Abstract
Purpose
To investigate the accuracy and reproducibility of hepatic lipid measurements using 1H MRS with T2 relaxation correction, in comparison to measurements without correction.
Materials and Methods
Experiments were conducted in phantoms of varying lipid and iron-induced susceptibility to simulate fatty liver with variable T2. Single-voxel 1H MRS was conducted with multiple TE values, and lipid content (lipid%) was determined at each TE to assess accuracy and TE dependency. Concurrently, T2 and equilibrium values of water and lipid were determined separately, and T2 effects on the lipid% were corrected. A similar procedure was conducted in 12 human subjects to determine susceptibility effects on water and lipid MRS signals and lipid%. Multiple measurements were used to test reproducibility.
Results
The use of T2-correction was found to be more accurate than uncorrected lipid% in phantom samples (<10% error). Uncorrected lipid% error increased with increasing TE (>20% when TE >24ms) and with increasing susceptibility effect. In humans, while measurement repeatability was high for both corrected and uncorrected MRS, uncorrected lipid% was sensitive to acquisition TE, with 83.6% of all measurements significantly different than T2-corrected measures (p<0.05).
Conclusion
Separate T2-correction of water and lipid 1H MRS signals provides more accurate and consistent measurements of lipid%, in comparison to uncorrected estimations.
Keywords: Spectroscopy, Hepatic Lipid, T2-correction
Introduction
Non-invasive liver fat measurement with MRI and magnetic resonance spectroscopy (MRS) is becoming an important alternative to liver biopsy for diagnosis of non-alcoholic fatty-liver disease (NAFLD) (1-6). NAFLD is now considered amongst the most concerning liver conditions in children and adults in developed and developing nations (7), and may lead to subsequent development of inflammation and fibrosis in a subset of individuals. With known inherent ambiguity of fat quantification on opposed-phase MRI (8), there is impetus to investigate the continued potential of MRS techniques. Single-voxel 1H MRS provides localized investigation of water and lipid species in liver, even when lipid amounts are low and difficult to distinguish above noise on MRI. An estimation of percent lipid content (lipid%) using MRS can be measured by relating the magnitude of the water and lipid spectra.
Recent investigations of hepatic MRS have transcended initial reports documenting its feasibility as a useful examination method of lipid content (1, 9), and evolved into a technique often used as a standard of reference against newly-developed advanced MRI acquisitions (4, 8, 10). Even though these latter studies report significant correlation with MRS, there are several inherent factors affecting the accuracy and reproducibility of lipid% measurements with MRS, such as field homogeneity, partial volume effects, physiologic motion, and signal decay due to relaxation. While the former issues can be ameliorated through optimized hardware or acquisition methods, signal decay from T1 and T2 are inherent to MR signals, and cause measurement inconsistencies, especially if true proton density information is desired either in magnitude, or in relative abundance, as with hepatic lipid% measures.
Recently, advanced MR imaging methods have been developed and investigated to address susceptibility effects in lipid quantification (11, 12). But even though some forms of MRS signal correction have been proposed (3, 9, 13, 14), few studies to date have provided quantitative and comparative insight into the role of T1 and T2 relaxation on the accuracy and precision of liver lipid% measurements using MR spectroscopy The absolute magnitude of water and lipid spectra within a voxel can be underestimated by both T1 saturation and T2 decay. Since TE is a finite positive value in Point-Resolved Spectroscopy (PRESS) and Stimulate Echo Acquisition Mode (STEAM), there will be inherent signal loss at the point of acquisition that is unique to both water and lipid protons. These factors must be considered when implementing lipid% calculations, especially if water and lipid T1 and T2 are very different from each other. In practice, T1 and T2 values are unknown in vivo; the MRS lipid% measurements, therefore, must incorporate correction methods to achieve consistent and accurate results.
With acquisition of hepatic MRS data normally achieved using TR > 2000ms, T1 saturation effects are lessened given global liver T1 below 700ms (15, 16, 17). However, individual spectral T1, especially the water peak, may differ experimentally and must be investigated. The T2 relaxation of liver is approximately 46ms at 1.5T (17), but is known to vary across individuals, primarily due to the presence of physiologically variable liver iron content. This unknown variability may drastically affect MRS measures of water and lipid peaks disproportionately, especially at standardized TEs greater than 20ms. In this regard, the influence of T2 decay on precise and accurate lipid% measures outweighs T1 effects.
The focus of this study is to investigate and demonstrate the influence of tissue relaxation effects on resultant lipid% measurements using MRS. This analysis is performed by comparing routine MRS lipid% measures at specific TE values against T2-corrected data in known lipid phantoms and selected human subjects.
Methods
Liver Phantoms
To initially test the influence of T1 and T2 relaxation on water and lipid spectral peaks, and subsequent lipid% measurements, 8 individual liver phantoms with known lipid content were constructed. The phantoms were comprised of water-lipid emulsions having two proportions of lipid (10% and 30% vegetable oil by volume), and doped with increasing amounts of iron (0, 0.1, 0.3, 0.5mM Ferridex, Berlex, NJ) to induce T2 variability. Tissue water space was simulated with 2% agar-water gels. Lipid micelles were produced by the addition of lecithin at a concentration of 2% wt/vol. Each mixture was contained within separate 200ml screw-top tubes.
Subjects
All in vivo experiments were HIPPA compliant and conducted with Internal Review Board approval, and each subject provided informed consent. A total of 12 subjects participated in this investigation. Four subjects (4 male; range: 26-31yrs) were volunteers and had no prior diagnostic history for fatty liver disease, but showed indication of abnormal lipid content from a separate study using standard opposed phase MR imaging. Eight subjects (7 males; 13.0 ± 2.4yrs; range: 11-17 yrs) were pediatric patients recruited in part of a larger cross-discipline NAFLD study being conducted at our institution.
MRS Acquisition
All experiments were performed on a Siemens 1.5T Avanto system with dedicated anterior-posterior phased-array surface coils. Planning for MRS was performed with the aid of 3 orthogonal, single-shot T2-weighted images (20 slices, 8mm thick, 0.8mm gap, TR/TE = 1500/86ms), and cross-reference lines as position indicators. MRS acquisition was performed with STEAM due to it ability for short TE. For T1 investigation, 3 phantoms (10% lipid/0.1mM iron, 10%/0.3mM, 30%/0.1mM) and two subjects were analyzed with STEAM using TE = 20ms, mixing time (TM) =10ms, and TR varied in 1000ms increments from 500ms to 7500ms in separate acquisitions. Other parameters were 30×30×30mm voxel, 2 preparation scans, 4 averages, 512 points, and bandwidth = 1200Hz/px. For longer TR acquisitions in vivo, averages were reduced to 2-3 for breath holding purposes.
For lipid% MRS measurements, phantom spectroscopy was performed with single-average (“single-shot”) STEAM with parameters as listed above, except with TR = 3000ms, 0 preparations, 1024 points, and TE varied from 12 to 72ms in 5 steps (12, 24, 36, 48 and 72ms). Each TE was collected in a separate acquisition. In vivo lipid% MRS was performed with STEAM in “single-shot” mode, and had equivalent voxel size (30×30×30mm), acquisition points (1024), preparation scans (0) and bandwidth (1200 Hz/px). Voxel placement was usually in the right lobe, with care taken to avoid large vessel contamination on the orthogonal scout images. The TE array for STEAM was as noted above for phantoms, with each TE acquired in separate, short breath hold acquisition of 3 seconds. Three repeated MRS measures of each TE were acquired in similar voxel positions to assess precision.
Data Analysis
All phantom MRS spectra were exported off-line for analysis using MATLAB software (MATLAB 7.04.365; The Mathworks, Natick, USA). Signal integrals were quantified for water and lipid at each echo time following Fourier transform, phase correction, and low-pass filtering with a cutoff of 240Hz. Baseline correction was implemented to normalize measurements. The algorithm performed signal integration of water (H2O at 4.7 ppm) and lipids (CH2 and CH3 at 1.3 ppm and 0.95 ppm, respectively) for each echo time. Human MRS spectra was exported off-line to a user-independent fitting routine package (LCModel version 6.1-A). The software was specially configured to fully analyze the spectra knowing that there were only lipid (and water) and possibly choline signals, plus possible signals in the 3.4–3.8 ppm region (sometimes attributed to glycogen and other metabolites). Commercial software was implemented for the in vivo data to demonstrate practical utility of fitting and correction algorithms for human MRS data. For both phantom and human data, the resultant signal integral values at each echo time were recorded for each sample, and represented as a time series of relative water and lipid content.
Water and lipid relaxation were quantified with a Matlab program. T1 was evaluated for water and lipid independently by means of a two-parameter least-squares curve fitting algorithm, with TR as the independent variable, and spectra integral values as the dependent variable. With TE assumed fixed, the fitting equation was reduced to S = S0*(1-exp(-R1*TR)), with S0 equivalent to the equilibrium spectral value (weighted by fixed T2 decay), and R1 representing the T1 relaxation rate (1/T1). Goodness-of-fit was expressed with an r-squared value.
T2 values of water and lipids were calculated by fitting the series of MRS echo data to a mono-exponential curve (S = S0*exp(-R2*TE)). T1 contributions were assumed absent due to the “single-shot” nature of the employed MRS technique. S0 (the equilibrium value) and R2 (1/T2) were estimated for all experiments, with goodness-of-fit represented by r-squared. Lipid content was calculated by Lipid% = SFat/(SFat + SWater)*100 at each measured TE, as well as “TE=0” using the calculated S0 values for water and lipid. Lipid% measurements using S0 represented the T2-corrected lipid%.
For phantoms lipid% measurements, the mean and standard deviation (mean ± SD) were calculated for 10% and 30% samples as a function of iron content (to assess precision and iron dependency) and TE value (to assess accuracy and TE dependency). This analysis was also performed for T2-corrected lipid% as a function of iron content. Accuracy as a function of TE and iron content in phantom experiments were represented by a percent error: (True Value – Measured)/(True Value). The resultant data were compared among individual groups using a two-tailed Student's t-test with p=0.05.
For in vivo comparative analysis, repeated lipid% measures were averaged, and uncorrected values at each TE were compared to T2-corrected values using a paired two-tailed Student's t-test with p=0.05. Similarly, the cumulative average lipid% from TE=12 to 72ms was compared to T2-corrected lipid%. As a measure of precision, the coefficient of variance (CV) was defined as: CV = SD/Avg, for both intra-subject repeated lipid% measurements and across the TE series. A t-test was used to determine significant variabilities between T2-correction and uncorrected lipid% measurements (p=0.05)
Results
The initial experiment investigating longitudinal relaxation revealed water and lipid T1 values of 821.7 and 680.3ms (rsq=0.98), respectively, for the “baseline” 10%/0.1mM lipid-iron phantom. For higher amounts of lipid and iron content, the measured T1 decreased measurably. An iron content increase to 0.3mM produced T1 of 340.8 (-58.5% from baseline) and 243.2ms (-64.2%) for water and lipid, respectively, while a lipid content increase to 30% from baseline produced a T1 of 591.7 (-28.0%) and 185.2ms (-72.8%). No observable trends between T1 and iron/lipid content could be realized with this data apart from inverse proportionality. One in vivo subject did not reveal significant amounts of lipid for useful T1 quantification; however, estimation of water spectral T1 was found to be 747.4ms (rsq=0.97). The second volunteer had measured T1 of 645.1 and 485.0ms for water and lipid, respectively. From the longer T1 of water spectra, the amount of T1 recovery between repetitions was estimated from Bloch equations to be 91% (for T1 = 821.7ms) and 93% (747.4ms) for short-echo MRS sequences of TR = 2000ms, and 97% and 98% for TR = 3000ms.
The spectra for water and lipid were sufficiently visible at all prescribed TEs using STEAM in single-shot mode. Figure 1a shows example MRS spectra of water and lipid from the 30%/0.1mM phantom, while Fig. 1b shows representative MRS spectra from one subject. All T2 curve fitting and calculation was achieved with high goodness-of-fit (rsq>0.95), and the results of phantom R2 calculations are given in Table 1. A global linear relationship was found between R2 of water and iron content, [Fe] (R2wat = 247.3 [Fe] + 20.0 s-1; rsq: 0.97), with no observable dependence on lipid content. For lipid R2, the slope of the linear regression (r2, relaxivity) is reduced (30% lipid: R2fat = 35.3 [Fe] + 20.0 s-1; rsq: 0.98); however, this R2fat trend exhibited some dependence on lipid content, with 10% lipid samples revealing minor iron dependence reflected by poor linear regression (10% lipid: R2fat = 8.2 [Fe] + 20.2s-1; rsq: 0.07). From this data, the r2 relaxivity of [Fe] in water is much greater than in lipid: r2wat/r2fat = 7.0. Furthermore, the baseline R2 (at 0mM iron) was found to be essentially equivalent for water and lipid (R20wat ≈ R20fat = 16.3 ± 4.0s-1).
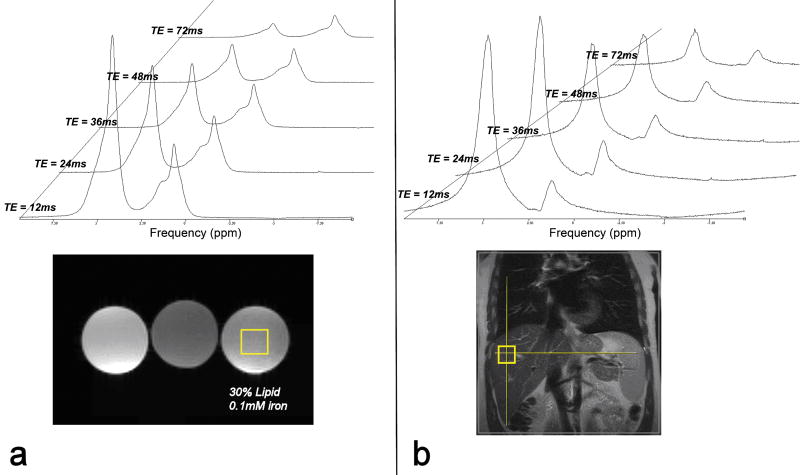
Figure 1.
Spectral peaks of water and lipid obtained from single-voxel, single-average STEAM in (a) a lipid-iron phantom and (b) one subject. Each MRS acquisition was obtained separately using TEs ranging from 12ms to 72ms. The relative ratio of lipid-to-water changes as TE increases, reflecting a TE-dependency of measured lipid%.
Table 1. Measured Transverse Relaxation Rates in Phantoms.
| Lipid (%) | Iron (mM) | R2 Relaxation Rate (s-1) | |
|---|---|---|---|
|
| |||
| Water | Lipid | ||
| 10% | 0 | 11.8 ± 0.5 | 14.0 ± 1.5 |
| 0.1 | 43.3 ± 0.3 | 30.4 ± 2.5 | |
| 0.3 | 93.6 ± 2.4 | 19.8 ± 2.7 | |
| 0.5 | 139.7 ± 4.0 | 24.3 ± 0.5 | |
|
| |||
| 30% | 0 | 20.1 ± 2.6 | 19.3 ± 1.5 |
| 0.1 | 43.1 ± 1.0 | 23.6 ± 0.1 | |
| 0.3 | 120.6 ± 8.3 | 32.2 ± 0.3 | |
| 0.5 | 132.5 ± 9.6 | 36.7 ± 0.1 | |
The resultant T2-corrected lipid%, along with all the measured lipid% at each TE are represented in Table 2 for the 10% and 30% phantoms with varying iron content. Overall, in terms of accuracy and precision, it was found that T2-correction of water and lipid MRS spectra for lipid% calculation produced low standard deviations and percent errors <15% (range: 1.0% to 14.5%) for 10 and 30% phantoms with varying iron content, whereas percent errors of uncorrected lipid% ranged greatly between <1% to >800%. Iron content was the primary contributor to inaccurate lipid% measurements without T2-correction, with the amount 0.3mM resulting in accuracy errors consistently over 75%, regardless of TE. For iron content of 0.1mM, accuracy was dependent on TE, with the lowest TE values producing the lowest percent errors. This dependency is reflected in the precision measurements for 0.1mM iron; standard deviation among the series of TE values (12 to 72ms) was found to be 6.3% (p<0.05) and 11.4% (p<0.01) for 10 and 30% phantoms, respectively. For 0mM iron content, there was no significant improvement in accuracy using T2-correction for any prescribed TE. However, T2-correction was insensitive to all degrees of iron content, and produced consistent results of 9.0 ± 0.3 (10% error) and 31.3 ± 3.4 (4.3%), which were significantly more precise than uncorrected lipid% using TE>12ms (p<0.05).
Table 2. Lipid% Measurements in Phantoms With and Without T2-correction.
| Lip % | Iron mM | Lipid% T2-cor | Lipid% TE=12ms | Lipid% TE=24ms | Lipid% TE=36ms | Lipid% TE=48ms | Lipid% TE=72ms | Avg Lipid% |
|---|---|---|---|---|---|---|---|---|
| 10% | 0 | 8.7 | 8.1 | 8.6 | 7.7 | 7.4 | 6.9 | 7.8± 0.6a |
| 0.1 | 8.7 | 10.1 | 12.2 | 13.4 | 16.2 | 26.2 | 15.6 ±6.3 | |
| 0.3 | 9.0 | 18.0 | 37.1 | 56.7 | 74.0 | 91.4 | 55.4±29.0a | |
| 0.5 | 9.3 | 28.8 | 61.2 | 81.7 | 88.2 | 93.4 | 70.6±26.4a | |
|
| ||||||||
| Avg (SD) | 9.0 ± 0.3 | 16.3 ± 9.4 | 29.8 ±24.5 | 39.9± 35.4 | 46.5± 40.6 | 54.5±44.7 | ||
|
| ||||||||
| 30% | 0 | 29.7 | 29.2 | 28.7 | 28.9 | 30.1 | 31.8 | 29.7 ±1.2 |
| 0.1 | 27.2 | 32.2 | 38.6 | 43.5 | 49.2 | 62.2 | 45.1±11.4a | |
| 0.3 | 34.4 | 57.4 | 80.6 | 90.6 | 94.7 | 97.3 | 84.1±16.2a | |
| 0.5 | 33.8 | 64.6 | 83.2 | 87.6 | 89.8 | 90.4 | 83.2±10.7a | |
|
| ||||||||
| Avg (SD) | 31.3± 3.4 | 45.8± 17.8 | 55.0 ±25.2 | 60.2 ±28.8 | 63.8 ±29.5 | 68.6± 28.5 | ||
p<0.05 vs T2-cor Lip%
One subject did not reveal detectable amounts of lipid on MRS, leaving 11 subjects for analysis. T2-corrected in vivo lipid% measures are given in Table 3, along with standard deviation representing measurement precision of 3 repeated acquisitions. Also shown are representative uncorrected lipid% measurements for TE=12, 24 and 48ms. The T2-corrected lipid% was significantly different (p<0.05) than uncorrected lipid% in 83.6% of all group-wise comparisons (46/55), while showing significant difference in 90.9% of comparisons (20/22) when uncorrected lipid% TE > 48ms. The dependency of lipid% measurement on TE is shown in the last column of Table 3, which is the average and deviation of all TE values (12 to 72ms). It is also depicted in Figure 2, which shows exponential decay of water and lipid in one subject, and subsequent lipid% calculations are each TE. Overall, in comparison to T2-corrected lipid%, uncorrected lipid% was significantly different (p<0.05) in 7 of 11 subjects. In two cases where no significant difference was found, the estimated lipid% was less than 10%. The overall average coefficient of variance for lipid% measures using TE=12 to 72ms was 22.3 ± 5.1%, signifying a lower degree of precision compared to T2-corrected CV, 7.5 ± 12.8% (p<0.05).
Table 3. Lipid% Measurements in Humans With and Without T2-Correction.
| Subj** | Lipid% T2-Cor | Lipid% TE=12ms | Lipid% TE=24ms | Lipid% TE=48ms | Avg* Lipid% |
|---|---|---|---|---|---|
| V1 | 13.8 ± 1.9 | 16.9 ± 0.9 | 18.9 ± 1.1a | 25.0± 0.8a | 23.3 ± 6.5a |
| V2 | 5.7 ± 0.2 | 6.5 ± 0.2a | 5.8 ± 0.6 | 8.2 ± 0.1a | 7.8 ± 2.5 |
| V3 | 3.5 ± 1.6 | 3.7 ± 1.4 | 2.7 ± 1.0 | 3.6 ±0.7 | 3.6 ± 0.8 |
| S4 | 23.5 ± 0.7 | 25.7 ± 0.4a | 26.5 ± 0.1a | 31.8 ±0.4a | 32.0 ± 8.5 |
| S5 | 18.2 ± 0.4 | 19.5 ± 0.3a | 18.8 ± 0.4 | 21.4 ±0.7a | 22.1 ± 4.5 |
| S6 | 31.6 ± 1.0 | 35.7 ± 1.0a | 38.0 ± 0.9a | 46.4 ± 1.1a | 43.4 ± 7.7a |
| S7 | 18.3 ± 0.3 | 21.1 ± 0.2a | 23.7 ± 0.4a | 29.5 ± 0.3a | 27.7 ± 6.5a |
| S8 | 20.9 ± 0.5 | 23.8 ± 0.2a | 25.5 ± 0.7a | 31.9 ±0.7a | 30.0 ± 6.5a |
| S9 | 19.6 ± 0.6 | 21.7 ± 0.6a | 22.5 ± 1.4a | 27.0 ±1.6a | 25.9 ± 4.8a |
| S10 | 18.5 ± 0.6 | 20.1 ± 0.6a | 21.2 ± 0.6a | 23.8± 0.2a | 23.1 ± 3.3a |
| S11 | 26.7 ± 0.5 | 30.3 ± 0.5a | 33.0 ± 0.1a | 40.9± 0.4a | 38.2 ± 8.0a |
Mean Lip% for TE=12 to 72ms.
V=volunteer; S=patient subject
p<0.05 vs. T2-cor Lip%
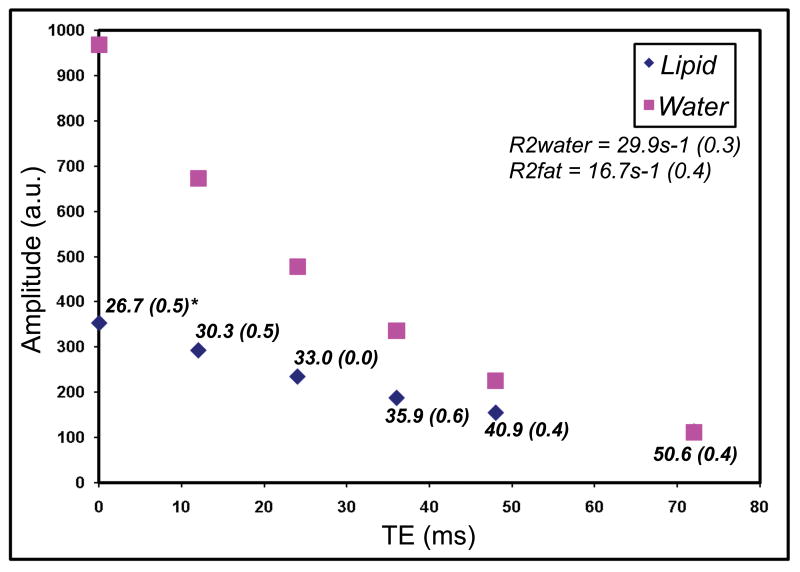
Figure 2.
Exponential T2 decay of water and lipid in one patient (subject #S11) is shown. Each point represents the integral amplitude of the respective spectrum at each TE. In addition, the calculated lipid% is given, with the T2-corrected measurement in bold script (TE=0). There is significant dependence of lipid% on the acquisition TE, as a result of R2 differences between water and lipid.
Measurement precision of 3 repeated MRS acquisitions with the prescribed TE series was sufficiently high in all subjects for both T2-corrected and uncorrected lipid%. The CV was less than 10% in all cases except volunteer #3.
Discussion
This study investigated the significance of T1 and T2 relaxation effects on hepatic lipid measures using MRS. It was found that for TR ≥ 3000ms, T1 recovery approaches equilibrium for MRS signal constituents (>95% recovery), while induced T2 shortening in phantoms demonstrates measurable variation on resultant lipid% calculations. The high accuracy of T2-corrected lipid% was revealed in phantoms, which gave consistent estimates despite variation in R2. Uncorrected lipid% measures, however, were found to be highly dependent on TE in both phantom and human subjects, with global coefficients of variation as high as 50%. With these described sensitivities, MRS lipid% measurement that incorporates T2-correction represents a more robust and consistently accurate analysis method compared to traditional single-echo acquisitions.
Several previous investigations of hepatic lipid measurement with MRS presented results accounting for T2 decay through serial TE measures (3, 9, 13, 14). However, these reports do not present specific data quantifying the difference with uncorrected lipid%, as described in this study. The use of STEAM or PRESS for hepatic lipid% measurement may be particularly sensitive to error because: a) TE is finite and on the order of T2 of liver; and b) T2 of water and lipid may not be equivalent in vivo. There are previous examinations of hepatic lipid% using MRS sequences with TE ≥ 30ms (8, 13, 18). Even though these studies demonstrated high measurement correlation against alternative measures of lipid, we are not able to ascertain the magnitude of T2 effects in these reports. It may be that the need for T2-correction was minor or absent in some or all of the subjects studied in these reports and/or the alternative measures of lipid had intrinsic variability obscuring T2-effects on the MRS results. In another report, Machann et al incorporated MRS as the reference standard, using a STEAM sequence with TE=10ms (without correction) for correlative studies with MRI opposed-phase imaging (4). However, the scope of their study did not lend to the discussion of either TE selection or T2-correction using MRS. Presumably, a TE=10ms used by this group would lessen the effects of T2 on lipid% measurements and, according to our findings, would yield reasonably accurate results for smaller T2 susceptibility effects.
In certain scenarios, however, these sequences will be prone to significant error and uncertainty. No study, to our knowledge, has systematically documented the sensitivity of MRS with TE > 0 to inherent T2 differences between water and lipid, as presented here. Our phantom studies, for example, at TE=24ms, produced accuracy errors ranging from -10.8% to above 500%, showing a significant sensitivity to T2 effects over a physiological range of iron. Similarly, the in vivo results, which were acquired in a setting of unknown T2 effects, revealed MRS lipid% at TE = 24 to be consistently different and higher than T2-corrected values.
It is well known that, depending on the T2 of liver water and lipid, the MRS signal for each constituent will be depressed from its equilibrium amount at a given TE. Apart from user-defined signal-to-noise requirements, the degree of signal loss at a particular TE is not crucial to the experiment per se; however, the subsequent lipid% calculation, which relies on the ratio between lipid and water amounts, can be significantly altered if the T2 values are very different. The lipid% equation can be expressed in terms of R2:
| [Eq. 1] |
With TE fixed, the independent variables are R2lip and R2wat. If these values are equivalent, as approximately found for 0mM iron (Table 1), lipid% calculations are unaffected by T2 relaxation, and any practical TE can be used. This is also proven from the phantom results (Table 2), which shows that T2-corrected lipid% is not significantly different from uncorrected results, regardless of TE. Even previous hepatic lipid% investigations have shown similar R2 for water and lipid (1, 13, 14, 19). However, for iron concentrations above 0mM, R2wat increases dramatically. This may be the result of iron localizing preferentially in the water compartment, causing greater susceptibility effects on water protons. The consequence is a disproportionate faster loss in liver water signal relative to lipid signal. T2-correction in this scenario would be very important. From the relationship between R2 and iron content (Table 1), the relative r2 relaxivity of iron favors liver water nearly ten-fold (r2wat/r2fat = 7.0). Since R2-vs-iron is linear and using R20fat ≈ R20wat, Eq 1 can be reduced to:
| [Eq 2] |
where A = r2fat · [Fe]. Thus, depending on iron content, the effective signal decay of water far exceeds lipid for a given TE, making the water spectra the primary contributor to accurate lipid% measurements. This was evident with both the phantom and human MRS data.
MRS lipid% measurement is also sensitive to differences in water and lipid T1 values. But this effect is dependent on TR, a value that typically exceeds 2000ms (2, 4, 8, 13, 14, 18, 20), which normally negates significant T1 effects. The longest T1 measured in this study (821.7ms) can be shown to recovery approximately 91% and 97% after 2000 and 3000ms, respectively. Since tissue water is associated with these longer T1, water spectral amount will be saturated successively over multiple TRs according to (21):
| [Eq. 3] |
which is valid for α = 90 and TR ≫ TE and TM. Mz represents the amount of longitudinal signal available for acquisition per TR. Hence, accuracy of lipid% is further improved with knowledge of water and lipid T1 values, especially for short TR. In contrast to Mz recovery normally exceeding 90%, a standard TE of 30ms produces magnetization less than 65% of equilibrium (using T2 < 70ms). Therefore, T2 effects typically outweigh T1 effects in almost all MRS applications. In this study, MRS acquisition was performed in single-shot mode, without dependence on TR. It has been shown with this study that this practice is a feasible and accurate option for breath hold MRS of large spectral peaks such as water and lipid.
A key benefit for utilizing a T2-correction strategy in MRS is improved accuracy and precision over uncorrected results. In a clinical setting, where T2 effects are unknown, it is vital to confidently produce consistent results. This may have particular implications in individuals with low lipid% levels, where significant T2 water spectral decay may falsely augment measured lipid% on traditional, uncorrected MRS sequences. However, it is noteworthy that low lipid% measured in this study was generally associated with less precision (Table 3). This is likely attributable to the low lipid% levels in these cases, as well as the use of single-shot MRS mode, both of which exacerbate baseline noise. The use of single-shot MRS mode was motivated by the need to acquire multiple TE in an expedited fashion. For higher lipid%, repeated measurements with the “single-shot” configuration produced excellent precision in all cases.
T2-correction of hepatic lipid measurement requires either prior assumption of water and lipid T2, or acquisition of spectra at multiple TE for T2 quantification on an individual basis. The latter approach is a more concise, since T2 can vary among individuals and between water and lipid. At minimum, two acquisitions are needed to fit a T2 curve, and optimization strategies can be developed around an understanding of expected constituent T2 values. If limited echoes are used in T2 quantification, adequate sampling of early T2 decay is more important, and includes using the shortest possible initial TE, and additional TEs spaced within the expected range of T2 (22). More echoes, however, reduce the uncertainty of T2 determination, since TEs can be allocated over the full duration of T2 decay. The current study used 5 TE values evenly spaced between 12 and 72ms, which effectively encompass the expected ranges of liver T2. Multiple-echo spectroscopy has been conducted in the brain for T2-correction of cerebral metabolites (23), but feasibility in the liver is limited by scan time and respiration compensation aspects. Individual T2 determination is also limited by post-processing requirements; one would need to implement a streamlined strategy to process and correct T2 information for the technique to be clinically viable. These steps were beyond the scope of this study, and will be the focus of future investigations.
In conclusion, 1H MRS technique is a known sensitive measure of hepatic lipid content, and has been shown to be feasible in vivo. For accurate lipid% measurement in a clinical setting, our study shows that a concise correction of relaxation effects is required.
References
- 1.Longo R, Ricci C, Masutti F, Vidimari R, Crocé LS, Bercich L, Tiribelli C, Dalla Palma L. Fatty infiltration of the liver. Quantification by 1H localized magnetic resonance spectroscopy and comparison with computed tomography. Invest Radiol. 1993;28:297–302. [PubMed] [Google Scholar]
- 2.Longo R, Pollesello P, Ricci C, et al. Proton MR spectroscopy in quantitative in vivo determination of fat content in human liver steatosis. J Magn Reson Imaging. 1995;5:281–285. doi: 10.1002/jmri.1880050311. [DOI] [PubMed] [Google Scholar]
- 3.Thomas EL, Hamilton G, Patel N, et al. Hepatic triglyceride content and its relation to body adiposity: a magnetic resonance imaging and proton magnetic resonance spectroscopy study. Gut. 2005;54:122–127. doi: 10.1136/gut.2003.036566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machann J, Thamer C, Schnoedt B, et al. Hepatic lipid accumulation in healthy subjects: a comparative study using spectral fat-selective MRI and volume-localized 1H-MR spectroscopy. Magn Reson Med. 2006;55:913–917. doi: 10.1002/mrm.20825. [DOI] [PubMed] [Google Scholar]
- 5.Schuchmann S, Weigel C, Albrecht L, et al. Non-invasive quantification of hepatic fat fraction by fast 1.0, 1.5 and 3.0 T MR imaging. Eur J Radiol. 2007;62:416–422. doi: 10.1016/j.ejrad.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Hussain HK, Chenevert TL, Londy FJ. Hepatic fat fraction: MR imaging for quantitative measurement and display--early experience. Radiology. 2005;237:1048–1055. doi: 10.1148/radiol.2373041639. [DOI] [PubMed] [Google Scholar]
- 7.Schwimmer JB, Deutsch R, Kahen T, Lavine JE, Stanley C, Behling C. Prevalence of fatty liver in children and adolescents. Pediatrics. 2006;118:1388–1393. doi: 10.1542/peds.2006-1212. [DOI] [PubMed] [Google Scholar]
- 8.Chang JS, Taouli B, Salibi N, Hecht EM, Chin DG, Lee VS. Opposed-phase MRI for fat quantification in fat-water phantoms with 1H MR spectroscopy to resolve ambiguity of fat or water dominance. Am J Roentgenol. 2006;187:W103–106. doi: 10.2214/AJR.05.0695. [DOI] [PubMed] [Google Scholar]
- 9.Thomsen C, Becker U, Winkler K, Christoffersen P, Jensen M, Henriksen O. Quantification of liver fat using magnetic resonance spectroscopy. Magn Reson Imaging. 1994;12:487–495. doi: 10.1016/0730-725x(94)92543-7. [DOI] [PubMed] [Google Scholar]
- 10.Chan DC, Watts GF, Ng TW, Hua J, Song S, Barrett PH. Measurement of liver fat by magnetic resonance imaging: Relationships with body fat distribution, insulin sensitivity and plasma lipids in healthy men. Diabetes Obes Metab. 2006;8:698–702. doi: 10.1111/j.1463-1326.2005.00543.x. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, McKenzie CA, Shimakawa A, et al. Multiecho reconstruction for simultaneous water-fat decomposition and T2* estimation. J Magn Reson Imaging. 2007;26:1153–1161. doi: 10.1002/jmri.21090. [DOI] [PubMed] [Google Scholar]
- 12.O'Regan DP, Callaghan MF, Wylezinska-Arridge M, et al. Liver fat content and T2*: simultaneous measurement by using breath-hold multiecho MR imaging at 3.0 T--feasibility. Radiology. 2008;247:550–557. doi: 10.1148/radiol.2472070880. [DOI] [PubMed] [Google Scholar]
- 13.Garbow JR, Lin X, Sakata N, Chen Z, Koh D, Schonfeld G. In vivo MRS measurement of liver lipid levels in mice. J Lipid Res. 2004;45:1364–1371. doi: 10.1194/jlr.D400001-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Szczepaniak LS, Nurenberg P, Leonard D, et al. Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab. 2005;288:E462–468. doi: 10.1152/ajpendo.00064.2004. [DOI] [PubMed] [Google Scholar]
- 15.Bluml S, Schad LR, Stepanow B, Lorenz WJ. Spin-lattice relaxation time measurement by means of a TurboFLASH technique. Magn Reson Med. 1993;30:289–295. doi: 10.1002/mrm.1910300304. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg MA, Hahn PF, Saini S, et al. Value of T1 and T2 relaxation times from echoplanar MR imaging in the characterization of focal hepatic lesions. Am J Roentgenol. 1993;160:1011–1017. doi: 10.2214/ajr.160.5.8470568. [DOI] [PubMed] [Google Scholar]
- 17.de Bazelaire CM, Duhamel GD, Rofsky NM, Alsop DC. MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology. 2004;230:652–659. doi: 10.1148/radiol.2303021331. [DOI] [PubMed] [Google Scholar]
- 18.Bernard CP, Liney GP, Manton DJ, Turnbull LW, Langton CM. Comparison of fat quantification methods: a phantom study at 3.0T. J Magn Reson Imaging. 2008;27:192–197. doi: 10.1002/jmri.21201. [DOI] [PubMed] [Google Scholar]
- 19.Heiken JP, Lee JK, Dixon WT. Fatty infiltration of the liver: evaluation by proton spectroscopic imaging. Radiology. 1985;157:707–710. doi: 10.1148/radiology.157.3.2997837. [DOI] [PubMed] [Google Scholar]
- 20.Szczepaniak LS, Babcock EE, Schick F, et al. Measurement of intracellular triglyceride stores by H spectroscopy: validation in vivo. Am J Physiol. 1999;276:E977–989. doi: 10.1152/ajpendo.1999.276.5.E977. [DOI] [PubMed] [Google Scholar]
- 21.Haacke EM, Brown RW, Thompson MR, Venkatesan R. Magnetic resonance imaging: physical principles and sequence design. 1st. New York: John Wiley and Sons Inc; 1999. p. 913. [Google Scholar]
- 22.Jackson JA, Schneiders NJ, Ford JJ, Bryan RN. Improvements in the clinical utility of calculated T2 images of the human brain. Magn Reson Imaging. 1985;3:131–143. doi: 10.1016/0730-725x(85)90250-4. [DOI] [PubMed] [Google Scholar]
- 23.Isobe T, Matsumura A, Anno I, et al. Quantification of cerebral metabolites in glioma patients with proton MR spectroscopy using T2 relaxation time correction. Magn Reson Imaging. 2002;20:343–349. doi: 10.1016/s0730-725x(02)00500-3. [DOI] [PubMed] [Google Scholar]