Abstract
Purpose
Genome-wide DNA methylation analyses have identified hundreds of candidate DNA-hypermethylated genes in cancer. Comprehensive functional analyses provide an understanding of the biologic significance of this vast amount of DNA methylation data that may allow the determination of key epigenetic events associated with tumorigenesis.
Experimental Design
To study mechanisms of cysteine dioxygenase type 1 (CDO1) inactivation and its functional significance in breast cancer in a comprehensive manner, we screened for DNA methylation and gene mutations in primary breast cancers and analyzed growth, survival, and reactive oxygen species (ROS) production in breast cancer cells with restored CDO1 function in the context of anthracycline treatment.
Results
DNA methylation-associated silencing of CDO1 in breast cancer is frequent (60%), cancer specific, and correlates with disease progression and outcome. CDO1 function can alternatively be silenced by repressive chromatin, and we describe protein-damaging missense mutations in 7% of tumors without DNA methylation. Restoration of CDO1 function in breast cancer cells increases levels of ROS and leads to reduced viability and growth, as well as sensitization to anthracycline treatment. Priming with 5-azacytidine of breast cancer cells with epigenetically silenced CDO1 resulted in restored expression and increased sensitivity to anthracyclines.
Conclusion
We report that silencing of CDO1 is a critical epigenetic event that contributes to the survival of oxidative-stressed breast cancer cells through increased detoxification of ROS and thus leads to the resistance to ROS-generating chemotherapeutics including anthracyclines. Our study shows the importance of CDO1 inactivation in breast cancer and its clinical potential as a biomarker and therapeutic target to overcome resistance to anthracyclines.
Introduction
Loss of proper tumor suppressor function leads to the initiation and progression of human cancer (1) and aberrant epigenetic alterations including DNA promoter hypermethylation can be responsible for such functional loss (2). Techniques to analyze genome-wide DNA methylation have become useful tools to identify hundreds of new candidate DNA-hypermethylated genes in cancer. Comprehensive functional analyses can provide an understanding of the biologic significance of the vast amount of DNA methylation data generated, allowing for the discovery of novel tumor suppressor genes and molecular mechanisms underlying tumor growth control, and biomarkers for early detection, prognosis, and response to therapeutic agents in cancer (3–7).
Cysteine dioxygenase type 1 (CDO1), recently identified as a candidate hypermethylated gene within the functional breast cancer hypermethylome (7), is a non–heme iron dioxygenase (8). CDO1 determines the flux between cysteine catabolism and glutathione synthesis (9) by catalyzing the oxidation of cysteine to cysteine sulfinic acid in the presence of molecular oxygen (10). Abnormal or deficient CDO1 activity has been implicated in a variety of neurologic and autoimmune diseases such as Parkinson’s, Alzheimer’s, rheumatoid arthritis, systemic lupus erythematosus (11–13), and recently in carcinogenesis. CDO1 is a promising prognostic biomarker in malignancies with loss of CDO1 expression being associated with relapse of Wilms tumor (14) and DNA methylation of the CDO1 promoter is associated with poor prognosis in patients with breast cancer (7, 15). CDO1 has also been implicated in several studies to play a role in the oxidative stress response of cancer cells (16, 17).
Many cancer cells, in particular at advanced stage, function with higher basal levels of endogenous oxidative stress than normal cells. Under persistently increased reactive oxygen species (ROS) production, cancer cells adapt to such stress to escape oxidative damage and ROS-induced apoptosis by developing an enhanced, endogenous detoxification capacity (18). The mechanisms of ROS stress adaptation involve the activation of ROS-scavenging enzymes and endogenous antioxidants (19, 20) such as glutathione (21). Although increased ROS stress promotes initiation and progression of cancer (22, 23), excessive levels of ROS can be toxic (24) and lethal if exceeding a threshold above cellular tolerability (25). This concept is of therapeutic interest, because it is thought that increased ROS production makes cancer cells more vulnerable to damage by further ROS insults induced by exogenous ROS-generating agents including the chemotherapeutic class of anthracyclines (24, 26). The redox adaptation of cancer cells, however, can provide a mechanism for resistance to anthracyclines (24, 27).
We present a comprehensive study that addresses the functional significance of silencing of CDO1 during breast tumorigenesis. We report the frequent inactivation of CDO1 by multiple mechanisms (60% DNA methylation, 20% missense mutations) in breast cancer and also across multiple other types of cancer. Cells with restored CDO1 function show reduced growth, viability, and ROS detoxification capacity and increased sensitivity to anthracyclines. Given these findings, we suggest that silencing of CDO1 is a critical event that drives tumorigenesis and contributes to the survival of oxidative-stressed breast cancer cells and their resistance to anthracyclines through reducing cellular ROS levels.
Materials and Methods
Cell culture and drug treatment
Cell lines were purchased from American Type Culture Collection and cultured in appropriate media (Mediatech) supplemented with 10% FBS (Atlanta Biologicals) and 1 × Penicillin–Streptomycin (Mediatech) at 37°C in 5% CO2 atmosphere. The HCT116 derivative cell line lacking the major DNA methyltransferases DNMT1 and 3b (DNMT1−/− and DNMT3b−/−; double knockout or DKO) was maintained as previously described (28). Drug treatment with 5-aza-2′-deoxycytidine (DAC) and trichostatin A (TSA) were carried out as previously described (7). For glutathione depletion, cells were treated with 0.5 mmol/L buthionine sulfoximine (BSO). Doxorubicin was supplemented in doses ranging from 0.078 to 20 μmol/L to determine LD50 dosage and 5-azacytidine in doses ranging from 1 to 5 μmol/L, chosen for maximal CDO1 expression.
Patient samples
Primary tumor specimen and normal breast tissues from cancer-free donors were obtained from the archives of the Department of Pathology, Johns Hopkins University (Baltimore, MD) and Department of Pathology, GROW-School for Oncology and Developmental Biology, Maastricht University Medical Center (the Netherlands) with Institutional review board approval and Health Insurance Portability and Accountability Art compliance. Genome-wide methylation and expression data of primary tissues were also used from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/).
Gene expression, methylation analysis, and ChIP
RNA was extracted using RNeasy Mini Kit (Qiagen) or purchased from Stratagene (normal tissues). For reverse transcriptase (RT) and real-time RT-PCR, 1 μg RNA was reverse transcribed into cDNA using Ready-To-Go You-Prime First-Strands Beads (GE Healthcare) with addition of pd(N)6 Random Hexamers (GE Healthcare) according to the manufacturer’s instructions. Genomic DNA was extracted following a standard phenol–chloroform extraction and bisulfite modified using the EZ DNA Methylation Kit (Zymo Research). Methylation-specific PCR (MSP) was conducted as previously described (29). PCR products for bisulfite sequencing were cloned using the TOPO TA Cloning Kit (Invitrogen), purified from single colonies using QIAprep Spin Miniprep Kit (Qiagen), and sequenced with M13 reverse primer by Johns Hopkins Medical Institutions Synthesis & Sequencing Facility. For chromatin immunoprecipitation (ChIP), cells were crosslinked in 1% formaldehyde as previously described (30). Nuclear extraction using CEBN and CEB (cytoplasmic extraction buffer), and ChIP on ~1 × 106 cells per IP was conducted as previously described (31). α-H3-K4me2 and α-H3-K27me3 antibodies from Millipore were used. IP-specific products were amplified using real-time PCR.
Mutation analysis
CDO1 coding exons were amplified and purified PCR products were bidirectionally sequenced using DNA Sequencing Kit BigDye-Terminator Cycle Sequencing Ready Reaction (Applied Biosystems). Sequencing products were separated with the Applied Biosystems ABI3730 Sequencing System and analyzed with Lasergene software (DNASTAR). Protein damaging scores for identified mutations were calculated using PolyPhen-2 software at: http://genetics.bwh.harvard.edu/pph2/.
Immunohistochemistry
CDO1 protein expression was detected on the sections of formalin-fixed and paraffin-embedded breast tissue (normal and tumors) using Vectastain blocking serum (Vector Laboratories), α-CDO1 primary antibody (Abcam), α-rabbit biotinylated secondary antibody (Vector Laboratories), horseradish peroxidase–labeled Vectastain Elite ABC Rabbit IgG Kit (Vector Laboratories), and 3,3′-diaminobenzidine (Sigma) as substrate. All slides were counterstained with DAKO hematoxylin and Scotts Blue.
Expression vectors
Wild-type CDO1 (NM_001801.2) or mutant CDO1 (Y157F; ref. 32) was cloned into pcDNA3.1/V5-His B expression vector (Invitrogen). Tetracycline-inducible CDO1-stable cells were generated using the T-REx System (Invitrogen). Expression of CDO1 was induced with 0.5 μg/ mL doxycycline.
Western blot analysis
CDO1 protein expression was detected in whole-cell protein extracts with either α-V5 (Invitrogen) or α-CDO1 (Abcam).
Colony formation and soft-agar assay
Cells, transiently transfected with CDO1, were harvested 24 hours after transfection, replated in 10 cm2 dishes in triplicates and selected with 0.8 mg/mL Geneticin/G418 (Invitrogen) for 15 days. Colonies were stained with Giemsa and counted. Soft-agar assays were started 48 hours following transfection. A total of 1.5 × 104 cells in complete media containing 0.4% agar were layered on top of 0.6% agar in 24-well plates in duplicates. Colonies were selected with 0.8 mg/mL Geneticin/G418 (Invitrogen) for 28 days and counted after staining with 0.005% crystal violet.
Cell viability assay
Cell viability was measured using CellTiter96 kit (Promega). Cells were incubated in MTS reagent for 4 hours at 37°C. Absorbance was measured at 490 nm.
Measurement of ROS production
ROS production was measured using CM-H2DCFDA probe (Invitrogen). Cells were loaded with 5 μmol/L CM-H2DCFDA probe in phenol red–free and serum-free media for 1 hour. Fluorescence was measured at 493 nm excitation and at 523 nm emission.
Statistical analysis
Tumor stage and gene methylation status were correlated using Pearson χ2. HR for prognostic value of gene methylation status was calculated using univariate and multivariate Cox regression analysis. Student t test and trend test were used to conduct group comparisons for colony formation, soft-agar, ROS, and cell viability assays. Gene expression and methylation status of TCGA data were correlated calculating a Spearman correlation coefficient. P values less than 0.05 were considered significant. All statistical analyses were conducted using the STATA 9.2 software package.
Results
Silencing of CDO1 is associated with DNA promoter hypermethylation or repressive chromatin structure
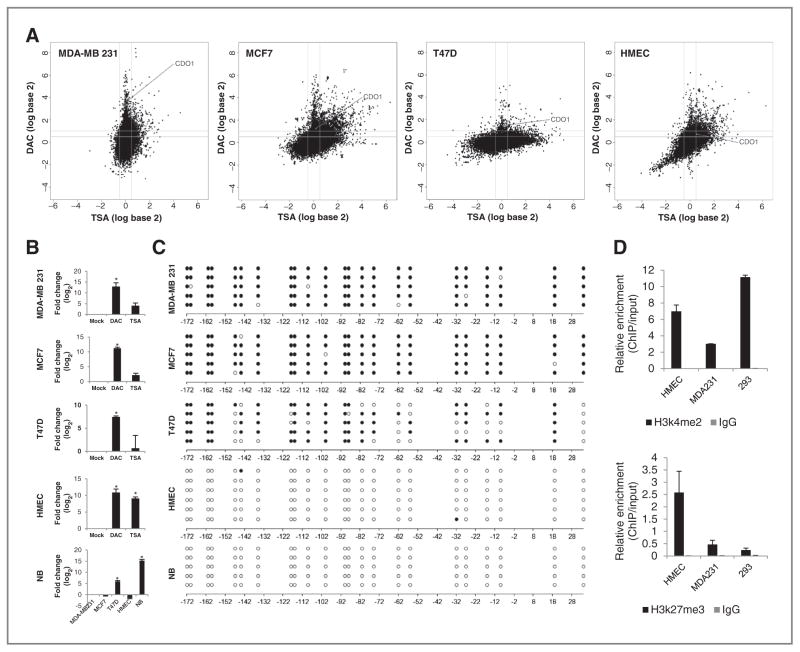
CDO1 (Supplementary Fig. S1 illustrates the genomic location, structure, and CpG island of the CDO1 gene) was identified as a candidate hypermethylated gene within the functional breast cancer hypermethylome (7), where potential DNA-hypermethylated genes appear in a zone in which gene expression was not detectable in untreated cells and cells treated with the histone deacetylase inhibitor TSA (<0.5 log-fold), but increased more than 0.5 log-fold in cells treated with the DNMT inhibitor DAC. CDO1 appeared in this characteristic spike of potentially DNA-hypermethylated genes in 3 of 4 tested invasive breast cancer cell lines (3.49 log-fold in MDA-MB-231, 1.24 log-fold in MCF7, and 1.48 log-fold T-47D; Fig. 1A). Conversely, CDO1 did not appear in the spike of potentially DNA-hypermethylated genes in nontransformed human mammary epithelial cells (HMEC), suggesting that CDO1 DNA methylation occurs specifically in cancer. In HMECs, CDO1 was silenced at basal level (untreated), but reexpressed with DAC (0.73 log-fold) and TSA (0.54 log-fold; Fig. 1A).
Figure 1.
Silencing of CDO1 is associated with DNA promoter hypermethylation or repressive chromatin structure. A, appearance of CDO1 within the breast cancer hypermethylome. Cell lines were treated with either 5 μmol/L DAC for 96 hours or 300 nmol/L TSA for 18 hours. Gene expression changes (analyzed on 4 × 44 K Agilent platform) are plotted by fold change (log scale) after DAC (y-axis) or TSA (x-axis) treatment. B, quantitative mRNA expression of CDO1 in DAC- (5 μmol/L 96 hours) or TSA- (300 nmol/L 18 hours) treated cells is shown in fold change (log2) relative to mock-treated cells. Expression of CDO1 in normal breast (NB) is shown in relation to basal expression levels of CDO1 in other cell lines. Group comparisons were carried out using Student t test. *, P < 0.05. C, bisulfite sequencing of the CDO1 promoter region from −192 bp to +60 bp relative to the transcription start site (TSS). White and black circles represent unmethylated and methylated CpG dinucleotides, respectively. D, ChIP at the CDO1 promoter region from −154 bp to −29 bp relative to TSS for α-H3k4me2 and α-H3k27me3. Data presented are the mean levels of enrichment relative to input obtained by real-time PCR from 2 independent experiments ± SEM.
The mRNA expression status of CDO1 was validated in untreated as well as DAC- and TSA-treated MDA-MB-231, MCF7, and T-47D breast cancer cells or HMECs (Fig. 1B). In addition, we detected CDO1 baseline transcription in normal breast tissue. Next, we examined the basal DNA methylation status of CDO1 using bisulfite sequencing. The CDO1 promoter is unmethylated in normal breast and HMECs, densely methylated in MDA-MB-231 and MCF7 cells, and partially methylated in T-47D cells (Fig. 1C). To investigate whether the loss of CDO1 expression in HMECs could be due to histone modifications, we next conducted ChIP for histone marks at the CDO1 promoter region in HMECs, MDA-MB-231, and 293 cells. CDO1-expressing 293 cells are enriched for the active H3K4me2 mark and have low levels of the repressive H3K27me3 mark, whereas MDA-MB-231 cells, which have CDO1 densely methylated, have low levels of the active and the repressive mark (Fig. 1D). In contrast, HMECs, which have CDO1 silenced but not methylated, display a bivalent chromatin pattern with the highest levels of the repressive H3K27me3 mark and relatively high levels of the H3K4me2 mark.
DNA methylation-associated silencing of CDO1 is cancer specific, frequent, and correlates with disease progression and outcome
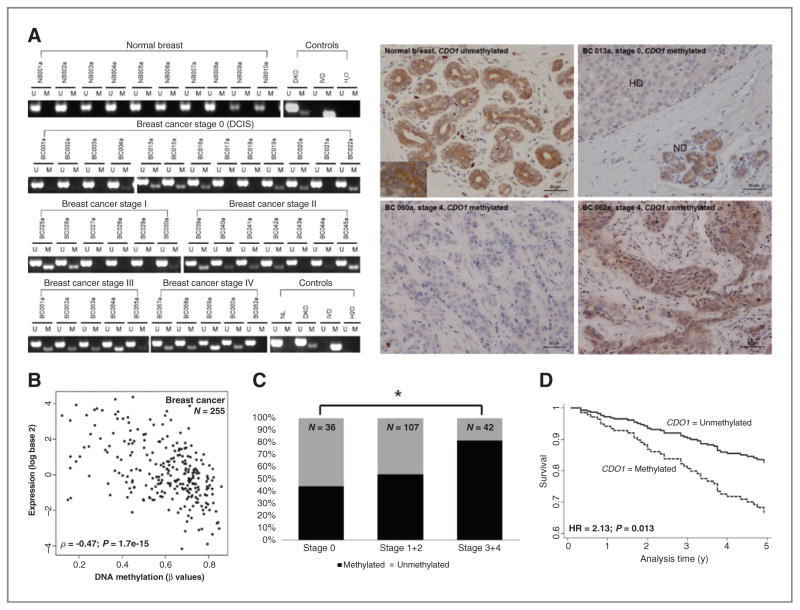
We studied DNA methylation of CDO1 in 20 normal breast specimens from cancer-free patients (Supplementary Table S1A) as well as in a cohort of 185 primary breast cancers including stages 0 [ductal carcinoma in situ (DCIS)] to 4 tumors (Supplementary Table S1B). DNA methylation of CDO1 is frequent and cancer-specific, that is, DNA methylation was detected in 108 of 185 breast cancers (~60%), but in none of the tested 20 normal breast samples (Fig. 2A left for selected samples). Next, we examined CDO1 protein expression by immunohistochemistry on selected tumor samples and in normal breast tissue. In normal breast, CDO1 is uniformly expressed in the cytoplasm of ductal termini cells (Fig. 2A right, top left). In DCIS, CDO1 expression is lost in hyperproliferative ductal termini where CDO1 is methylated but not in normal differentiated ductal termini (Fig. 2A right, top right and Fig. 2A left, sample breast cancer 013a). In higher stage breast cancers, we also observed a correlation between CDO1 expression and CDO1 methylation status. A methylated CDO1 promoter was associated with the loss of CDO1 expression, (breast cancer 060a, Fig. 2A left and 2A right, bottom left), whereas an unmethylated promoter correlated with retained expression of CDO1 (breast cancer 062a, Fig. 2A left and 2A right, bottom right). We next used data of 255 primary breast cancers from the TCGA database and determined a significant inverse correlation (ρ = −0.47, P = 1.7e-15) between DNA methylation and expression of CDO1 (Fig. 2B).
Figure 2.
DNA methylation-associated silencing of CDO1 is cancer specific, frequent, and correlates with disease progression and outcome. A, DNA methylation and protein expression status of CDO1. CDO1 promoter region from −168 bp to −45 bp relative to the TSS was assayed by MSP in a cohort of 20 normal breast tissues from non–cancer patients and 185 primary breast cancers (BC) of stages 0 (DCIS) to 4 with U and M marking unmethylated and methylated bands, respectively. Representative examples are shown for each cohort and each tumor stage. In vitro methylated DNA (IVD), DKO cells, normal lymphocytes (NL), and H2O controls were assayed along with samples. Protein expression status of CDO1 was assayed by immunohistochemistry in normal breast and selected primary breast cancers. Note, BC013a, a DCIS sample, displays loss of CDO1 expression in hyperproliferative ductuli (HD) but expression of CDO1 in normal ductuli (ND). B, scatter plot depicting correlation between expression log2 values (y-axis; analyzed on Agilent 244 K Custom Gene Expression G4502A-07 platform) and DNA methylation β values (x-axis; analyzed on Illumina HumanMethylation 27k platform) of CDO1 in 255 primary breast cancers from TCGA data portal. A Spearman correlation coefficient of ρ = −0.47 and P value of 1.7e-15 were calculated. A P < 0.05 was considered statistically significant. C, DNA methylation frequency (in%) of CDO1 plotted by tumor stage of 185 primary breast cancers. CDO1 methylation status and tumor stage were correlated using Pearson χ2 test. *, P < 0.05. D, survival curve depicting prognostic value of CDO1 methylation status for outcome prediction in 185 primary breast cancers. HR for prognostic value was calculated using univariate Cox proportional hazards regression model.
We next tested whether the presence of CDO1 methylation altered prognosis of women with breast cancer. Common prognostic clinicopathologic variables were compared with CDO1 methylation status in our cohort of 185 patients with breast cancer. The frequency of CDO1 methylation significantly increased with tumor stage, i.e., 44% in stage 0/ DCIS, 53% in stage 1 and 2, and 81% in stage 3 and 4 tumors (Fig. 2C and Supplementary Table S2). In addition, CDO1 was significantly more frequently methylated in tumors with lymphovascular invasive (lvi)/perinodal invasive (pni) breast cancers (P = 0.011; 52% in lvi/pni neg breast cancers and 73% in lvi/pni pos breast cancers). Furthermore, a methylated CDO1 promoter status was associated with an unfavorable patient outcome [HR 2.13, 95% confidence interval (CI) 1.17–3.86, P = 0.013], but not independently of age and stage (HR 1.10, 95% CI 0.57–2.12, P = 0.771). A survival curve based on the univariate Cox regression model is shown in Fig. 2D.
DNA methylation-associated silencing of CDO1 occurs in multiple cancer types
We expanded our efforts to analyze the DNA methylation and expression status of CDO1 in other tumor types. We assayed the CDO1 expression status in normal tissues as well as in 3 corresponding cancer cell lines (ovary, lung, pancreas, and liver). CDO1 was expressed in all tested normal tissues but not expressed in association with DNA promoter methylation in the corresponding cancer cell lines with the exception of HEPG2 liver cancer cells, in which the CDO1 promoter is unmethylated (Supplementary Fig. S2A). We further determined CDO1 methylation status in primary tumor specimens of these tumor types. We found CDO1 to be commonly methylated (>60%) in ovary, lung, and pancreas cancer (Supplementary Fig. S2B), but not in hepatocellular cancer (9%). Again, using data from the TCGA database, analysis of 104 primary lung cancers and 584 primary ovarian cancers confirmed an inverse relationship (lung cancer: ρ = −0.60, P = 1e-63 and ovarian cancer: ρ = −0.62, P = 1.6e-63) between DNA methylation and expression of CDO1 (Supplementary Fig. S2C). As in breast cancer, we observed a correlation between tumor stage and CDO1 methylation frequency in TCGA lung and ovarian cancers (Supplementary Fig. S2D).
Tumor-specific point mutations within the CDO1 gene have a predicted protein-damaging effect
To test whether CDO1 may be inactivated by ways other than epigenetic mechanisms, we screened for mutations within the CDO1 gene in 60 primary breast cancers (unmethylated CDO1 promoter status). We found 9 single-nucleotide polymorphism in 10 patients (17%; ref. Table 1) leading to amino acid substitutions (missense mutations). The identified mutations did not associate with an unfavorable patient outcome. To evaluate the functional significance of these mutations, the PolyPhen-2 software was used to calculate a protein damage score. Three mutations (T4I, L62F, and E79K) reached a score of approximately 1, predicting for a protein damaging effect (values near 1 are predicted to be deleterious) with the highest possible probability. The Y157F mutation was introduced as a control into this assay. This mutation, within the catalytic center of the CDO1 enzyme, has been shown to reduce the enzymatic activity to up to approximately 95% (32). A calculated protein damaging score of 0.999 reliably predicted for the experimental proven loss-of-function caused by this mutation. Subsequently, we confirmed the 3 mutations with the highest damage scores as tumor-specific by screening normal tissue of the 4 patients that harbored these mutations in their tumor. Supplementary Figure S3 provides chromatograms of the tumor and matching normal tissue. Overall, we identified 4 of 60 patients (7%) that harbor tumor-specific and protein-damaging point mutations within the CDO1 gene.
Table 1.
Missense mutations in CDO1 gene
| Mutation | Patient | PolyPhen-2 damage score |
|---|---|---|
| T4I | 177a | 0.907 |
| G25S | 203a | 0 |
| D26N | 085a | 0.001 |
| V28I | 166a | 0 |
| E41K | 210a | 0 |
| L62F | 162a; 048a | 1 |
| M73I | 210a | 0.09 |
| E79K | 087a | 1 |
| G195D | 164a | 0.009 |
| Y157Fa | control | 0.999 |
Reduces enzymatic activity of CDO1 to up to 95%.
Restoration of CDO1 function reduces growth and viability of cancer cells and their capacity to detoxify ROS
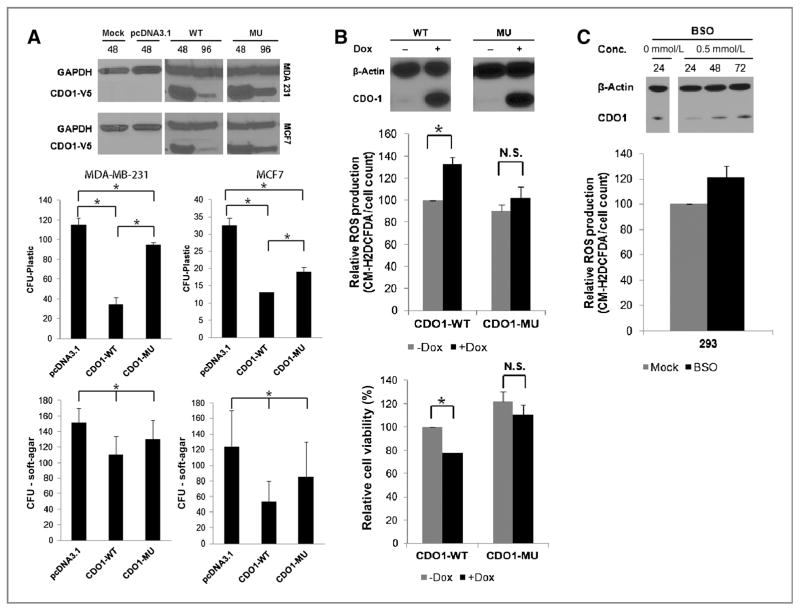
To test whether CDO1 can alter cancer cell growth, we conducted colony formation and soft-agar assays after transient expression of CDO1 in MCF7 and MDA-MB-231 cells, in which endogenous expression of CDO1 is silenced by DNA methylation. Following expression of enzymatic active wild-type CDO1 (CDO1-WT) in MCF7 and MDA-MB-231 cells, cells formed markedly fewer colonies (Fig. 3A and Supplementary Fig. S4A and S4B) on plastic and in soft-agar than cells transfected with empty vector (pcDNA3.1) or mutant CDO1 protein (CDO1-MU). Notably, expression of enzymatic impaired CDO1-MU protein suppressed growth of cells compared with cells expressing empty vector, possibly due to the incomplete catalytic loss of the Y157F mutation (32). We confirmed reexpression of CDO1 at protein level by Western blot analysis (Fig. 3A).
Figure 3.
Restoration of CDO1 function reduces growth and viability of cancer cells and their capacity to detoxify ROS. A, tumor cell clonogenicity was assessed on plastic and in soft-agar. Cells were transiently transfected with pcDNA3.1 (empty vector), pcDNA3.1-CDO1-WT (wild-type CDO1), or pcDNA3.1-CDO1-MU (mutant CDO1), and replated 24 hours posttransfection for selection with Geneticin/G418. After 18 days of selection, colonies were stained with Giemsa and counted. Data presented are the mean of 2 independent experiments ± SEM. Group comparisons were carried out using Student t test and trend test. *, P < 0.05. Reexpression of CDO1 was confirmed 48 and 96 hours posttransfection by Western blot analysis using α-V5 antibody, targeting the V-5-His tag of recombinant CDO1 protein, and α-GAPDH as a control. ROS production and cell viability were assayed in tetracycline-inducible CDO1-stable MDA-MB-231 cells before and after treatment with 0.5 μg/mL doxycycline (Dox; B), and 293 cells before and after the treatment with 0.5 mmol/L BSO (for depletion of glutathione; C) by fluorescence of the CM-H2DCFDA probe. Obtained values were normalized to untreated or treated empty vector controls and plotted as % relative to untreated MDA-MB-231-CDO1-WT or untreated 293 cells. Data presented are the mean of 3 independent experiments ± SEM. Group comparisons were carried out using Student t test. *, P < 0.05; n.s., not significant. Reexpression of CDO1 in MDA-MB-231 cells 48 hours after doxycycline treatment or downregulation of CDO1 expression in 293 cells 24, 48, and 72 hours after glutathione depletion with BSO was assessed by Western blot analysis using α-CDO1 antibody and α-β-actin as a control.
Given CDO1’s key role in the cysteine and glutathione metabolism (9, 17), we next studied ROS levels and cell viability in MDA-MB-231 cells having inducible expression CDO1-WT or CDO1-MU. ROS production was 33% higher in cells having inducible CDO1-WT expression as compared with mock cells and these cells were 20% less viable (Fig. 3B). ROS production was slightly, but not significantly, increased between mock cells or those expressing CDO1-MU protein without a change in cell viability. These results suggest that expression of enzymatic active CDO1 reduces viability of MDA-MB-231 cells through decreasing their ROS detoxification capacity.
Next, we treated 293 cells, which endogenously express CDO1, with the oxidative-damaging and glutathione-depleting agent BSO (33). Interestingly, 24 hours upon treatment with BSO, 293 cells show decreased levels of CDO1 protein and increased levels of ROS production that return to baseline at 48 hours posttreatment (Fig. 3C). This suggests that CDO1 protein level may decrease in response to increasing ROS production as an antioxidant adaptive mechanism.
CDO1-induced reduction in ROS detoxification sensitizes breast cancer cells to anthracycline treatment
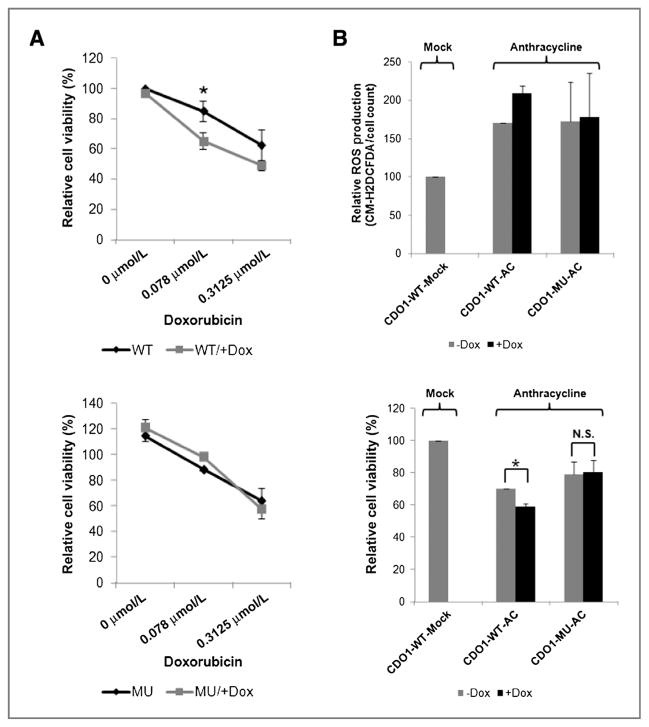
Anthracyclines, such as doxorubicin, are cytotoxic ROS-generating chemotherapeutic agents (27) widely used in the treatment of breast cancer. Resistance to these agents is believed to be conferred by the upregulation of the ROS detoxification capacity in adaptation to intrinsic oxidative stress in cancer cells (27). To test whether inactivation of CDO1, as observed above, might contribute to the resistance of breast cancer cells to doxorubicin therapy, we treated CDO1 inducible MDA-MB-231 cells with different doses of doxorubicin. Cells expressing enzymatic active CDO1-WT, but not CDO1-MU, were significantly less viable at doxorubicin doses of 0.078 and 0.3125 μmol/L than mock cells (Fig. 4A and B). As expected, doxorubicin treatment increases ROS production as compared with untreated cells. Expression of CDO1-WT, but not CDO1-MU, further increased the doxorubicin-induced ROS production compared with mock cells (Fig. 4B).
Figure 4.
CDO1-induced reduction in ROS detoxification sensitizes breast cancer cells to anthracycline treatment. A, cell viability of doxycycline-induced/not doxycycline-induced CDO1-stable MDA-MB-231 cells before and 48 hours after treatment with different doses of anthracycline (doxorubicin) was measured by MTS assay. Data presented are the mean of 2 independent experiments ± SEM. Group comparisons were carried out using Student t test. *, P < 0.05. B, ROS production, using CM-H2DCFDA probe and cell viability of same cells before and 48 hours after treatment with 0.078 μmol/L doxorubicin was measured. Data presented are the mean of 2 independent experiments ± SEM. Obtained values for A and B were normalized to anthracycline-untreated or -treated doxycycline-induced/not doxycycline-induced empty vector control cells and plotted as % relative to anthracycline-untreated and not doxycycline-induced MDA-MB-231-CDO1-WT cells. Group comparisons were carried out using Student t test. *, P < 0.05; n.s., not significant.
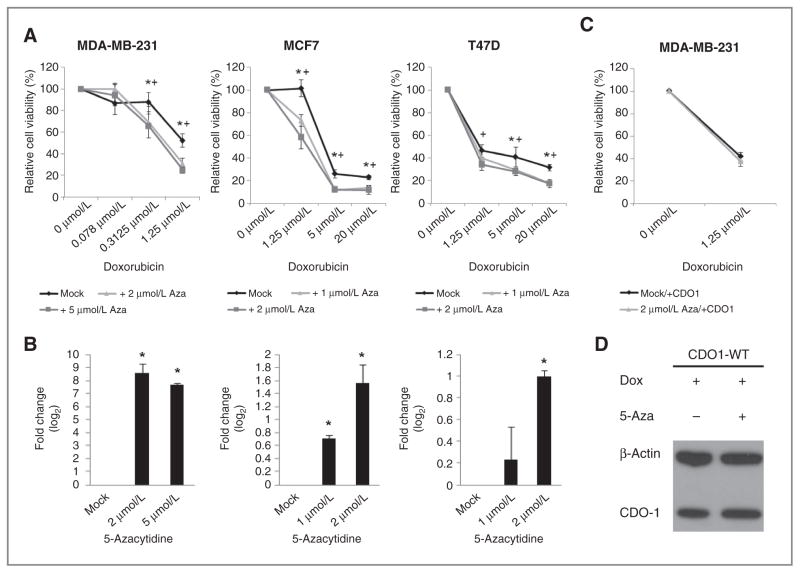
To determine whether the restoration of CDO1 expression through treatment with 5-azacytidine was a viable strategy to sensitize breast cancer cells to doxorubicin therapy, we pretreated MDA-MB-231, MCF7, and T-47D cells with 5-azacytidine for 72 hours. This treatment resulted in reexpression of CDO1 and an up to 40% decreased cell viability when cells were subsequently treated with doxorubicin compared with cells not pretreated with 5-azacytidine (Fig. 5A and B). To further implicate the reexpression of CDO1 in this synergistic effect, we pretreated MDA-MB-231 cells with doxycycline-induced CDO1 expression with 5-azacytidine and subsequently with doxorubicin. We observed no difference in viability between cells that over-expressed CDO1 and were pretreated with 5-azacytidine and cells that overexpressed CDO1 and were not pretreated with 5-azacytidine (Fig. 5C and D).
Figure 5.
Reactivation of epigenetically silenced CDO1 through priming with 5-azacytidine contributes to the sensitization of breast cancer cells to anthracycline treatment. A, cell viability of MDA-MB-231, MCF7, and T47-D cells primed with 5-azacytidine at doses ranging from 1 μmol/L to 5 μmol/L for 72 hours and subsequently treated with doxorubicin at doses ranging from 0.078 μmol/L to 20 μmol/L for 48 hours. Obtained values are plotted as % relative to doxorubicin-untreated cells. Group comparisons were carried out using Student t test. *, P < 0.05 for 1 μmol/L 5-azacytidine in MCF7 and T47-D cells or 2 μmol/L in MDA-MB-231 cells. +, P < 0.05 for 2 μmol/L 5-azacytidine in MCF7 and T47-D cells or 5 μmol/L in MDA-MB-231 cells. B, quantitative reexpression of CDO1 in 5-azacytidine (1 μmol/L, 2 μmol/L, or 5 μmol/L for 72 hours)-treated cells prior doxorubicin treatment is shown in fold change (log2) relative to mock-treated cells. Group comparisons were carried out using Student t test. *, P < 0.05. C, cell viability of doxycycline-induced CDO1-stable MDA-MB-231 cells (doxycyline was supplemented every 24 hours throughout the entire experiment) primed with 5-azacytidine at a dose of 2 μmol/L for 72 hours and subsequently treated with doxorubicin at a dose of 1.25 μmol/L for 48 hours. Obtained values are plotted as % relative to doxorubicin-untreated cells. As a control, cell viability was measured in CDO1-stable MDA-MB-231 cells that were not treated with 5-azacytidine. D, restoration of CDO1 protein expression in MDA-MB-231 cells 72 hours posttreatment with doxycycline and with or without 5-azacytidine was confirmed by Western blot using α-CDO1 antibody and α-β-actin as a control.
Discussion
In the current study, we show that aberrant DNA methylation of CDO1 is a tumor-specific and frequent (~60%) event in breast cancer that is associated with gene silencing. We observed a stage-dependent increase in CDO1 methylation frequency that significantly correlates with disease progression and outcome. Loss of CDO1 expression by DNA methylation is also a frequent event in multiple other cancer types. In addition to other studies that uncovered CDO1 as aberrantly methylated and silenced in colorectal cancer (34) and malignant glioma (35), we found promoter hypermethylation of CDO1 in association with gene silencing in ovary, lung, pancreas, and hepatocellular cancer. We further show that CDO1 function can also be lost by other mechanisms. In HMECs, we correlated silenced expression of CDO1 with a decrease of the active H3K4me2 histone mark and an increase of the repressive H3K27me3 mark, indicative of bivalent chromatin, suggesting that a repressive chromatin structure at the CDO1 promoter can adequately suppress the expression of CDO1, similar to the poised state of embryonic stem cells (36). In addition to aberrant epigenetic regulation of CDO1 gene expression, we discovered genetic aberrations that potentially alter CDO1 function. Screening of primary breast cancers with an unmethylated CDO1 status revealed missense mutations in 17% of these tumors and when tested for functional significance, half of these mutations predicted for protein damage. Unlike CDO1 methylation, the identified mutations within the CDO1 gene did not correlate with disease outcome, potentially due to the small number of samples screened.
Under persistent increased ROS production, cancer cells adapt to such stress to escape oxidative damage and cell death by developing an enhanced, endogenous antioxidant capacity (18, 21). We find that cells that reexpress enzymatic active CDO1 harbor more ROS and are less viable than cells expressing enzymatic impaired CDO1. Restoration of CDO1 function in breast cancer cells may shift the flux from glutathione synthesis toward cysteine catabolism resulting in a decreased antioxidant capacity that is not sufficient to keep ROS levels below a toxic threshold. Similar findings have been made by Dominy and colleagues (17). Overexpression of CDO1 resulted in reduced levels of cysteine and glutathione and in enhanced sensitivity to a glutathione-dependent stressor, suggesting that glutathione levels and cellular redox capacity change in response to CDO1 expression through the limitation of cysteine, the substrate for glutathione synthesis. On the basis of these and our data, we suggest that epigenetic silencing of CDO1 may occur in cancer cells with increased ROS production and that this event may contribute to the survival of these oxidative stressed cancer cells through an increased ROS detoxification.
It is thought that increased ROS production makes cancer cells more vulnerable to damage by further ROS insults induced by exogenous ROS-generating agents such as anthracyclines (24, 26). However, an enhanced antioxidant capacity not only enables cancer cells to survive under increased ROS stress and contributes to cancer cell transformation and metastasis (37–39) but also leads to resistance to ROS-generating agents (24, 27). In this respect, we observed that MDA-MB-231 cells, upon treatment with the anthracycline doxorubicin, are more sensitive when they reexpress enzymatic active CDO1 as compared with cells which express functional impaired CDO1. This finding is particularly interesting when taking into account that hypermethylation of CDO1 is an outcome predictor in anthracycline-treated, estrogen receptor-positive, and lymph node-positive patients with breast cancer (15). Our finding, that breast cancer cells with loss of CDO1 function are less sensitive to doxorubicin treatment, provides a mechanism for the predictive value of CDO1 methylation in anthracycline-treated patients and expands our understanding of how cancer cells escape the damage of ROS-generating chemotherapeutics. Anthracyclines are key components of the treatment of patients with breast cancer and loss of CDO1 expression might be a useful marker for prediction of resistance to this therapy and for selection of patients for priming therapy with 5-azacytidine to overcome resistance. Our data support that priming with 5-azacytidine of breast cancer cells with epigenetically silenced CDO1 may sensitize them to anthracycline therapy partly through the reexpression of CDO1.
Given the inactivation of CDO1 by multiple mechanisms across multiple types of cancer, a pattern that has been observed for important tumor growth–suppressive genes, the reduced growth, viability, and ROS detoxification capacity of cells with restored CDO1 function, we suggest that CDO1 may have tumor-suppressive function and that silencing of CDO1 may contribute to the survival of oxidative-stressed cancer cells and their resistance to anthracy-clines through increased ROS detoxification. Dependence of cancer cell survival has recently been shown to rely on the methylation of CDO1 as one of the driver epigenetic events (40). Our findings not only support these results, but also explore in detail the functional significance of epigenetic silencing of CDO1 during breast tumorigenesis.
Supplementary Material
Supplementary Table 1. Baseline characteristics of NB donors and BC patients
Supplementary Table 2. Statistical association between clinocopathologic characteristics of BC patients and DNA methylation frequency (%) of CDO1
Supplementary Table 3. Primer information
Translational Relevance.
Breast cancer is a heterogeneous disease driven by molecular changes of genetic and epigenetic nature. By screening for genome-wide DNA methylation changes in breast cancer, we identified cysteine dioxygenase type 1 (CDO1) as a DNA-hypermethylated gene. We show that CDO1 is frequently DNA methylated in breast primary tumors and that this event is associated with adverse clinical features and poor prognosis. On the basis of the suggested role for CDO1 in the oxidative stress response of cancer cells, we examined its role in the resistance to the reactive oxygen species (ROS)-generating chemo-therapeutic class of anthracyclines. We found that restoration of CDO1 function in breast cancer cells alters the oxidative stress response in a way that it leads to the sensitization to anthracylines. We further provide potential clinical implications for this finding by showing that priming with 5-azacytidine of breast cancer cells with epigenetically silenced CDO1 increases the sensitivity to anthracycline therapy. This finding provides a potential clinical strategy to overcome resistance to this drug and DNA methylation of CDO1 may be useful as a marker to select patients for priming with 5-azacytidine.
Acknowledgments
The authors thank Sharon Metzger and Theresa Sanlorenzo-Caswell from the Johns Hopkins Tumor Registry for their help with clinicopathologic patient information as well as Kathy Bender and Rick Moore for administrative support.
Grant Support
This study was supported by the Susan G. Komen Foundation, German Academic Exchange Service (DAAD), and Dr. Jost Henkel Stiftung.
Footnotes
Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: N. Ahuja, J. Jeschke, H.M. O’Hagan
Development of methodology: N. Ahuja, J. Jeschke, H.M. O’Hagan, W. Zhang, R. Vatapalli, K.E. Schuebel
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): N. Ahuja, J. Jeschke, R. Vatapalli, M.F. Calmon, C. Nelkenbrecher, M. Van Engeland, E.W. Gabrielson, A. Winterpacht
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): N. Ahuja, J. Jeschke, H.M. O’Hagan, R. Vatapalli, L. Danilova, L. Van Neste, I.T.G.W. Bijsmans, M. Van Engeland, A. Winterpacht
Writing, review, and/or revision of the manuscript: N. Ahuja, J. Jeschke, H.M. O’Hagan, L. Van Neste, S.B. Baylin, J.G. Herman
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): N. Ahuja, J. Jeschke, I.T.G.W. Bijsmans
Study supervision: N. Ahuja, J. Jeschke, H.M. O’Hagan, W. Zhang
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2:S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 3.Glockner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–9. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Glockner SC, Guo M, Machida EO, Wang DH, Easwaran H, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–72. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill VK, Ricketts C, Bieche I, Vacher S, Gentle D, Lewis C, et al. Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer Res. 2011;71:2988–99. doi: 10.1158/0008-5472.CAN-10-4026. [DOI] [PubMed] [Google Scholar]
- 7.Jeschke J, Van Neste L, Glockner SC, Dhir M, Calmon MF, Deregowski V, et al. Biomarkers for detection and prognosis of breast cancer identified by a functional hypermethylome screen. Epigenetics. 2012;7:701–9. doi: 10.4161/epi.20445. [DOI] [PubMed] [Google Scholar]
- 8.Joseph CA, Maroney MJ. Cysteine dioxygenase: structure and mechanism. Chem Commun. 2007:3338–49. doi: 10.1039/b702158e. [DOI] [PubMed] [Google Scholar]
- 9.Schuller-Levis GB, Park E. Taurine: new implications for an old amino acid. FEMS Microbiol Lett. 2003;226:195–202. doi: 10.1016/S0378-1097(03)00611-6. [DOI] [PubMed] [Google Scholar]
- 10.Stipanuk MH, Ueki I, Dominy JE, Jr, Simmons CR, Hirschberger LL. Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino Acids. 2009;37:55–63. doi: 10.1007/s00726-008-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradley H, Gough A, Sokhi RS, Hassell A, Waring R, Emery P. Sulfate metabolism is abnormal in patients with rheumatoid arthritis. Confirmation by in vivo biochemical findings. J Rheumatol. 1994;21:1192–6. [PubMed] [Google Scholar]
- 12.Gordon C, Bradley H, Waring RH, Emery P. Abnormal sulphur oxidation in systemic lupus erythematosus. Lancet. 1992;339:25–6. doi: 10.1016/0140-6736(92)90144-r. [DOI] [PubMed] [Google Scholar]
- 13.Heafield MT, Fearn S, Steventon GB, Waring RH, Williams AC, Sturman SG. Plasma cysteine and sulphate levels in patients with motor neurone, Parkinson’s and Alzheimer’s disease. Neurosci Lett. 1990;110:216–20. doi: 10.1016/0304-3940(90)90814-p. [DOI] [PubMed] [Google Scholar]
- 14.Maschietto M, Piccoli FS, Costa CM, Camargo LP, Neves JI, Grundy PE, et al. Gene expression analysis of blastemal component reveals genes associated with relapse mechanism in Wilms tumour. Eur J Cancer. 2011;47:2715–22. doi: 10.1016/j.ejca.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Dietrich D, Krispin M, Dietrich J, Fassbender A, Lewin J, Harbeck N, et al. CDO1 promoter methylation is a biomarker for outcome prediction of anthracycline treated, estrogen receptor-positive, lymph node-positive breast cancer patients. BMC Cancer. 2010;10:247. doi: 10.1186/1471-2407-10-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, et al. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118:781–94. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 17.Dominy JE, Jr, Hwang J, Stipanuk MH. Overexpression of cysteine dioxygenase reduces intracellular cysteine and glutathione pools in HepG2/C3A cells. Am J Physiol Endocrinol Metab. 2007;293:E62–9. doi: 10.1152/ajpendo.00053.2007. [DOI] [PubMed] [Google Scholar]
- 18.Young TW, Mei FC, Yang G, Thompson-Lanza JA, Liu J, Cheng X. Activation of antioxidant pathways in ras-mediated oncogenic transformation of human surface ovarian epithelial cells revealed by functional proteomics and mass spectrometry. Cancer Res. 2004;64:4577–84. doi: 10.1158/0008-5472.CAN-04-0222. [DOI] [PubMed] [Google Scholar]
- 19.Ray G, Batra S, Shukla NK, Deo S, Raina V, Ashok S, et al. Lipid peroxidation, free radical production and antioxidant status in breast cancer. Breast Cancer Res Treat. 2000;59:163–70. doi: 10.1023/a:1006357330486. [DOI] [PubMed] [Google Scholar]
- 20.Oltra AM, Carbonell F, Tormos C, Iradi A, Saez GT. Antioxidant enzyme activities and the production of MDA and 8-oxo-dG in chronic lymphocytic leukemia. Free Radic Biol Med. 2001;30:1286–92. doi: 10.1016/s0891-5849(01)00521-4. [DOI] [PubMed] [Google Scholar]
- 21.Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–52. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Behrend L, Henderson G, Zwacka RM. Reactive oxygen species in oncogenic transformation. Biochem Soc Trans. 2003;31:1441–4. doi: 10.1042/bst0311441. [DOI] [PubMed] [Google Scholar]
- 23.Wu WS. The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. 2006;25:695–705. doi: 10.1007/s10555-006-9037-8. [DOI] [PubMed] [Google Scholar]
- 24.Pelicano H, Carney D, Huang P. ROS stress in cancer cells and therapeutic implications. Drug Resist Updat. 2004;7:97–110. doi: 10.1016/j.drup.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Fruehauf JP, Meyskens FL., Jr Reactive oxygen species: a breath of life or death? Clin Cancer Res. 2007;13:789–94. doi: 10.1158/1078-0432.CCR-06-2082. [DOI] [PubMed] [Google Scholar]
- 26.Schumacker PT. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell. 2006;10:175–6. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–91. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 28.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 29.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–8. [PubMed] [Google Scholar]
- 31.Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, et al. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012;40:4334–46. doi: 10.1093/nar/gks031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye S, Wu X, Wei L, Tang D, Sun P, Bartlam M, et al. An insight into the mechanism of human cysteine dioxygenase. Key roles of the thioether-bonded tyrosine-cysteine cofactor. J Biol Chem. 2007;282:3391–402. doi: 10.1074/jbc.M609337200. [DOI] [PubMed] [Google Scholar]
- 33.Griffith OW, Meister A. Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine) J Biol Chem. 1979;254:7558–60. [PubMed] [Google Scholar]
- 34.Mossman D, Scott RJ. Long term transcriptional reactivation of epigenetically silenced genes in colorectal cancer cells requires DNA hypomethylation and histone acetylation. PLoS ONE. 2011;6:e23127. doi: 10.1371/journal.pone.0023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- 36.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider BL, Kulesz-Martin M. Destructive cycles: the role of genomic instability and adaptation in carcinogenesis. Carcinogenesis. 2004;25:2033–44. doi: 10.1093/carcin/bgh204. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Sanchez G, Giuliani A. Cellular redox status regulates hypoxia inducible factor-1 activity. Role in tumour development. J Exp Clin Cancer Res. 2007;26:39–50. [PubMed] [Google Scholar]
- 39.Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, et al. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–86. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 40.De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–67. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Baseline characteristics of NB donors and BC patients
Supplementary Table 2. Statistical association between clinocopathologic characteristics of BC patients and DNA methylation frequency (%) of CDO1
Supplementary Table 3. Primer information