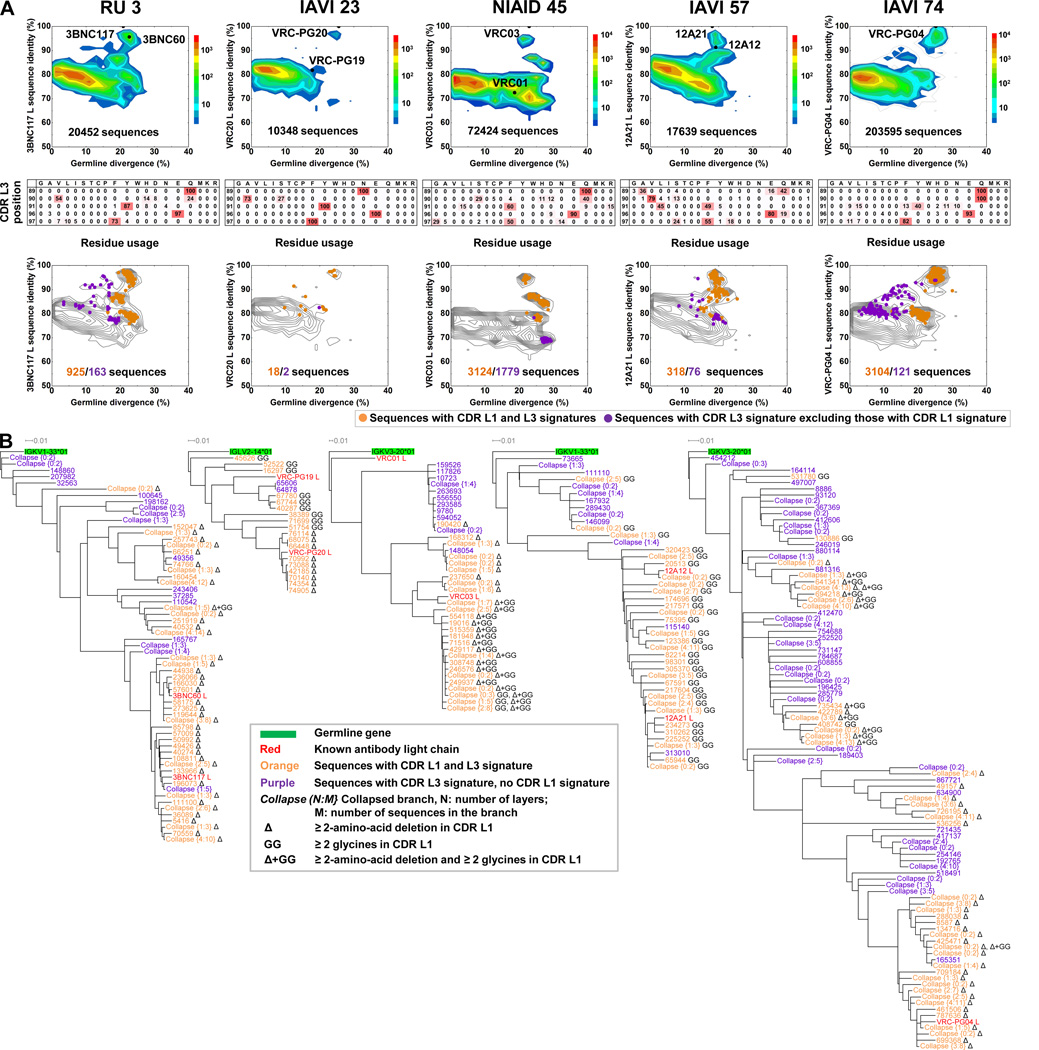
Figure 4. Genetic Records for the Development of CDR L1 and CDR L3 Signatures for the VRC01 Class.
(A) Light-chain antibodyomes of donors with VRC01-class antibodies. Top row shows that for each donor, light-chain sequences are plotted as a function of sequence identity to an antibody of the VRC01 class identified in that donor (vertical axis) versus the sequence divergence from their assigned genomic V-gene alleles. Color coding indicates the number of sequences. Middle row shows heat map display of the amino-acid usage at each position of the CDR L3, for all light chains with 5-amino acid CDR L3s and greater than 15% germline divergence. Frequencies are colored from dark red (100% usage) to white (0%). Bottom row shows sequences with 5-residue CDR L3s with sequences that contain amino acids with at least 10% usage are plotted as dots. Orange (or purple) indicates the presence (or absence) of a CDR L1 deletion or mutation to glycine. Total numbers of sequences are listed in the corresponding colors.
(B) Maximum-likelihood phylogenetic trees of light-chain sequences with mature genetic CDR L3 signature as defined in (A), colored orange (or purple) for sequences with (or without) CDR L1 signature. The nature of the CDR L1 signature is defined by “Δ” (deletion) or “GG” (mutation to glycine). See also Figure S3 and Table S2.
To identify light chains of the VRC01 class, we used a criterion of 5-amino acid CDR L3 length as a partial signature because it is present in all previously identified VRC01-class antibodies but in only ~1% of general human antibodies (West et al., 2012). We further restricted our analysis to light chains of the same assigned germline IgVK or IGVL gene as the template antibody. In addition, we chose those transcripts that displayed greater than 15% nucleotide divergence from germline V-gene because we expected VRC01-class antibodies to have a substantial degree of somatic mutation (Wu et al., 2011). This analysis identified just a few sequences in donor IAVI 23, hundreds in donors RU3 and IAVI 57, and thousands in donors IAVI 74 and NIAID 45. The CDR L3s of these sequences showed remarkable similarity, within each donor and between donors, with the frequency of residue usage for each position of the CDR L3 clearly nonrandom, suggesting a specific signature (Figure 4A, middle row). To confirm functionality of these identified sequences, we synthesized diverse sequences from donors IAVI 57 and IAVI 74, reconstituted with the heavy chains from each of these donors, and confirmed HIV-1 neutralization (Tables S2B and S2C). A combined “mature CDR L3 genetic signature”—defined from the five donors and based on observed amino acid frequencies of >20% for each CDR L3 position—was (CDR L3 position 1) residue 89LC Q, N, or A, (position 2) residue 90LC Q, V, A, or S (position 3), residue 91LC F, L, or Y, (position 4) residue 96LC E, and (position 5) residue 97LC F, G, or S. We applied this genetic signature to all transcripts (Figure 4A, bottom row). Notably, the mature genetic CDR L3 signature was absent in light-chain transcripts of less than 5% germline divergence in all donors, except donors RU3 and IAVI 74 (Figure 4A, bottom row), indicating that in some donors the CDR L3 of the initial recombinant either differs or contains amino acids with less than 20% frequency in the mature antibody population.
To identify the sequence of the initial unmutated light chain, we used the less restrictive structural signature (5-amino acid CDR L3, hydrophobic residue 91LC, and glutamine or glutamic acid residue 96LC). For all donors, except IAVI 23 for which the genetic record was sparsely sampled, transcripts with the structural signature were observed with low V-gene divergence. We analyzed five sequences with the lowest V-gene divergence from each donor to provide insight into germline recombinants (Figures S3F and S3G). In donor RU3, an unmutated V-gene with CDR L3 sequence QQYEF was identical in both initial recombinant and mature antibodies. In donor IAVI 57, sequences with four of the mature genetic CDR L3 signature residues, but a Leu at residue 90LC were identified, and in donor NIAID 45 and IAVI 74, several sequences with four of the mature genetic CDR L3 signature residues were observed, but with a Thr at residue 97LC. These results show that recombination produces a CDR L3 with the elements critical for recognition: 5-amino acid length, hydrophobic-91LC, and Glu or Gln-96LC. Somatic mutation can then alter the CDR L3, but essential structurally-defined elements are retained.
The use of a “mature genetic CDR L3 signature” to identify likely VRC01-class antibody transcripts allowed us to follow CDR L1 maturation in these sequences. We created and analyzed phylogenetic trees for transcripts containing the “mature genetic CDR L3 signature”—with the additional criterion of whether a characteristic CDR L1 signature, either deletion or mutation to glycine, was present or absent (Figure 4B). The CDR L1 signature was observed to occur or not to occur in nearly identical sequences throughout these phylogenetic trees, in all donors except for IAVI 23, where the number of VRC01-class transcripts was limited (Figure 4B). To compare functionality of paired sequences, we synthesized light-chain pairs of donors NIAID 45 and IAVI 57, reconstituted with heavy chains from each of these donors, and observed virtually identical HIV-1 neutralization in those that expressed (Figures S3H–S3K; Table S2D). Overall, these results indicate that the maturational alterations required to produce the characteristic deletion or mutation to glycine in CDR L1 might occur with minimal alteration elsewhere; apparently, characteristic developmental features might not always have substantial functional impact.
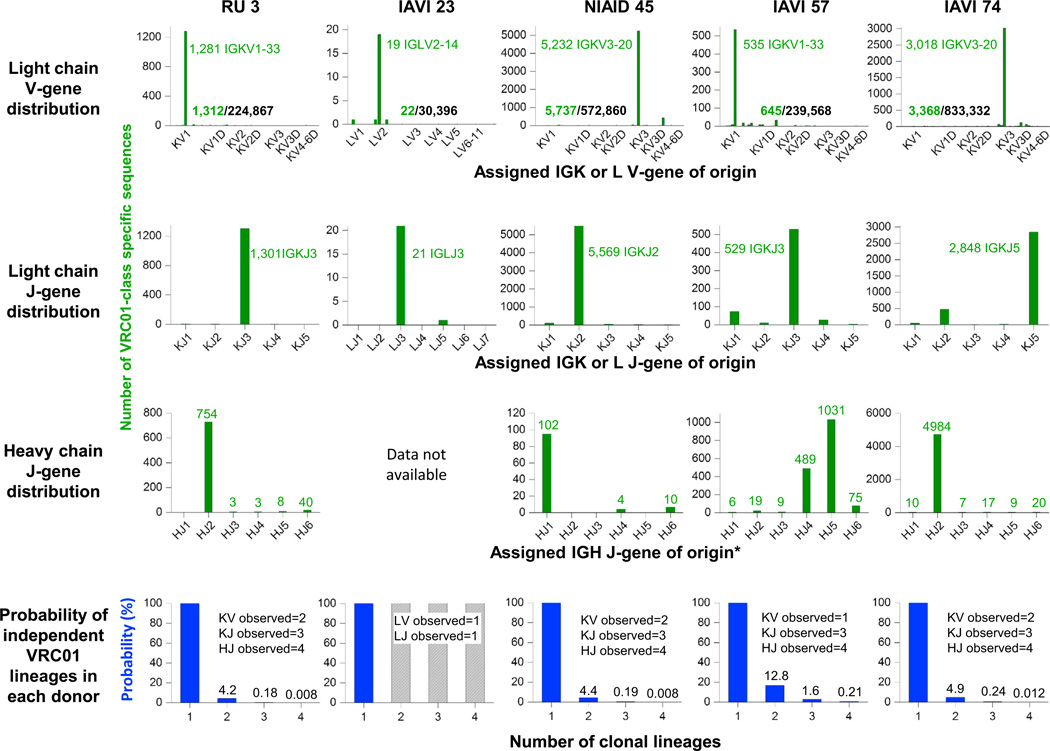
The genetic record for heavy chains of the VRC01 class in six donors derives primarily from probe-identified monoclonal antibodies, supplemented in donors NIAID 45 and IAVI 74 with VRC01-class heavy-chain transcripts identified by a phylogenetic method (Scheid et al., 2011; Wu et al., 2011). We therefore applied this phylogenetic method to donors RU 3 and IAVI 57 and identified 809 and 1632 VRC01-class transcripts, respectively (Figures S3L and S3M), to supplement the genetic record for these donors. To confirm functionality, we synthesized ten diverse sequences from donor IAVI 57, reconstituted with the 12A21 light chain from donor IAVI 57, and confirmed HIV-1 neutralization (Tables S2B and S2E). As with previous analyses (Scheid et al., 2011; Wu et al., 2011), VRC01-class transcripts from these donors included diverse CDR H3 sequences, diverse variable-region sequences, and highly skewed germline-gene usage (Figure 5, Table S3A).
Figure 5. 454 Pyrosequencing-Derived Genetic Record of V- and J-Gene Usage for the VRC01 Class and Lineage Implications.
Histograms of the assigned V-gene origin (top row) or J-gene origin (rows 2 and 3) are shown for VRC01-class transcripts identified from donor antibodyomes with the specific signatures provided in Figure 6B. Comparison of origin-gene assignments for VRC01-class transcripts (rows 1–3) versus those for all non-VRC01-class transcripts provides a measure of the skewing of origin genes used by the VRC01 class. Observations from six donors define genes compatible with the class (Figure 6B) and allow for an estimate of the skewing associated with functional selection. For skewing associated with clonal amplification from a limited number of ancestor-B cells, we estimated probabilities (bottom row) that different numbers of clonal lineages could have given rise to the 454-observed VRC01- class transcripts in each donor. Probability calculations based on both Bayesian and frequentist analyses are provided in Supplemental Experimental Procedures. See also Figure S4 and Table S3. (*Heavy chain J-gene distributions extracted from published antibody sequences for NIAID 45 and IAVI 74.)
Overall, next-generation sequencing and associated bioinformatics analyses provide a genetic record for developmental processes of recombination and somatic mutation. For heavy chain, recombination selects the IGHV1-2 gene, but D and J gene choice does not appear to be restricted. For the light chain, the genetic record shows that the signature CDR L3 of the VRC01 class is generated primarily through recombination, which selects elements critical for recognition, and that a characteristic CDR L1 deletion or mutation to glycine generally appears later in the maturation process, and then becomes common, often interleaved on multiple phylogenetic branches.
Highly Skewed Germline V- and J-Gene Usage
The thousands of VRC01-class sequences now known in multiple donors allows us to address questions such as which germline antibody genes are compatible with VRC01-class recognition, and how frequently B cells with requisite class features are produced by recombination. One source of insight into these questions is through statistical analysis of genetic features, where biases or skewing of gene usage can help to reveal mechanisms governing selection (Gilbert et al., 2011; Rolland et al., 2012). Such skewing can derive from two dominant sources: (1) functional selection, whereby only certain genes are compatible with the VRC01 class, or (2) clonal amplification, due to there being only a limited number of ancestor-B cells responsible for the observed VRC01-class sequences in each donor.
For VRC01 class-heavy-chain sequences, preferential use of the IGHV1-2 gene has been attributed to particular residues (e.g., Trp50HC, Asn58HC, and Arg71HC) present in VH1-2 (West et al., 2012). Not all VRC01-class antibodies, however, include these residues (e.g., 12A21 has Leu50HC and VRC03 and VRC-PG19 have Ser58HC), suggesting that the identified three-residue signature is beneficial to recognition, but variation is permitted. We incorporated the newly available sequence and structural information into a sequence and structural analyses of heavy-chain characteristics of the VRC01 class. The analysis indicated a number of additional signature residues (Figure S4A–S4G). Some of these (e.g., Gly55HC) were common to a number of germline genes, while others (e.g., Met or Leu69HC) were more specific to VH1-2. In terms of heavy chain-J genes, we observed all confirmed neutralizers of the VRC01 class to use IGHJ-genes from four different JH-gene families, IGH-J1, IGH-J2, IGH-J4, and IGH-J5. One requirement for the CDR H3 interaction for VRC01-class members is the presence of a large hydrophobic residue at position 100B (generally Trp, but Tyr and other hydrophobic residues are also observed) (Table S3A). These observations suggest that the heavy-chain-germline V-gene choice derives from functional selection, whereas the J-gene choice does not. Substantial skewing of J-gene usage in VRC01-class antibodies would thus likely be related to clonal amplification.
The common use of IGVK3-20 and IGVK1-33 alleles in different donors indicates light-chain V-gene preference or selection. To understand the basis of this selection, we examine the two regions of the light chain identified structurally as being critical to recognition: the CDR L1 and CDR L3. With CDR L1, we observed no substantial differences with respect to the length or to the number of glycines in the CDR L1 region, between the light-chain-germline genes of VRC01-class antibodies and the rest of the human variable lambda or kappa V-genes. Moreover, a number of additional germlines were found to exhibit short CDR L1s or a number of glycines, suggesting that these CDR L1 features were not the dominant factors influencing the compatibility of light-chain V-genes (Figures S5A and S5B). With CDR L3, analysis of the C termini of the light-chain-germline V-genes revealed only ~38% to have a hydrophobic (e.g., Y, F, L, I, or V) at residue 91LC, and this might serve as a limiting factor for germline selection (Figures S5C and S5D). Notably, only two kappa genes, IGKV1-33 and IGKV3-20, had a guanine base immediately after residue 91LC that could potentially facilitate the emergence of the critical Glu96LC upon recombination (Figures S5C and S5D), and these appeared to be preferentially observed as precursor light-chain V-genes for the VRC01 class.
Application of Bayesian and frequentist analyses to observed light-chain V-genes indicates either 3–17 (Bayesian) or 3–12 (frequentist) are compatible with the VRC01 class (Figures S4H–S4J). For light-chain J-genes, we observed four different J-genes (LJ2, KJ2, KJ3, and KJ5), indicating little preference, despite stringent CDR L3 requirements.
Despite the compatibility of multiple light-chain V-gene and J-genes and multiple heavy-chain J-genes within the VRC01 class of antibodies, next-generation sequencing in five donors (RU 3, IAVI 23, NIAID 45, IAVI 57, and IAVI 74) indicated highly donor-specific gene usage. Indeed, in any given donor, all VRC01-class antibodies appear to arise from a single set of origin genes. The observation of a single V-gene and a single J-gene for VRC01 class-light chains of each donor, and a single J-gene for VRC01 class-heavy chains of each donor, an outcome that statistical analysis suggests is < 5%probable for more than a single lineage in three donors (Figure 5; Tables S3B and S3C). The use of a single JH, VL, and JL origin for VRC01-class antibodies in each donor thus suggests that these antibodies derive from a very limited—likely a single—ancestor-B cell.
VRC01-Class Ontogeny
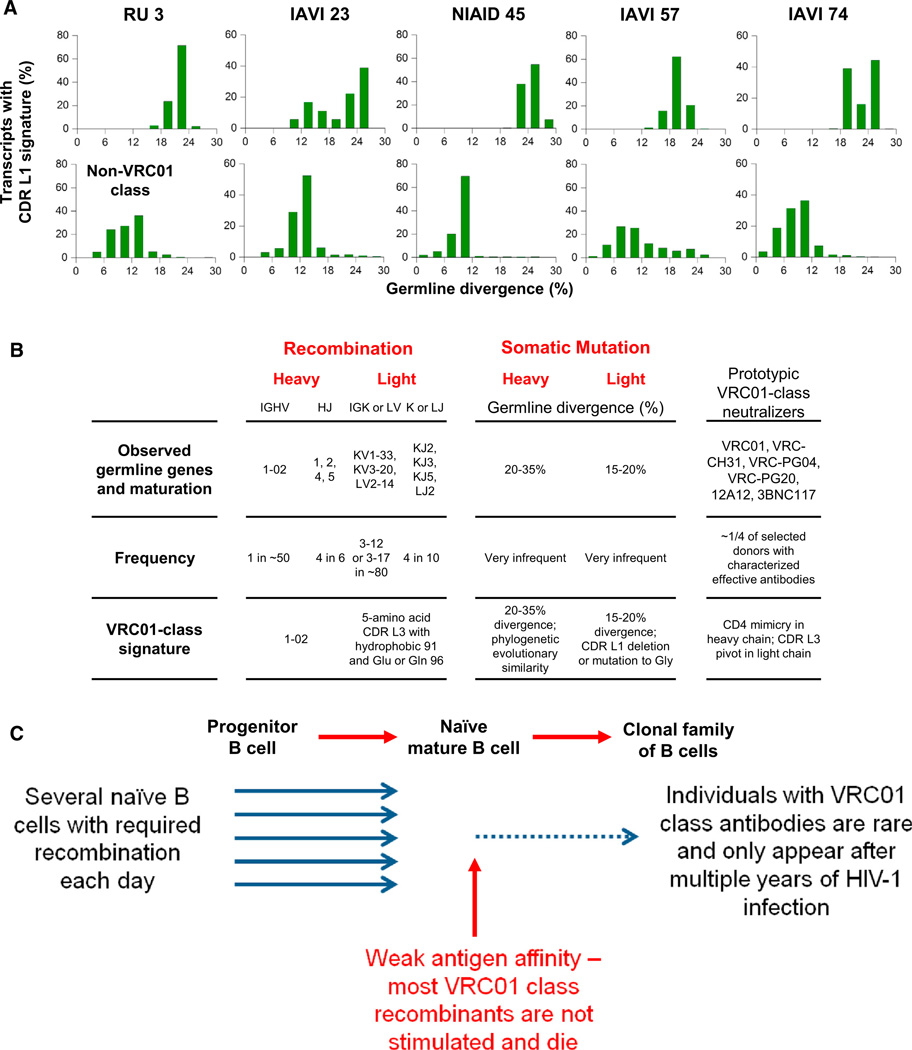
The identification of VRC01-class antibodies in six donors coupled to the determination of bioinformatics signatures for the VRC01 class provides an unprecedented opportunity to characterize the B cell ontogeny of a highly effective class of HIV-1 neutralizing antibodies. This class-wide B cell ontogeny analysis includes antibody lineages from several HIV-1 donors and thus differs from the analysis of individual B cell lineages such as that carried out for HIV-1-neutralizing antibody CH103 in donor CH505 (Liao et al., 2013) or for hen egg white lysozyme- recognizing antibodies (Li et al., 2003). The antibodyomics analyses described here are from cross-sectional sampling; longitudinal sampling should in principle allow a more complete understanding of the development of antibody responses. Nonetheless, our results show that it is possible to trace the B cell ontogeny of the VRC01 class from unmutated B cell transcripts to a clonal family of mature B cell transcripts corresponding to neutralizing antibodies (Figure 6; Figures S5E and S5F). These results include outcomes that appear stochastic, such as substantial sequence variation between class members, and outcomes that appear deterministic, such as the remarkable similarity in gp120 recognition or the development of the CDR L1 signature, which occurs in all six donors at similar levels of V-gene divergence (Figure 6A; Tables S4A and S4B).
Figure 6. B Cell Ontogeny of the VRC01 Class of HIV-1-Neutralizing Antibodies.
(A) CDR L1 signature as a function of V-gene divergence in VRC01-class transcripts (top row) or non-VRC01-class transcripts (bottom row). Additional data provided in Table S4.
(B) Observed B cell ontogeny in donors RU3, IAVI 23, NIAID 45, IAVI 57, and IAVI 74. Frequency is calculated from observed versus total number of elements. For example only 1 IGHV gene is observed (VH1-2) out of the ~50 VH-germline alleles in each human. For IGKV or IGLV, frequentist analysis indicates 3–12 and Bayesian analysis 3–17 out of a total of ~80 VL/K-germline alleles in each human. Specific recombination (to select the IGHV1-2 gene and a 5-amino acid CDR L3 with hydrophobic-Glu or Gln residue pair) and somatic mutation (to evolve heavy chain and CDR L1 signature) are observed in antibodies of the VRC01 class.
(C) Schematic of B cell ontogeny of the VRC01 class. See also Figure S5 and Table S4.
While the precise frequencies of antibody gene recombination are dependent on a number of variables (Paul, 1999) including the genes being combined, the frequency of required sequence elements, and other factors (see for example, Feeney et al., 2000), one can nonetheless provide a rough estimate of frequency based on the assumption that each V, D, and J gene has an equal probability of recombining. With this approximation, an estimate of the frequency of antibody recombinants with VRC01-class characteristics is roughly 1 in 150,000: VH-gene of IGHV1-2 (1 in 50), CDR L3 of 5 residues (1 in 100) with hydrophobic at 91LC (1 in 3) and Gln or Glu at 96LC (1 in 10) (Figure 6B). If specific VL- or VK-genes are required (e.g., the 3 that are observed among known tzhou VRC01 class antibodies out of the total 77 known VL or VK genes, with an upper estimates of 12 or 17 as described in Figures S4K and S4L and Table S3B), then this would reduce probability to be about 1 in roughly 1–4 million. These estimates suggest that, among the many millions of naive mature B cells in each individual, a number with requisite VRC01-class characteristic are produced each day. This frequent occurrence of appropriate recombinants reflects the relatively few specific genetic elements required for the class.
In general, selection from a small library can appear deterministic, whereas selection from a large library is more likely to appear stochastic. Thus, in the case of the VRC01 class, because of the limited number of structural solutions (for example, the two solutions observed for CDR L1, Figures 3A–3E), selection yields an apparently deterministic outcome. Such outcomes might pertain in particular to antibodies that require individual V-gene-encoded interactions such as the VH1-2-derived VRC01 class or the VH1-69-derived influenza-stem antibodies. By contrast, antibodies that recognize antigen through CDR H3-encoded mechanisms might have many more structural solutions, because of the far greater diversity of the combinatorial V(D)J-derived library of sequences.
DISCUSSION
A detailed understanding of the B cell ontogeny for the VRC01 class provides a framework by which to define vaccine strategies and to monitor the maturation process. Recent structure-based immunogen design has created multimeric Env capable of binding VRC01 class-germline precursors (Jardine et al., 2013); although these achieve in vitro B cell activation, in vivo activation has not yet been demonstrated. If immunization were to stimulate naive mature B cells with VRC01-class characteristics, then one should observe an increase in antibodies with heavy chain derived from the VH1-2 germline and light chains with 5-amino acid CDR L3, hydrophobic91LC and Glu or Gln96LC. Such an increase should be apparent in the genetic record, through use of the next-generation sequencing techniques.
Overall, our structural and sequencing findings define a reproducible B cell ontogeny for a highly effective antibody class: the VRC01 class. Such reproducibility enables class-specific strategies of elicitation, and the specific genetic signature defined here allows for efficient detection. We note that the observation of one or a very few ancestor-B cells for VRC01-class antibodies in each donor parallels the observation of a single “founder” virus that predominates for cases of HIV-1 infection involving mucosal transmission (Keele et al., 2008). It will be interesting to see whether alleviating blocks in VRC01 class-B cell development, perhaps through immunizing with a modified gp120 or series of gp120 immunogens with affinity to initial germline-recombinants and maturation intermediates (Jardine et al., 2013; McGuire et al., 2013) will allow multiple VRC01-class lineages to flourish in the context of a vaccine.
EXPERIMENTAL PROCEDURES
Human Specimens
The peripheral blood mononuclear cells (PBMCs) of donor 45 (Li et al., 2007; Wu et al., 2010), donor RU3 (Scheid et al., 2011), and donors 23, 57, and 74 from the international AIDS-vaccine initiative (IAVI) protocol G (Simek et al., 2009; Walker et al., 2010) have been described previously. All human samples were collected with informed consent under clinical protocols approved by the appropriate institutional review board (IRB).
Isolation of Antibodies VRC-PG19 and VRC-PG20
Fluorescence-activated cell sorting of antigen-specific IgG+ B cells from donor 23 PBMC and the amplification and cloning of immunoglobulin genes were carried out with previously described protocols (Wu et al., 2010).
Expression and Purification of Antibodies and Fab Fragments
Expression plasmids for heavy and kappa chains were constructed as described previously (Wu et al., 2010), as was the expression and purification of antibody IgGs and preparation of Fab fragments.
ELISA
The binding of VRC-PG19 and VRC-PG20 to HIV-1 gp120 proteins and competition with other antibodies were accessed with previously described protocols (Wu et al., 2010).
Assessment of HIV-1 Neutralization
Neutralization by VRC-PG19 and VRC-PG20 was measured with single-round of-infection HIV-1 Env-pseudoviruses containing a luciferase reporter gene to infect TZM-bl target cells that stably expresses high levels of CD4 and the coreceptors CCR5 and CXCR4 (Shu et al., 2007). Neutralization curves were fit by nonlinear regression and the 50% and 80% inhibitory concentrations (IC50 and IC80) were reported as the antibody concentration required to inhibit infection by 50% and 80%, respectively.
Surface Plasmon Resonance
The effects of antibody light-chain CDR L3 mutations on binding constants to gp120 were assessed by surface plasmon resonance on a Biacore T-200 instrument (GE Healthcare).
Crystallization, X-Ray Data Collection, Structure Determination, and Refinement of VRC01-Class Antibodies in Complex with HIV-1 gp120
The extended core (coreE) gp120 proteins from different HIV-1 stains were constructed, and binary complexes of antibody Fab and HIV-1gp120 were purified and crystallized with protocols as described previously (Zhou et al., 2010). X-ray diffraction data were integrated and scaled with the HKL2000 suite (Otwinowski and Minor, 1997). Crystal structures were solved by molecular replacement with Phaser (McCoy et al., 2007) in the CCP4 Program Suite (Collaborative Computational Project, 1994). Further refinements were carried out with PHENIX (Adams et al., 2002). Starting with torsion-angle simulated annealing with slow cooling, iterative manual model building was carried out on COOT (Emsley and Cowtan, 2004) with maps generated from combinations of standard positional, individual B-factor, TLS refinement algorithms. Detailed crystallization conditions, structure solution, and refinement protocols are provided in Supplemental Experimental Procedures.
454 Pyrosequencing
454 pyrosequencing libraries of donor 45, donor RU3, and IAVI donors 23, 57, and 74 were prepared, and 454 pyrosequencing of the PCR products were performed as previously described (Wu et al., 2011).
Bioinformatics Analysis
Bioinformatics analyses of 454 pyrosequencing generated data were performed with antibodyomics methods and consisted of two phases. In phase one, the raw sequencing data were filtered, annotated, and processed with a previously described pipeline (Wu et al., 2011) to improve sequence quality, as well as to determine critical antibody characteristics such as V(D)J gene origin, rate of somatic hypermuation, and complementarity-determining regions (CDRs). In phase two, a suite of bioinformatics tools was used to analyze the antibodyomes to identify light chains with specific signatures, to construct phylogenetic trees, and to trace the evolution of sequence lineages of interest. Details of these methods are described in Supplemental Experimental Procedures.
Probability Analysis of Transcript Frequencies
Both frequentist and Bayesian analyses were used to analyze donor antibodyomes with VRC01-class antibodyomes to understand the B cell ontogeny of the VRC01 class. These analyses focused on defining the number of genetic elements that contribute to the VRC01 class, as well as the number of independent lineages that contribute to the VRC01 class in each donor. Details are provided in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Coleman, M. Park, B. Schmidt, and A. Young for 454 pyrosequencing at the NIH Intramural Sequencing Center (NISC), J. Stuckey for assistance with figures, and the study participants and research staff at each of the Protocol G clinical centers, the Protocol G team members, the IAVI Human Immunology Laboratory and the Protocol G clinical investigators, including George Miiro, Anton Pozniak, Dale McPhee, Olivier Manigart, Etienne Karita, Andre Inwoley,Walter Jaoko, Jack DeHovitz, Linda-Gail Bekker, Punnee Pitisuttithum, Robert Paris, Jennifer Serwanga, and Susan Allen. We also thank Anthony West and members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for discussions or comments on the manuscript. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases and the National Human Genome Research Institute, National Institutes of Health, and by grants from the International AIDS Vaccine Initiative’s Neutralizing Antibody Consortium and by the Center for HIV/AIDS Vaccine Immunology Grant AI 5U19 AI 067854-06 from the National Institutes from Health. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors for the nine antibody-HIV-1 gp120 complex structures have been deposited with the Protein Data Bank under accession codes 4JPV, 4JPW, 4LSP, 4LSQ, 4LSR, 4LSS, 4LST, 4LSU, 4LSV. Raw Roche 454 pyrosequencing data sets used in this study were deposited to NCBI Sequence Read Archive (SRA) with accession number of SRA072279. Verified neutralizing heavy- and light-chain variable-region sequences were deposited to NCBI GenBank with accession numbers: KC848679–KC848710.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures, four tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2013.04.012.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D. Antibody-based protection against HIV infection by vectored immuno-prophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 2008;82:11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dhillon AK, Donners H, Pantophlet R, Johnson WE, Decker JM, Shaw GM, Lee FH, Richman DD, Doms RW, Vanham G, Burton DR. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 2007;81:6548–6562. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Feeney AJ, Tang A, Ogwaro KM. B-cell repertoire formation: role of the recombination signal sequence in non-random V segment utilization. Immunol. Rev. 2000;175:59–69. [PubMed] [Google Scholar]
- Gilbert PB, Berger JO, Stablein D, Becker S, Essex M, Hammer SM, Kim JH, Degruttola VG. Statistical interpretation of the RV144 HIV vaccine efficacy trial in Thailand: a case study for statistical issues in efficacy trials. J. Infect. Dis. 2011;203:969–975. doi: 10.1093/infdis/jiq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc. Natl. Acad. Sci. USA. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. USA. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. 5 edn. Bethesda, MD: U.S. Department of Health and Human Service, National Institutes of Health; 1991. [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li H, Yang F, Smith-Gill SJ, Mariuzza RA. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nat. Struct. Biol. 2003;10:482–488. doi: 10.1038/nsb930. [DOI] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. NISC Comparative Sequencing Program. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, McTamney PM, Yassine HM, Whittle JR, Guo X, Boyington JC, Wei CJ, Nabel GJ. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paul WE. Fundamental Immunology. Fourth Edition. Philadelphia, PA: Lippincott Raven; 1999. [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Winfrey S, Yang ZY, Xu L, Rao SS, Srivastava I, Barnett SW, Nabel GJ, Mascola JR. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine. 2007;25:1398–1408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl. Acad. Sci. USA. 2012;109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. NISC Comparative Sequencing Program. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang C, O’Dell S, Li Y, Keele BF, Yang Z, Imamichi H, Doria-Rose N, Hoxie JA, Connors M, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J. Virol. 2012;86:5844–5856. doi: 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.