Abstract
The eukaryotic RNA exosome is an essential multi-subunit ribonuclease complex that contributes to the degradation or processing of nearly every class of RNA in both the nucleus and cytoplasm. Its nine-subunit core shares structural similarity to phosphorolytic exoribonucleases such as bacterial PNPase. PNPase and the RNA exosome core feature a central channel that can accommodate single stranded RNA although unlike PNPase, the RNA exosome core is devoid of ribonuclease activity. Instead, the core associates with Rrp44, an endoribonuclease and processive 3′→5′ exoribonuclease, and Rrp6, a distributive 3′→5′ exoribonuclease. Recent biochemical and structural studies suggest that the exosome core is essential because it coordinates Rrp44 and Rrp6 recruitment, RNA can pass through the central channel, and the association with the core modulates Rrp44 and Rrp6 activities.
Introduction
The RNA exosome is a ubiquitous endo- and 3′→5′ exoribonuclease in eukaryotic cells that collaborates with multiple co-factors for processing, quality control and degradation of virtually all classes of RNA. 3′→5′ RNA decay pathways are conserved in all kingdoms of life and perform a multitude of tasks including regulating the abundance of RNAs, eliminating dysfunctional or mis-folded RNAs, and processing of precursor RNAs to their mature form [1] [2] [3]. Three major enzyme classes catalyze 3′→5′ exoribonuclease activity in bacteria, archaea, and eukaryotes. One catalyzes processive hydrolytic RNA decay: (bacterial RNase II & RNase R and eukaryotic Rrp44); another catalyzes distributive hydrolytic RNA decay (bacterial RNase D and eukaryotic Rrp6), and one catalyzes processive phosphorolytic exoribonuclease activity (bacterial and mitochondrial PNPase and the archaeal exosome).
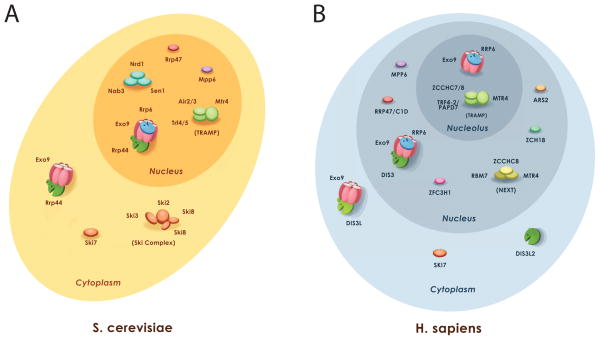
The eukaryotic RNA exosome core (Exo9) is similar in architecture to PNPase but it lacks phosphorolytic activity and has instead evolved to interact with and regulate the RNase activities of Rrp44 and Rrp6 [4,5]. The eukaryotic RNA exosome is observed in two main forms: a cytoplasmic RNA exosome that includes the nine-subunit core and Rrp44 (Exo1044) and a nuclear RNA exosome that includes Exo9, Rrp44 and Rrp6 (Exo1144/6) [5–9]. An additional nucleolar form has been hypothesized based on cellular co-localization studies in human cells that contains Exo9 and Rrp6 [9]. Each of these exosome complexes interacts with an array of co-factors to process or degrade different RNA substrates (Figure 1).
Figure 1. Exosome function in the eukaryotic cell.
A) Budding yeast. Nuclear and cytoplasmic forms of the exosome have been detected that use the exosome core (Exo9). Co-factors and co-factor complexes that are either functionally or directly associated with the S. cerevisiae exosome are depicted in either the cytoplasm (Exo10) or nucleus (Exo11). B) Human. Evidence for three different forms of the exosome existed: cytoplasmic, nuclear, and nucleolar. Each associates with different classes of co-factors. Figure prepared by Abigail C. Wasmuth.
The exosome core
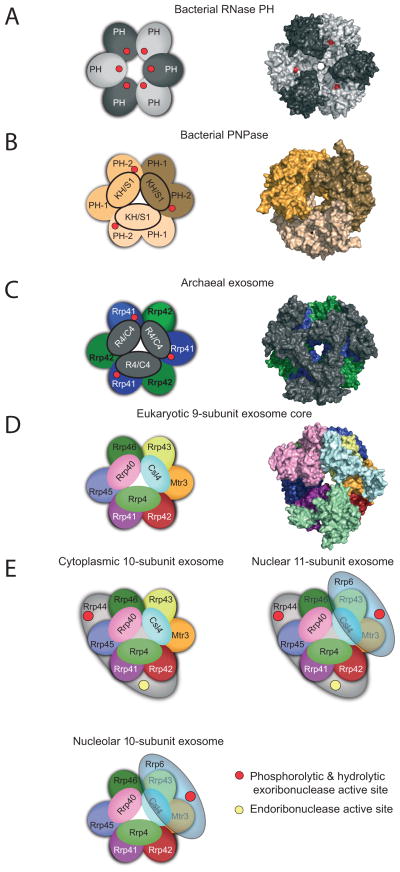
The global architecture of the RNA exosome core is conserved throughout prokaryotic, archaeal, and lower and higher eukaryotic phylogeny. A principal feature illustrated by structures from each group includes a ring composed of six RNase PH domains that form a central channel just large enough to accommodate single stranded RNA (Figure 2). Archaeal and bacterial RNase PH form a ring with six individual protomers arranged head to tail (Figure 2A) [10–12]. The multi-domain bacterial PNPase forms a ring with three PNPase protomers that contain a N-terminal RNase PH-like domain (PH-1), an alpha helical domain, a second RNase PH-like domain (PH-2), and the canonical RNA binding domains: KH (ribonucleoprotein K Homology) and S1 (ribosomal protein S1) that form a cap-like structure on top of the ring (Figure 2B) [4]. Archaeal exosomes form a ring with three Rrp41 and Rrp42 heterodimers that share similarity to PNPase PH-1 and PH-2 domains, respectively (Figure 2C) [13–16]. Archaeal Csl4 or Rrp4 contain S1 or S1 and KH domains, respectively, that form a ‘cap’ on top of the ring. While PNPase projects the KH domain toward the central channel [17], the S1 domains of archaeal Csl4 or Rrp4 line the central channel of the archaeal exosome.
Figure 2. Conserved architecture of exosome core from bacteria, archaea, and eukaryotes.
Structures of complexes reveal a six-component ring architecture with or without phosphorolytic active sites (shown as red dots). The left panel shows the cartoon representation, and the right panel shows the x-ray structure surface representation. A) RNase PH. The Aquifex aeolicus RNase PH structure (PDB ID = 1UDN) forms a homohexamer of PH subunits (colored dark grey and light grey). B) PNPase. The S. antibioticus PNPase structure (PDB ID = 1E3P) forms a homotrimer. Each of PNPase protomers are colored differently: light yellow, dark yellow, and light brown to emphasize the homotrimer of RNase PH 1-like (PH 1) and RNase PH 2-like (PH 2) domains. C) Archaeal exosome. The S. solfataricus archaeal exosome (PDB ID = 2JE6) is shown with Rrp41 subunits (blue) and Rrp42 subunits (green). Rrp41 and Rrp42 form a heterodimer; the resulting heterodimer can trimerize, culminating in a six-component ring. Either Csl4 or Rrp4 (shown in grey) form a trimeric cap above the ring. The surface representation of the structure depicts the Rrp4-bound form. D) Eukaryotic exosome: 9-subunit core. Human subunits are labeled and color-coded and include the PH-like ring subunits Mtr3 (orange), Rrp42 (red), Rrp41 (purple), Rrp45 (blue), Rrp46 (green) and Rrp43 (yellow); the S1/KH domain proteins Csl4 (light blue), Rrp4 (green) and Rrp40 (pink). E) Cytoplasmic, nuclear and nucleolar exosome architectures. The non-catalytic exosome core interacts with additional hydrolytic enzymes to form: the 10 subunit cytoplasmic exosome (panel 1, Exo9 + Rrp44), the 11 subunit nuclear exosome (panel 2, Exo9 + Rrp44 + Rrp6), and the 10 subunit nucleolar exosome (panel 3, Exo9 + Rrp6). The S1/KH protein ring is shown on the top of the PH-like ring with Rrp44 shown below the PH-like ring to reflect structural models of the complex. Rrp6 is shown on the other side of the complex below the PH-like ring, although there is no definitive structural data for this complex. The exoribonuclease active sites are depicted with red circles in Rrp44 and Rrp6, and the endoribonuclease active site of Rrp44 is shown as a yellow circle. Graphics generated with Pymol [59].
The composition of eukaryotic exosome cores (Exo9) is more complex because it consists of nine unique subunits [5]. The RNA exosome is present in all eukaryotic cells; however, it has been most extensively studied in budding yeast and therefore the yeast nomenclature will be referred to for eukaryotic RNA exosomes in which RrpX stands for “Ribosomal RNA processing protein X”. A pseudo-hexameric ring is formed by three heterodimeric pairings, Rrp41-Rrp45, Rrp46-Rrp43, and Mtr3-Rrp42, that share structural and sequence similarity with PNPase PH-1 and PH-2 domains, respectively (Figure 2D). Csl4, Rrp4, and Rrp40 include S1 or S1 and KH domains that cap the eukaryotic PH-domain ring. Analogous to archaeal exosomes, eukaryotic S1 domains project toward the central channel (Figures 3C & D).
Figure 3. Structures of ‘exosome’ domains and exosome associated exoribonucleases.
Residues in red indicate phosphate binding regions and residues in yellow highlight RNA binding surfaces. RNA is shown as green ribbon. Structures depicted in cartoons with helices as tubes and β-strands as arrows. A) C. crescentus PNPase RNase PH 1/RNase PH 2 domain binding interface (PDB ID = 4AM3). Phosphate binding residues include: H405, S440, and S441. RNA binding interface residues: R93, R97, R100, and R401. A second RNA binding site includes residues F77, F78, K79, and R80. B) S. cerevisiae exosome Rrp41/Rrp45 domain binding interface (PDB ID = 4IFD). Rrp41 RNA binding interface residues are R3, K62, S63, T67, R95, and R119. Rrp45 RNA binding site includes residues Y68, R71, R86, R106, R113, and R114. C) C. crescentus PNPase KH/S1 protein ring + RNA (PDB ID = 4AM3). Ribbon and surface diagram of the cap of PNPase bound to RNA. Solely the KH and RNA are represented (because no electron density was detected fro the S1 domains). A circle indicates the central pore. D) S. cerevisiae exosome KH/S1 protein ring + RNA (PDB ID = 4IFD). Ribbon and surface diagram of the budding yeast S1/KH proteins shown from the “top” with subunits labeled and color coded as in previous figures with Csl4 (light blue), Rrp4 (green), and Rrp40 (pink). Note that the S1 domains from each S1/KH protein face the central channel for the eukaryotic exosome complexes, while the KH domains face the channel for PNPase. E) Domain structure of the Rrp44 and Rrp6. Rrp44 contains five domains: a PIN (PIlus N terminal) domain with a Cysteine-Rich sequence (CR3), two Cold Shock Domains (CSD1 and CSD2), a Ribo Nuclease Binding (RNB) domain, and an S1 domain. The hydrolytic endoribonucleolytic active site is located within the PIN domain (yellow circle), and the processive 3′ to 5′ hydrolytic exoribonucleolytic active site is in the RNB domain (red circle). Rrp6 contains three domains: PMC2NT, EXO (EXOribonuclease domain), and HRDC (Homology to RNase D domain C-terminal). A second putative HRDC domain (HRDC2) may also exist similar to one detected in RNase D. The 3′ to 5′ distributive hydrolytic exoribonucleolytic active site is located within the EXO domain (red circle). F) Structures of Rrp6 and RNase D. Structures depicted in cartoons with helices as tubes and β-strands as arrows. Left panel depicts a cartoon ribbon representation of budding yeast Rrp6 catalytic domain structure (PDB ID = 2HBK). Middle panel depicts a cartoon ribbon representation of human Rrp6 catalytic domain structure (PDB 3SAF). Left panel depicts a cartoon ribbon representation of RNase D catalytic domain structure (PDB ID = 1YT3). For all three panels, domains are colored and labeled as in the domain schematic. The EXO active sites are colored red and shown in stick representation; the magnesium ions are shown as green spheres. G) Structure of RNase II + RNA. E. coli RNase II bound to RNA (PDB ID = 2IX1). H) Structure of yeast Rrp44 without PIN + RNA. Budding yeast Rrp44 bound to RNA (PDB ID = 2VNU). I) Structure of yeast Rrp44 from trimer. Structure of full-length budding yeast Rrp44 in complex with Rrp41 and Rrp45 (PDB ID = 2WP8) J) Structure of yeast Rrp44 + RNA from Exo1144/6CTD. Structure of full-length yeast Rrp44 in the yeast core exosome and the caboxy terminus of Rrp6 (PDB ID = 4IFD) For G–I, the domains are labeled and color-coded as in the schematic. The exoribonuclease active site residues are colored red in stick and surface representation. RNA is shown as a green ribbon with the 5′ end labeled; the 3′ end is buried in the exoribonuclease active site. The endoribonuclease active site residues are colored yellow in stick and surface representation. Red and yellow arrows are drawn in H–I on two helices to represent the 100–120° rigid body rotation conformational change of Rrp44 within Exo1144/6CTD.
Bacterial RNase PH, PNPase, and archaeal exosomes utilize a phosphorolytic mechanism to cleave RNA (reviewed in [18]). In these enzymes multiple phosphorolytic active sites (3 or 6) are located within the central channel in the interface between PH or PH-1/PH-2 domains (Figure 3A). It is not clear if each active site is utilized in sequential fashion to degrade a single RNA; however, processivity is likely achieved by holding the RNA in the central channel by contacts to the KH domains and respective active sites, at least for PNPase [17]. While some of the RNA binding residues are present in the central channel, the catalytic residues required for phosphorolysis are not conserved in eukaryotic exosome PH-domain subunits (Figure 3B). In fact, no detectable phosphorolytic activity has been demonstrated for the eukaryotic exosome core based on studies in budding yeast and human systems [5] [7]. The eukaryotic exosome core has instead evolved to interact with and coordinate the activities of the processive hydrolytic exoribonuclease, Rrp44, and the distributive hydrolytic exoribonuclease, Rrp6[19,20].
Rrp44 and the Exo1044 Exosome
Rrp44 includes five domains: an N-terminal Pilus-forming N-terminus (PIN) domain that contains an active site for endoribonuclease activity, two cold-shock domains (CSD1 and CSD2), a central ribonuclease domain (RNB) that contains an active site for exoribonuclease activity, and a C-terminal S1 domain (Figure 3E) [7,21–24]. The Rrp44 PIN domain structure was first revealed in a complex between Rrp44 and two exosome core subunits, Rrp41 and Rrp45 (Figure 3I) [25]. The PIN domain active site is structurally homologous to T4 RNase H, and it is predicted to utilize a two metal ion-dependent catalytic mechanism for hydrolysis[26,27].
The RNB domain active site coordinates two magnesium ions that are required for hydrolytic 3′→5′ exoribonuclease activity, resulting in the release of NMP [28]. Rrp44 is related to bacterial exoribonucleases RNase II and RNase R[29], however the arrangement of individual domains differs (Figure 3G and H). A structure of RNase II showed that CSD2 and S1 domains make contacts to the 5′ end of the RNA before engaging the RNB active site (Figure 3G), contacts that are important for achieving processivity. In contrast, a structure of Rrp44 in complex with single stranded RNA (Figure 3H) showed that RNA contacts CSD1 with a different orientation for the three OB-containing domains (CSD1, CSD2 and S1). As a result, RNase II and Rrp44 structures reveal two distinct paths that guide RNA toward the exoribonuclease active site via an interface that can contact either 12 or 9 nucleotides, respectively. These alternative pathways may reveal different methods to achieve processivity for RNAs larger than >9 nucleotides; where RNase II CSD2 and S1 domains hold onto a single RNA substrate for multiple rounds of catalysis, Rrp44 binds RNA and projects it in an orthogonal direction to present it to the exosome core, presumably to ensure processivity on longer RNAs. It remains unclear if this is the only way Rrp44 can engage RNA substrates or if alternative paths are possible, such as the one observed for RNase II.
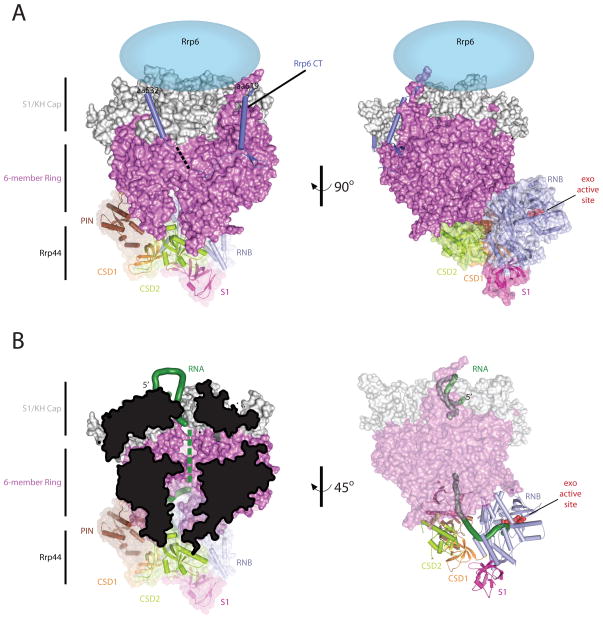
Rrp44 and Exo9 form Exo1044 and this association has been shown to decrease Rrp44 RNA binding and exoribonucleolytic activities in a manner dependent on the integrity of the central channel [25,30–31]. Previous models for Rrp44 bound to Exo9 based on x-ray structures of the human nine-subunit exosome core[5] and budding yeast Rrp41-Rrp45-Rrp44[25], or negative-stain and cryo-EM structures of budding yeast Rrp44 bound to the core exosome (Exo1044) [32] were confirmed and trumped by a recent x-ray structure of yeast Exo9 in complex with Rrp44 and a C-terminal domain of Rrp6 (6CTD) (Exo1144/6CTD) bound to a RNA substrate[33]. This model reveals several important features: Rrp44 is anchored at the bottom of Exo9 through contacts between the Rrp44 PIN domain and CSD1 and the Rrp41/Rrp45 and Rrp43 core subunits, respectively. The 6CTD is positioned at that top of Exo9 through an extensive interface via contacts to Csl4, Mtr3 and Rrp43. Finally, RNA passes through the central channel before emerging to engage in contacts within the Rrp44 exoribonuclease active site (Figures 3I and 4) [33].
Figure 4. The 10 component exosome with CT of Rrp6.
A) Overall architecture of the Exo1144/6CTD. Orthogonal view of the budding yeast 10-subunit exosome in complex with the carboxy terminus of Rrp6 (PDB ID = 4IFD). The S1/KH cap is shown in light gray, the six-membered ring is shown in dark pink, the Rrp6 CT is shown as a light blue cartoon, the location of the EXO and HRDC are currently unknown but they are predicted to be above the cap (light blue ellipse), and each of the Rrp44 domains are colored as discussed in Fig 3. B) RNA ingress into Rrp44 exo active site. The RNA ingress via the central channel is highlighted in two different views: Left, the central channel is viewed by cutting the core in half, and right, the RNA ingress is viewed by making the core transparent.
Extensive conformational changes were observed for Rrp44 in the Exo1144/6CTD complex compared to the Rrp44/Rrp41/Rrp45 complex (Figures 3I and J). The most striking change was a ~100° rigid body rotation of the RNB and S1 domains that flips these two domains orthogonal to the PIN domain and the exosome core. This movement distorts CSD2, and it creates a contiguous L-shaped RNA binding surface to facilitate RNA recruitment to the RNB active site. Although it is likely that other conformations exist, this structure shows one route of RNA ingress to the Rrp44 active site: the 5′ RNA end is coordinated by the cap proteins, RNA then threads through the central channel, and after emerging from the channel, RNA turns ~90° toward the RNB domain before making a final turn into the Rrp44 exoribonuclease active site (Figure 4). The importance of the central channel is supported by biochemical and in vivo data that characterized channel-occluding mutations [25,30,31]; however, channel integrity may only be important for RNA molecules long enough (>35 nt) to traverse the Exo9 channel and Rrp44.
Budding yeast has only one Rrp44, but other eukaryotes encode two or three versions of Rrp44 that harbor different activities or are restricted to different subcellular locations. In humans these are called DIS3, DIS3L, and DIS3L2 [8,9,34,35]. DIS3 is primarily nuclear, DIS3L and DIS3L2 are restricted to the cytoplasm, and all of these proteins appear excluded from nucleoli[9]. DIS3 and DISL associate with the Exo9 core, albeit weakly, and contain all five domains observed in yeast Rrp44 although DIS3L lacks detectable endoribonuclease activity [8,9]. DIS3L2 lacks a PIN domain altogether and no interaction with Exo9 has been observed; interestingly DIS3L2 exhibits specificity for RNA substrates containing 3′ polyU tails such as the polyuridylated pre-let-7 microRNA [8,34].
Rrp6 and the Exo106 and Exo1144/6 exosome
Rrp6 includes the following domains: an N-terminal PMC2NT domain, an EXO domain that catalyzes distributive 3′→5′ exoribonuclease activity, a HRDC domain (helicase and RNase D carboxy terminal domain), a predicted HRDC2 domain, and a C-terminal domain (CTD) (Figure 3E). The PMC2NT domain is required for interaction with a nuclear cofactor, Rrp47[36]. The EXO domain contains a DEDD-Y amino acid motif that coordinates two metals to catalyze distributive, hydrolytic 3′→5′ exoribonuclease activity as illustrated by structures obtained for budding yeast Rrp6 and human RRP6 (Figure 3F) [37–39]. Human and yeast Rrp6 are related to RNase D however several differences exist between these structures: the active site of human RRP6 is more solvent exposed than yeast Rrp6 and RNase D includes a structured HRDC2 that limits access to the active site (Figure 3F).
Rrp6 associates with Exo9 and Rrp44 to form the canonical eleven-subunit nuclear exosome (Exo1144/6) although an additional exosome complex has been suggested in human cells that contains Exo9 and RRP6 (Exo106) [9]. Rrp6 is not essential, but budding yeast strains lacking Rrp6 exhibit a temperature-sensitive growth phenotype and accumulate nuclear RNA precursors such as snRNAs, snoRNAs, pre-rRNAs, and cryptic unstable transcripts[40–43]. Rrp6 possesses activities that are both dependent and independent of Exo9, however Exo9-independent activities have only been detected for processing a subset of nuclear RNAs[44].
The budding yeast Rrp6 C-terminal domain (6CTD) interacts with Exo9 via an extensive interface that contacts Csl4, Mtr3 and Rrp43 (Figure 4). The 6CTDs location on the core suggests the Rrp6 catalytic domain would reside near the top of Exo9 [33]. Consistent with this model, a 35-Å resolution negative-stain EM structure of the Leishmania tarentolae exosome places the Rrp6 EXO and HRDC domains above the exosome core via interactions with the cap subunits[45]. How Rrp6 engages RNA alone or in complex with the exosome remains unknown.
Exosome co-factors
The exosome interacts with a multitude of co-factors to facilitate interaction with different RNAs (Figure 1). The exosome associates with the SKI complex (composed of Ski3p, Ski8p, and the DEVH ATPase Ski2p) via Ski7p in the yeast cytoplasm to participate in 3′→5′ mRNA degradation through association with the translation apparatus [46–48]. While a homolog of Ski7 exists in higher eukaryotes, it is unclear if a similar mechanism is employed to recruit the exosome and DIS3L1 to promote decay.
The nuclear form of the exosome interacts with a co-factor Rrp47/C1D via Rrp6 to process RNAs including the 3′ extended form of the 5.8S rRNA precursor [36,49,50]. Mpp6/MPP6 is another nuclear co-factor that associates with the exosome to assist in processing structured RNAs[51,52] as loss of MPP6 accumulates 3′ extended forms of 5.8S rRNA. The nuclear exosome also interacts with the TRAMP complex (Mtr4 helicase, Trf4/Trf5 poly(A) polymerases, and Air1/Air2 Zn-knuckle RNA binding proteins) to promote surveillance and degradation of aberrant RNA [53]. The human exosome may also interact with the Nuclear Exosome Targeting (NEXT) complex (hMTR4, the Zn-knuckle protein ZCCHC8, and the putative RNA binding protein RBM7) to promote degradation of promoter upstream transcripts [54]. In addition, the NRD complex (Nrd1, Nab3, and Sen1) recruits the exosome to degrade or process certain sn-and snoRNA polymerase II transcripts [55,56]. Additional co-factors have been identified in both human and budding yeast systems, although their functional significance is less clear (Figure 1).
Conclusions
Structural models obtained thus far suggest that the non-catalytic Exo9 core channels RNA and is flanked on either end by the catalytic subunits Rrp44 and Rrp6. This is interesting when compared to RNases such as PNPase or protease complexes (such as the proteasome) because in these complexes the catalytic sites lie within the central channel or chamber, respectively. In contrast, the eukaryotic exosome appears inside out, with a non-catalytic core and central channel sandwiched between the Rrp6 and Rrp44 catalytic subunits.
Although Rrp44 and Rrp6 are not in direct contact, their activities appear co-dependent when associated with Exo9[57,58]. For instance, Rrp6 can stimulate endo- and exoribonucleolytic activities of Rrp44 in a manner independent of Rrp6 catalytic activity[31]. Conversely, an Rrp44 isoform lacking exoribonuclease activity severely inhibits Rrp6 exoribonucleolytic activity. These and other studies suggest that Rrp44 utilizes the full extent of the central channel to engage RNA substrates while Rrp6 utilizes a portion of this channel comprised by the cap proteins to engage its RNA substrates. A shared path may ensure that a particular exosome remains engaged with one substrate, degrading or processing it via Rrp44 or Rrp6, before it can bind another. In other words, when Rrp44 is engaged with a substrate that occupies the channel, Rrp6 cannot bind a substrate. The converse may be true although this has not yet been demonstrated.
It remains unclear how a path is selected to feed RNA to Rrp6 and/or Rrp44; but it is likely that exosome co-factors influence this by competing for interactions with the exosome core or by influencing activities of Rrp6 and/or Rrp44. Further investigation in this area will be required to fully understand how the activities of the eukaryotic RNA exosome are coordinated to facilitate RNA processing and degradation.
Highlights.
Eukaryotic exosome cores and phosphorolytic exoribonucleases are structurally similar
Exosome cores include a central channel but are catalytically inert
Central channel directs single-stranded RNA to the exoribonucleases Rrp44 and Rrp6
Rrp6 and Rrp44 exoribonuclease activities are modulated by the exosome core
Rrp6 and Rrp44 bind at opposite ends of the exosome core and central channel
Acknowledgments
Special thanks to Abigail C. Wasmuth for preparing graphics in Figure 1 and Elizabeth Wasmuth for advice during preparation of Figure 1. Research reported in this publication was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under award R01GM079196 (C.D.L). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. C.D.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Porrua O, Libri D. RNA quality control in the nucleus: the Angels’ share of RNA. Biochim Biophys Acta. 2013;1829:604–611. doi: 10.1016/j.bbagrm.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Schneider C, Tollervey D. Threading the barrel of the RNA exosome. Trends Biochem Sci. 2013 doi: 10.1016/j.tibs.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chlebowski A, Lubas M, Jensen TH, Dziembowski A. RNA decay machines: the exosome. Biochim Biophys Acta. 2013;1829:552–560. doi: 10.1016/j.bbagrm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 6.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 7.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 8.Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013 doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. Interaction studies conducted in human cells reveal two different homologs of Rrp44 (DIS3 and DIS3L) bind Exo9. DIS3 is predominantly located in the nucleus, while DIS3L is only detected in the cytoplasm. Both DIS3 and DIS3L were shown to have exoribonucleolytic activity; however, only DIS3 contains endonucleolytic activity. Localization studies further suggest that a nucleolar exosome complex exists that contains solely Exo9 and Rrp6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Z, Yang WZ, Lin-Chao S, Chak KF, Yuan HS. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA. 2008;14:2361–2371. doi: 10.1261/rna.1244308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlow LS, Kadziola A, Jensen KF, Larsen S. Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis and implications for its quaternary structure and tRNA binding. Protein Sci. 2004;13:668–677. doi: 10.1110/ps.03477004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii R, Nureki O, Yokoyama S. Crystal structure of the tRNA processing enzyme RNase PH from Aquifex aeolicus. J Biol Chem. 2003;278:32397–32404. doi: 10.1074/jbc.M300639200. [DOI] [PubMed] [Google Scholar]
- 13.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 16.Navarro MV, Oliveira CC, Zanchin NI, Guimaraes BG. Insights into the mechanism of progressive RNA degradation by the archaeal exosome. J Biol Chem. 2008;283:14120–14131. doi: 10.1074/jbc.M801005200. [DOI] [PubMed] [Google Scholar]
- 17.Hardwick SW, Gubbey T, Hug I, Jenal U, Luisi BF. Crystal structure of Caulobacter crescentus polynucleotide phosphorylase reveals a mechanism of RNA substrate channelling and RNA degradosome assembly. Open Biol. 2012;2:120028. doi: 10.1098/rsob.120028. Crystal structures of RNA-bound and apo- Caulobacter crescentus PNPase are solved that reveal the mechanism for coordinating RNA. A model is proposed in which the apo- form recruits RNA via its S1 domains and delivers the RNA to the three KH domains to direct the 3′ end of RNA towards the central channel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Januszyk K, Lima CD. Structural components and architectutres of RNA exosomes. In: Jensen TH, editor. RNA Exosome. Vol. 702 Landes Bioscience and Springer Science; 2010. pp. 9–28. [Google Scholar]
- 19.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 21.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 23.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 26.De Vivo M, Dal Peraro M, Klein ML. Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. J Am Chem Soc. 2008;130:10955–10962. doi: 10.1021/ja8005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 30.Drazkowska K, Tomecki R, Stodus K, Kowalska K, Czarnocki-Cieciura M, Dziembowski A. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013;41:3845–3858. doi: 10.1093/nar/gkt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wasmuth EV, Lima CD. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol Cell. 2012;48:133–144. doi: 10.1016/j.molcel.2012.07.012. Functional and in vivo studies on the budding yeast exosome demonstrate that Exo9 and its central channel regulate all three yeast exosome ribonuclease activities. An additional significant finding for the Exo116/44 was that Rrp6 can stimulate both endo- and exoribonucleolytic activities, while Rrp6 activity is decreased by a dysfunctional Rrp44; therefore, these results suggest that Rrp44 and Rrp6 activities are interdependent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci U S A. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makino DL, Baumgartner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495:70–75. doi: 10.1038/nature11870. The x-ray structure of budding yeast Exo-10 bound to a carboxy-terminal region of Rrp6 bound to RNA is solved which reveals: a conformational change in Rrp44 to recruit RNA into its active site, the minimal interaction surface for Rrp6, and the RNA binding surfaces in the cap, hexameric ring, and Rrp44. [DOI] [PubMed] [Google Scholar]
- 34.Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stead JA, Costello JL, Livingstone MJ, Mitchell P. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 2007;35:5556–5567. doi: 10.1093/nar/gkm614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA. 2011;17:1566–1577. doi: 10.1261/rna.2763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci U S A. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo Y, Wang Y, Malhotra A. Crystal structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure. 2005;13:973–984. doi: 10.1016/j.str.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 44.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cristodero M, Bottcher B, Diepholz M, Scheffzek K, Clayton C. The Leishmania tarentolae exosome: purification and structural analysis by electron microscopy. Mol Biochem Parasitol. 2008;159:24–29. doi: 10.1016/j.molbiopara.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Halbach F, Reichelt P, Rode M, Conti E. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell. 2013;154:814–826. doi: 10.1016/j.cell.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 47.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Lewis MS, Johnson AW. Domain interactions within the Ski2/3/8 complex and between the Ski complex and Ski7p. RNA. 2005;11:1291–1302. doi: 10.1261/rna.2060405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costello JL, Stead JA, Feigenbutz M, Jones RM, Mitchell P. The C-terminal region of the exosome-associated protein Rrp47 is specifically required for box C/D small nucleolar RNA 3′-maturation. J Biol Chem. 2011;286:4535–4543. doi: 10.1074/jbc.M110.162826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milligan L, Decourty L, Saveanu C, Rappsilber J, Ceulemans H, Jacquier A, Tollervey D. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol Cell Biol. 2008;28:5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schilders G, Raijmakers R, Raats JM, Pruijn GJ. MPP6 is an exosome-associated RNA-binding protein involved in 5. 8S rRNA maturation. Nucleic Acids Res. 2005;33:6795–6804. doi: 10.1093/nar/gki982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. The Nuclear Exosome Targeting (NEXT) complex is identified and contains: hMTR4, ZCCHC8, and RBM7. This complex works as a co-factor for the exosome to degrade upstream transcripts (PROMPTs) [DOI] [PubMed] [Google Scholar]
- 55.Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 57.Gudipati RK, Xu Z, Lebreton A, Seraphin B, Steinmetz LM, Jacquier A, Libri D. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell. 2012;48:409–421. doi: 10.1016/j.molcel.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. Transcriptome-wide analysis of exosome targets. Mol Cell. 2012;48:422–433. 56–57. doi: 10.1016/j.molcel.2012.08.013. Using different techniques and combinations of Rrp6p and Dis3p catalytic mutants, the transcriptome was analyzed and it was determined that these proteins have both overlapping and unique functions for degradation of different RNA substrates. An interesting observation was that large quantities of tRNA precursors are degraded via the exosome without any detectable processing defects, suggesting that large quantities of RNAs are produced and then eliminated by the exosome before entering into a functional pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]