Abstract
Excess superoxide and nitric oxide (NO•) forms peroxynitrite (ONOO−) during cardiac ischemia reperfusion (IR) injury, which in turn induces protein tyrosine nitration (tyr-N). Mitochondria are both a source of and target for ONOO−. Our aim was to identify specific mitochondrial proteins that display enhanced tyr-N after cardiac IR injury, and to explore whether inhibiting during IR decreases mitochondrial protein tyr-N and consequently improves cardiac function. We show here that IR increased tyr-N of 35 and 15 kDa mitochondrial proteins using Western blot analysis with 3-nitrotyrosine antibody. Immunoprecipitation (IP) followed by LC–MS/MS identified 13 protein candidates for tyr-N. IP and Western blot identified and confirmed that the 35 kDa tyr-N protein is the voltage-dependent anion channel (VDAC). Tyr-N of native cardiac VDAC with IR was verified on recombinant (r) VDAC with exogenous ONOO−. We also found that ONOO− directly enhanced rVDAC channel activity, and rVDAC tyr-N induced by ONOO− formed oligomers. Resveratrol (RES), a scavenger of , reduced the tyr-N levels of both native and recombinant VDAC, while L-NAME, which inhibits NO• generation, only reduced tyr-N levels of native VDAC. and ONOO− levels were reduced in perfused hearts during IR by RES and L-NAME and this was accompanied by improved cardiac function. These results identify tyr-N of VDAC and show that reducing ONOO− during cardiac IR injury can attenuate tyr-N of VDAC and improve cardiac function.
Keywords: Mitochondria, Protein tyrosine nitration, Cardiac injury, VDAC, ROS/RNS scavengers
1. Introduction
Mitochondrial dysfunction during cardiac ischemia (I) and reperfusion (R) injury is associated with Ca2+ overload, excess emission of reactive oxygen species (ROS) [1–3] and reactive nitrogen species (RNS) [4,5] that can lead to deleterious post-translational modifications (dPTMs) of mitochondrial proteins [6]. Superoxide , the origin of most O2-derived free radicals, is overproduced and under scavenged during cardiac IR injury [1,7]. Under hypoxic or ischemic conditions, the mitochondrial electron transport chain (ETC) is initially in a highly reduced state [1], which results in electron leak so that O2 is reduced to . During reperfusion, production, initiated during ischemia, is further exacerbated [1] because of diminished ADP levels, abundant O2, and poor ROS scavenging capability. Nitric oxide (NO•), a precursor of RNS, may also increase during IR [8,9]. When is in excess it reacts with available NO• to generate peroxynitrite (ONOO−) [2,10], a highly reactive non-radical and a strong marker of RNS-induced cell cytotoxicity. Indeed, a marked increase in ONOO− occurs during cardiac IR injury [4,5] and a reduction in injury is associated with decreased release of ONOO− [4,5]. is a charged and diffusion limited molecule generated in mitochondria, whereas NO• is an uncharged, and diffusible molecule that easily traverses mitochondrial membranes. ONOO− has a half-life of about 3–5 ms in mitochondria and is a strong oxidant of mitochondrial proteins and lipids lying in close proximity [3]. Thus mitochondria can be both a site for ONOO− production and a target of ONOO−-induced dPTMs [3]. For example, ONOO− mediates inactivation of mitochondrial manganese superoxide dismutase (MnSOD) through nitration and oxidation of critical tyrosine residues [11,12]. However, the most vulnerable protein sites, the mechanisms by which ONOO− mediates damage to mitochondria, and the cardiac functional consequences are not well known.
In general, ONOO− can directly oxidize molecules, decompose to NO2• and •OH, or react with CO2 to form and NO2• [13]. These free radicals induce protein nitration, especially of exposed tyrosine (tyr) residues to form 3-nitrotyrosine (3-NT) [6]. Tyrosine nitration (tyr-N) is an established marker not only for RNS-induced stress resulting from ONOO− and its radical products, but also for ONOO− mediated cytotoxicity; once formed, 3-NT is relatively stable and the nitration can result in deleterious protein structural and functional changes [3,13]. Tyr-N may itself cause protein dysfunction and it may also prevent tyr phosphorylation by tyr kinases [14]. Although tyr-N is widely used as a biological assay indicative of ONOO− generation, it is acknowledged that ONOO− can also nitrate tryptophan and phenylalanine and that it also oxidizes other sites, e.g. FeS centers, protein thiols, and lipids [10]. Moreover, tyr-N may be induced by ONOO− independent oxidants like the gas NO2• formed by oxidation of (itself an oxidized product of NO•) in the presence of H2O2 by peroxidase, but high concentrations of this labile NO2• product are likely required to induce tyr-N [15].
Both in vivo and in vitro evidence indicates that proteins involved in oxidative phosphorylation and apoptosis can be nitrated in various diseases and disorders [16]. Nitration of proteins has been observed after IR injury [17–22], but the effect of IR on inducing irreversible dPTMs that alter the structure and function of key mitochondrial proteins has not been well examined. Liu et al. [17] indirectly identified 10 mitochondrial proteins including 4 subunits from the oxidative phosphorylation system, five enzymes in the matrix, and the voltage-dependent anion channel (VDAC) that were nitrated after IR injury using two-dimensional gel electrophoresis (2-DE) gel and mass spectrometry (MS) analysis. However, the identification of these tyr-N proteins using specific antibodies, and the determination of the specific sites of tyr-N by MS have not been undertaken; nor is it known if nitration of mitochondrial proteins during cardiac IR could be reduced by ROS/RNS scavenging or by inhibiting nitric oxide synthase (NOS), and if identified nitrated mitochondrial proteins exhibit altered function.
We proposed that some key mitochondrial proteins undergo tyr-N during IR injury and that attenuating formation of ONOO− reduces tyr-N of these proteins and leads to improve cardiac function. Thus our first objective was to identify specific mitochondrial proteins that undergo enhanced tyr-N after cardiac IR injury. We identified VDAC as a tyr-N protein, and so pursued VDAC as a focus for additional studies. Although other proteins can be nitrated during IR injury, we focused on VDAC based on our initial results. We probed ONOO− induced tyr-N of recombinant (r) VDAC and its effect on changing rVDAC channel activity. In addition we tested if resveratrol (RES, trans-3, 4′, 5-trihydroxystilbene), a putative and ONOO− scavenger, and NG-nitro-L-arginine methyl ester (L-NAME), an unspecific NOS inhibitor, could reduce the extent of tyr-N in mitochondrial proteins in association with reduced levels of and ONOO− and improve functional recovery after IR injury in intact hearts. Our focus on the molecular aspect in this study provides amore mechanistic insight into how ROS/RNS impacts mitochondrial proteins and leads to the functional changes in myocardial function during IR. Such an in depth approach could provide for better strategies to mitigate IR injury.
2. Methods
2.1. Isolated heart preparation and protocols
Hearts in situ cannot be used to detect and ONOO−, so isolated hearts from guinea pigs were utilized to directly correlate changes in total cardiac oxidative stress and function in isolated mitochondria after IR injury. Our animal protocols conformed to the Guide for the Care and Use of Laboratory Animals (National Institutes of Health No. 85-23, Revised 1996) and were described previously [4,7,23,24] (see detailed descriptions in the Online Supplement Materials and Methods). After 30 min of global ischemia, or after 10, 30, or 60 min reperfusion, hearts were removed and mitochondria were immediately isolated. In the treated groups either 100 µmol/L L-NAME or 10 µmol/L RES was perfused for 10 min up to initiation of global ischemia so that the drug remained in the hearts during ischemia. RES, 10 µmol/L, is cardioprotective in isolated rat hearts [25] and 100 µmol/L L-NAME blocks increased ONOO− production in 17 °C-perfused isolated hearts [24].
2.2. Detection of and ONOO− in isolated hearts
The intracellular fluorescent probe dihydroethidium (DHE) (Molecular Probes) was used to assess emission in isolated hearts as described previously (see Online Supplement Materials and Methods) [7,23,24,26]. After attaining stability, hearts were loaded with 10 µmol/L DHE for 20 min followed by washout of residual DHE for another 20 min. Dityrosine (DiTyr) was used to detect ONOO− formation indirectly as described previously (see Online Supplement Materials and Methods) [24]. ONOO− reacts with continuously perfused L-tyrosine (0.3 mmol/L) to form the fluorescent dimer DiTyr [9], a stable product at tissue pH [27]. DHE loaded/L-tyrosine perfused hearts were treated with/without RES/L-NAME before ischemia, followed by IR.
2.3. Isolation of mitochondria
Cardiac mitochondria were isolated at the end of each functional study using well-established methods in our laboratory [28–30] with minor modifications. All isolation procedures were carried out at 4 °C. Hearts were minced into very small pieces in a chilled isolation buffer containing 200 mmol/L mannitol, 50 mmol/L sucrose, 5 mmol/L KH2PO4, 5 mmol/L MOPS, 0.1% fatty acid free bovine serum albumin (BSA) and 1 mmol/L EGTA, at pH 7.15, then homogenized in 2.65 ml isolation buffer containing protease (5 U/ml, Sigma Cat: P-5495) for 15 s with an ultrasonic homogenizer. After this, 15 ml of isolation buffer containing protease inhibitors (Roche, Complete protease inhibitor cocktail tablets) was added rapidly to the homogenate followed by another 15 s of homogenization. The slurried homogenates were centrifuged at 10,000 g for 5 min, and then the pellet was resuspended in isolation buffer containing protease inhibitors and centrifuged at 8000 g for another 10 min. After that, the pellet was resuspended in isolation buffer containing protease inhibitors and spun at 750 g for 10 min; then the supernatant was collected and centrifuged again at 8000 g. The resulting mitochondrial pellet was resuspended in isolation buffer containing protease inhibitors and stored at −80 °C for later use. The amount of mitochondrial protein was assessed using Bio-Rad protein assay with BSA as the standard.
2.4. Immunoprecipitation of mitochondrial proteins
Immunoprecipitation (IP)was performed as described before [17,31] with minor changes. Mitochondria were pelleted and then lysed in RIPA buffer containing 50 mmol/L Tris-Cl pH 7.4, 150 mmol/L NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS and protease inhibitors on ice for 30 min. The sample was pre-cleared with 30 µl protein G Sepharose-4B beads (Invitrogen) for 1h at 4°C with constant end-over-end shaking and then centrifuged at 3000 g for 5 min. The supernatant was collected, adjusted to 2 mg/ml protein concentration with RIPA buffer containing protease inhibitors, and subjected to IP with an anti-3-nitrotyrosine monoclonal antibody (anti-3-NT, Millipore) or anti-VDAC antibody (Sigma) at 4 °C with constant end-over-end shaking overnight. The next day, after adding 30 µl protein G Sepharose-4B beads, the mixture was incubated for another 2 h under the same conditions as above. The beads were collected and exhaustively washed (five times) with RIPA buffer. The final pellet was re-suspended in 50 µl sample buffer containing 50 mmol/L Tris-Cl, 10% glycerol, 500 mmol/L β-mercaptoethanol, 2% SDS, 0.01% w/v bromophenol blue and protease inhibitors at pH 7.4 and boiled at 95 °C for 5 min. The immunoprecipitated proteins were separated using SDS-PAGE. The gels were silver stained or subjected to Western blot analysis.
2.5. Two-dimensional gel electrophoresis (2-DE)
2-DE was performed with the IEF/Criterion gel system (Bio-Rad). Mitochondrial pellets were lysed in lysis buffer containing 7.8 mol/L urea, 2.2 mol/L thiourea, 4% CHAPS, and protease inhibitors on ice for 30 min, then desalted twice with Amicon 3 kDa (Millipore). After adding 65 mmol/L DTT and 1% Bio-Lyte ampholyte (pH3-10), about 800 µg of protein from each sample was immediately loaded onto an 11-cm nonlinear pH 3–10 IPG strip (Bio-Rad). The strips were rehydrated at 50 V for 12 h and then focused under the following conditions: voltage was first increased to 250 V over 20 min followed by a linear increase to 8000 V over 70 min and then held at 8000 V until 26 kVh was attained. The focused IPG strips were then equilibrated for 15 min in 0.375 mol/L Tris-Cl, pH 8.8, 6 mol/L urea, 20% glycerol, 2% SDS and 2% DTT followed by 15 min in 0.375 mol/L Tris-Cl, pH 8.8, 6 mol/L urea, 20% glycerol, 2% SDS and 2.5% iodoacetamide. The equilibrated strips were loaded on pre-cast gels (Criterion™10%, 11 cm; Bio-Rad) for the second dimension separation. The gels were run at 150 V for 70 min and then transferred onto polyvinylidene difluoride (PVDF) membranes for Western blot analysis with anti-VDAC and anti-3-NT antibodies.
2.6. Western blot analysis
Mitochondrial pellets were lysed in sample buffer and boiled at 95 °C for 5 min. Mitochondrial samples (100 µg) or immunoprecipitates were resolved by SDS-PAGE and transferred onto PVDF membranes. Membranes were incubated with specific primary antibodies against 3-NT, VDAC (Cell Signaling), and ANT (adenine nucleotide translocator, Santa Cruz). Washed membranes were incubated with the appropriate secondary antibody conjugated to horseradish peroxidase, then immersed in an enhanced chemiluminescence detection solution (GE Healthcare) and exposed to X-Ray film for autoradiography. For monitoring loaded protein amounts, VDAC or ANT protein was used as loading controls because VDAC and/or ANT are mitochondrial membrane proteins and their detection confirms mitochondrial isolation. Blots were stripped with buffer (62.5 mmol/L Tris-Cl, 2% SDS, and 100 mmol/L β-mercaptoethanol at pH 6.8), and then probed with specific antibodies against VDAC or ANT.
2.7. In-gel tryptic digestion of protein bands
Protein bands from silver-stained gels were excised and cut into small pieces and then subjected to in-gel digestion by trypsin as described by Newman et al. [32]. In brief, after destaining silver ions with a 1:1 solution of 100 mmol/L Na2S2O3 and 30 mmol/L K3Fe9(CN)6, the gel pieces were dehydrated with 50% acetonitrile and then 25 mmol/L (NH4)HCO3/50% acetonitrile (pH 8.0). The dehydrated gel pieces were dried and then digested with 40 ng trypsin in 100 µl of 25 mmol/L (NH4)HCO3 (pH 8.0) overnight at 37 °C. Tryptic fragments were extracted from gel slices with acetonitrile/trifluoroacetic acid (TFA)/water (70:0.1:29.9 v/v/v). Extracted samples were dried and prepared either for LC–MS/MS or MALDI-TOF MS analysis.
2.8. Protein identification by LC–MS/MS mass spectrometry
Extracted tryptic fragments were subjected to LC–MS/MS analysis using a LTQ (Thermo-Fisher) MS coupled to a Surveyor HPLC system equipped with a Micro AS auto sampler. Separation of tryptic peptides was achieved online before MS/MS analysis on an Aquasil, C18 PicoFrit capillary column (75 mm × 100 mm; New Objective) with the following conditions: mobile phase A, 0.1% formic acid containing 5% acetonitrile; and mobile phase B, 0.1% formic acid in 95% acetonitrile, respectively. A 180 min linear gradient was applied. The ions eluted from the column were electro-sprayed at a voltage of 1.75 kV. MS/MS spectra data were analyzed using the SEQUEST search engine and analysis carried out via the Visualize software program [33]. The SEQUEST search parameters were set with no fixed modification, one missed trypsin cleavage site, a peptide tolerance of 1.2 Da, and a MS/MS tolerance of 0.8 Da, and then searched against the rodent subset of the Uniprot database. All proteins identified had to meet a False Discovery Rate (FDR) of 1.5% or less.
2.9. Protein identification by MALDI-TOF mass spectrometry
MALDI-TOF MS was also performed as described by Newman et al. [32]. Briefly, the dried extracted samples were resuspended in 20 µl 0.1% TFA and subjected to purification using Micro-C18 Zip-Tips (Millipore). The purified samples were eluted in 1–2 µl volume of α-cyano-4-hydroxycinnamic acid in 60% acetonitrile/0.1% TFA and then directly loaded on the sample plate for analysis using a Voyager DE PRO MALDI-TOF mass spectrometer in positive ion, reflector, delayed-extraction mode (Applied Biosystems). Peptide masses were analyzed using the Protein Prospector software (http://prospector.ucsf.edu/prospector/mshome.html).
2.10. Nitration of recombinant VDAC induced by exogenous ONOO− in vitro
Because VDAC was identified as a key tyr-N protein after cardiac IR injury, we further explored for specific VDAC tyr-N residues and determined if tyr-N of VDAC affects its structure and function. To do so we exposed recombinant VDAC (rVDAC) to different concentrations of exogenous ONOO− to induce concentration-dependent nitration of rVDAC in vitro. Rat VDAC-1 full-length cDNA (GenBank: BC072484, purchased from Open Biosystems) was cloned into pET-21a vector (Novagen) and expressed in BL21 cells. rVDAC proteins were extracted with BugBuster Master Mix (Novagen) and purified with Ni-NTA His·Bind Resins (Novagen). Purified rVDAC was refolded at 4 °C by dropwise dilution of one volume protein with 10 volumes refolding buffer containing 20 mmol/L Tris-Cl, 100 mmol/L NaCl, 1% lauryldimethylamineoxide (LDAO), and 1 mmol/L DTT at pH 7.4 [34,35]. It has been reported that using the detergent LDAO resulted in a 90% refolding of rVDAC compared to 75% and 30% for the detergents DDM or C8E4, respectively [35]. After refolding, the refolded rVDAC protein was dialyzed against refolding buffer, without DTT, at 4 °C to remove the DTT. The reconstituted rVDAC was used for in vitro nitration by ONOO−. ONOO− (Calbiochem) concentration was determined by absorbance at 302 nm with an extinction coefficient of 1670 M−1 cm−1 in 100 mmol/L of NaOH prior to use [36]. For exposure of rVDAC to ONOO−, 500 µg of rVDAC at 3 mg/ml was incubated with 0–400 µmol/L ONOO− at 23 °C for 30 min. The ONOO−-treated rVDAC was subjected to SDS-PAGE or 2-DE, followed by Western blot with antibodies against 3-NT and VDAC, or gels were stained with silver staining and VDAC bands were cut for MALDI-TOF MS.
2.11. Reconstitution of rVDAC into planar lipid bilayers for channel activity
Channel activity of rVDAC was monitored by incorporating rVDAC into planar lipid bilayers (PLB), as previously described [37]. Briefly, phospholipids were prepared by mixing phosphatidylethanolamine, phosphatidylserine and phosphatidylcholine (Avanti Polar Lipids, Alabaster, AL) in a ratio of 5:4:1 (v/v); the phospholipids were dried under N2 and re-suspended in n-decane to a final concentration of 25 mg/ml. The cis/trans chambers contained identical solutions of 10 mmol/L HEPES, 500 mmol/L NaCl and 1 mmol/L CaCl2 at pH 7.4. The cis chamber was held at virtual ground and the trans chamber was held at the command voltages. rVDAC protein was added into the cis chamber. After channel incorporation into the bilayer, ONOO− was added to the cis chamber and diluted proportionally to achieve the desired concentration of 50 µmol/L. Currents were sampled at 5 kHz and low pass filtered at 1 kHz using a voltage clamp amplifier (Axopatch 200B, Molecular Devices) connected to a digitizer (DigiData 1440, Molecular Devices), and recorded in 1 min segments. The pClamp software (version 10, Molecular Devices) was used for data acquisition and analysis. Additional analyses were conducted using Origin 7.5 (OriginLab).
2.12. Statistical analysis
All results are expressed as means ± SEM and were analyzed by one-way ANOVA followed by a post-hoc analysis (Student–Newman–Keuls' test) to determined statistically significant differences of means among groups. A value of p < 0.05 was considered significant (two-tailed).
3. Results
3.1. and ONOO− production increase in isolated hearts during IR injury
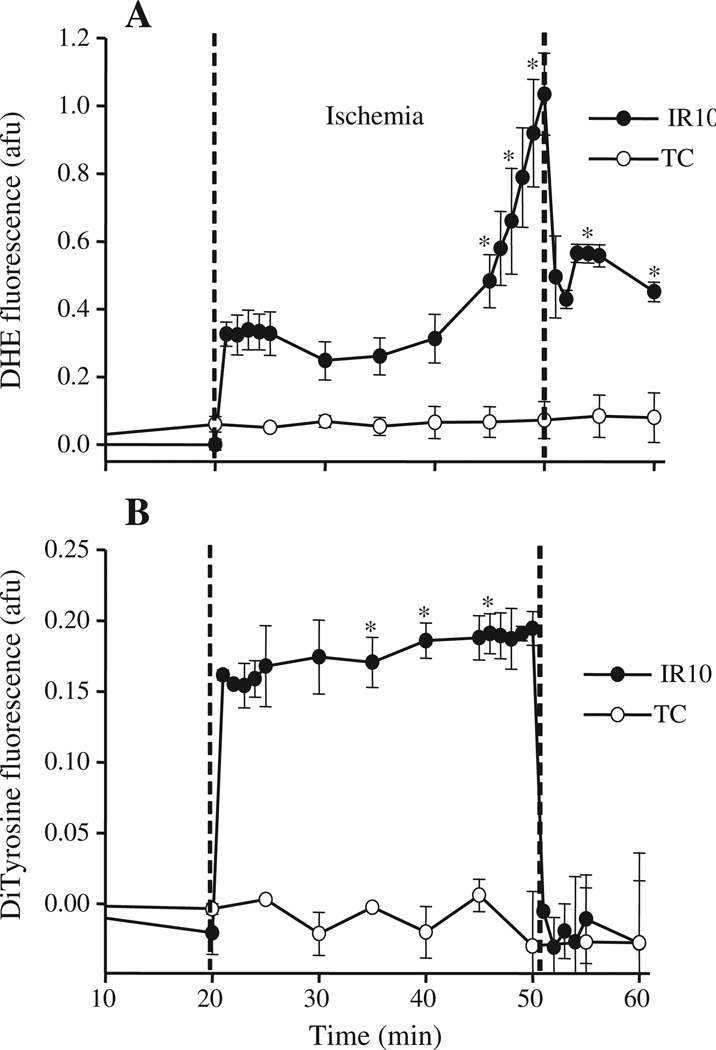
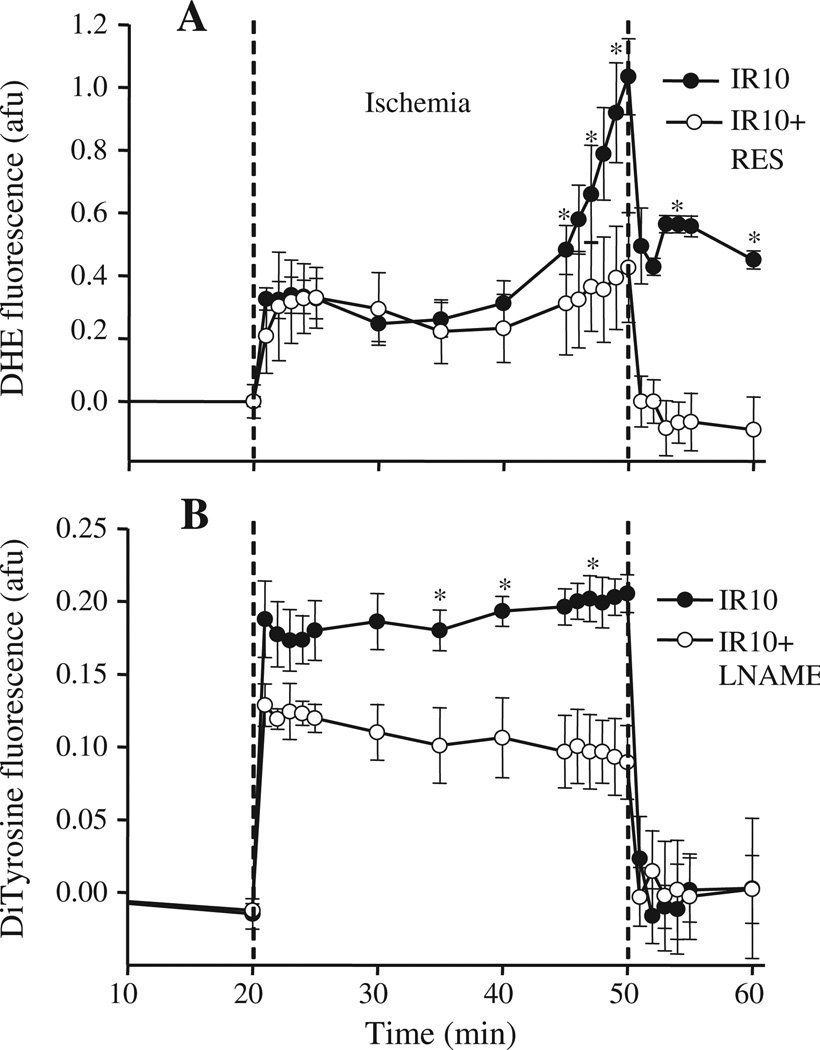
DHE fluorescence ( emission) increased during ischemia; on reperfusion, it declined abruptly, but remained higher than baseline values (Fig. 1A, IR10), consistent with our previous findings [7,23,26]. Similarly, DiTyr fluorescence (ONOO−) increased and remained elevated during ischemia; on reperfusion it decreased to baseline levels (Fig. 1B, IR10). Time control (TC) groups showed no significant changes in DHE and DiTry (ONOO−) fluorescence during the duration of the perfusion protocol (Fig. 1A and B, TC).
Fig. 1.
Time course of changes in superoxide and peroxynitrite (ONOO−) levels during IR. A: The fluorescence probe DHE, via its stable (ethidium) or unstable and labile 2-hydoxyethidium (2-OH-E+) product was used to assess intracellular emission online in isolated hearts at the LV free wall. B: Dityrosine (DiTyr) fluorescence formed from L-tyrosine in the presence of ONOO− was used to monitor ONOO− online similar to that described for the DHE fluorescence probe. TC: Time control; IR10: 30 min ischemia and 10 min reperfusion; N=4–5 hearts/group/probe; Values are mean ± SEM. *: p < 0.05 vs. TC. See Table 1 for functional data.
These changes in and ONOO− generation also correlated with the changes of function in isolated hearts. Systolic–diastolic left ventricular pressure (dLVP), indices of contractility and relaxation (dLVP/dtmax and dLVP/dtmin) respectively decreased, while diastolic LVP increased during ischemia (Table 1, IR10, IR60); during reperfusion, dLVP/dtmax, dLVP/dtmin and dLVP recovered to almost half the pre-ischemia values while diastolic LVP remained significantly higher than the pre-ischemia level (Table 1, IR10, IR60). Cardiac function was unchanged over time in the TC groups (see Online Supplement Table S3). Heart rate was unchanged on reperfusion compared to pre-ischemia in all groups (Table 1).
Table 1.
Cardiac function in IR10, 60, resveratrol and L-NAME treated IR10 groups during baseline, ischemia 30 min and reperfusion 10 and 60 min.
| Variable | Pre-ischemia | Ischemia 30 min |
Reperfusion 10 min |
Reperfusion 60 min |
|---|---|---|---|---|
| dLVP/dtmax (mm Hg/s) | ||||
| IR10, 60 | 2440 ± 119 | 23 ± 1 | 1450 ± 172 | 1550 ± 151 |
| IR10 + RES | 2429 ± 104 | 25 ± 1 | 1913 ± 119* | |
| IR10 + L-NAME | 2778 ± 191 | 23 ± 1 | 1799 ± 119* | |
| dLVP/dtmin (mm Hg/s) | ||||
| IR10, 60 | −2178 ± 128 | −32 ± 2 | −1151 ± 162 | −1255 ± 140 |
| IR10 + RES | −2140 ± 83 | −29 ± 1 | −1944 ± 190* | |
| IR10 + L-NAME | −2290 ± 134 | −29 ± 1 | −1722 ± 83* | |
| Sys-dia LVP (mm Hg) | ||||
| IR10, 60 | 88 ± 4 | 0.1 ± 0 | 38 ± 5 | 39 ± 4 |
| IR10 + RES | 90 ± 4 | 0.1 ± 0 | 57 ± 7* | |
| IR10 + L-NAME | 95 ± 5 | 0.1 ± 0 | 64 ± 4* | |
| Diastolic LVP (mm Hg) | ||||
| IR10, 60 | 0 ± 0 | 10 ± 3 | 17 ± 3 | 16 ± 3 |
| IR10 + RES | 0 ± 0 | 1 ± 1* | 3 ± 2* | |
| IR10 + L-NAME | 0 ± 0 | 3 ± 2* | 4 ± 2* | |
| Coronary flow (ml/min) | ||||
| IR10, 60 | 21.2 ± 2.2 | 0 ± 0 | 18.6 ± 2.4 | 19.3 ± 2.2 |
| IR10 + RES | 21.6 ± 1.8 | 0 ± 0 | 19.4 ± 1.4 | |
| IR10 + L-NAME | 16.3 ± 2.2* | 0 ± 0 | 15.4 ± 1.6* | |
| Heart rate (beats/min) | ||||
| IR 10, 60 | 215 ± 5 | 0 ± 0 | 212 ± 6 | 213 ± 4 |
| IR10 + RES | 210 ± 3 | 0 ± 0 | 214 ± 7 | |
| IR10 + L-NAME | 215 ± 4 | 0 ± 0 | 220 ± 4 |
IR10: 30 min of ischemia and 10 min of reperfusion; IR60 only in non-treated group; sys–dia LVP, systolic–diastolic left ventricular pressure; dLVP/dtmax, index of contractility; dLVP/dtmin: index of relaxation; N = 8–10 hearts/group. Values are mean ± SEM.
p < 0.05: IR10 + RES, IR10 + L-NAME vs. IR10, 60.
3.2. Mitochondrial protein tyr-N increases after cardiac IR injury
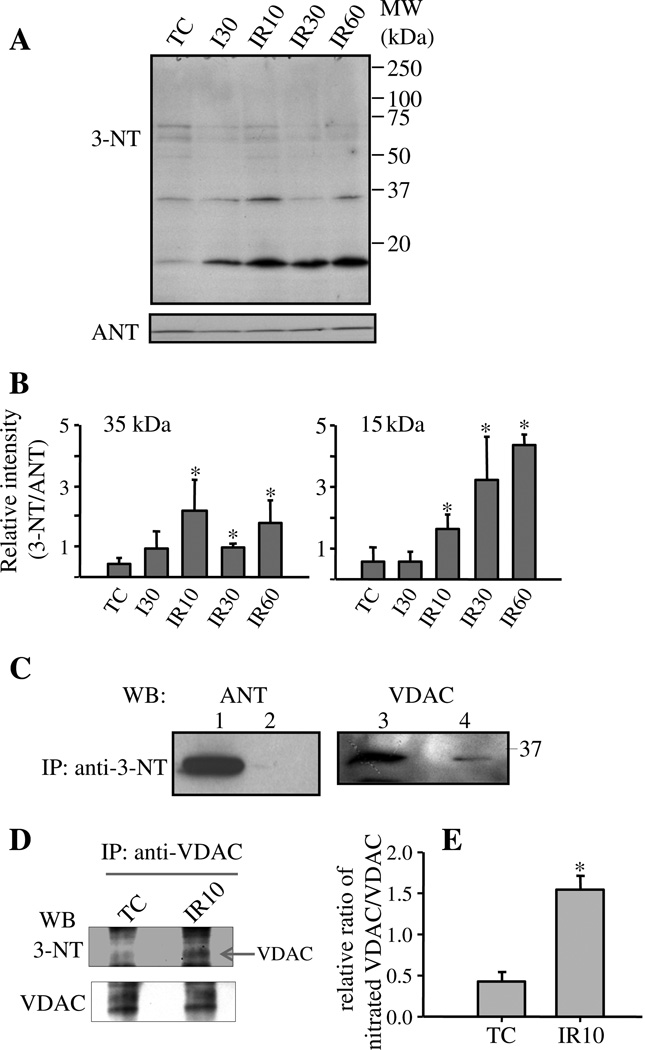
The isolated heart experiments strongly implicated the potential for cardiac protein tyr-N, as a marker of ONOO−-induced dPTMs. Because mitochondrial dysfunction plays a key role in IR injury, we assessed concurrently the extent of mitochondrial protein tyr-N. Mitochondria isolated from TC, I30, IR10, IR30 and IR60 groups were subjected to Western blotting with anti-3-NT antibody. Several immunoreactive bands were detected in all groups (Fig. 2A). Compared to TC, IR caused an increase in anti-3-NT band signal intensity at 35 kDa and 15 kDa. To exclude the possibility that stronger anti-3-NT signals were derived from large amounts of protein loaded, we chose ANT (~35 kDa) protein as the loading control, which was similar in all groups (Fig. 2A). Comparison of the ratio of densities of anti-3-NT bands to anti-ANT bands DHE fluorescence (afu) among all groups (Fig. 2B) clearly confirmed that enhanced anti-3-NT immunoreaction occurred in the IR groups.
Fig. 2.
Assessment of tyr-N of mitochondrial proteins isolated from hearts and identity of tyr-N mitochondrial proteins. A: Assessment of tyr-N of cardiac mitochondrial proteins fromTC and IR groups hearts by Western blot with anti-3-NT antibody. Amounts of loaded mitochondrial proteins were monitored with anti-ANT antibody. B: Densitometric analysis of Western blot of tyr-N of mitochondrial proteins. The densities of anti-3-NT bands at about 35 kDa and 15 kDa, and ANT bands were measured using ImageJ system, after which % density of each anti-3-NT bands was normalized to % density of their corresponding ANT bands. A is a representative blot and B is the mean ± SEM of four experiments. *: p < 0.05 vs. TC group. TC: time control; I: Ischemia; R: Reperfusion. MW: molecular weight. C: Mitochondrial proteins isolated from IR groups were subjected to IP with anti-3-NT antibody followed by Western blots with indicated antibodies. Lanes 1 and 3: total mitochondrial protein lysates (input); lanes 2 and 4: IP with anti-3-NT antibody. MW: molecular weight; WB: Western blot. D: VDAC from mitochondrial protein of TC and IR10 groups was first individually immunoprecipitated with anti-VDAC antibody. The immunoprecipitates were subjected to Western blotting with anti-3-NT antibody and then the membrane was stripped and reblotted with anti-VDAC antibody. E: Densities of anti-3-NT bands and VDAC bands in TC and IR10 groups were measured using ImageJ system, after which % density of each anti-3-NT bands was normalized to % density of their corresponding VDAC bands. WB: Western blot. D is a representative blot and E is the mean ± SEM of three experiments. *: p < 0.05 IR10 vs. TC.
3.3. Identity of tyr-N mitochondrial proteins with specific antibodies and mass spectrometry
To identify the enhanced tyr-N proteins implicated by the Western blot above (Fig. 2A), we subjected the total mitochondrial protein extracts from IR groups to IP with an anti-3-NT antibody. After IP, the immunoprecipitates were subjected to SDS-PAGE to remove the detergent and collect sample for LC–MS/MS assay. After the immunoprecipitates entered the separating gel part of the SDS-PAGE gel from loading wells, the gel was run for 5 min and then silver stained. The silver stained protein areas on the gel that include all sizes of proteins were collected and cut into two parts for LC–MS/MS (Fig. S1 lane 1, Online Supplement). Initially, 13 mitochondrial proteins were obtained from immunoprecipitates of the anti-3-NT (Table S1, Online Supplement). Four of these were polypeptides from the ETC and five were components of the TCA cycle. Notably, two proteins, VDAC and ANT, with molecular weights of about 35 kDa, which are similar to the above Western blot 35 kDa tyr-N protein band size (Fig. 2A), were also obtained. This indicated that these two proteins might be candidates of tyr-N mitochondrial proteins.
We used protein specific antibodies to further identity tyr-N proteins displayed on the Western blots. Based on molecular weights of the candidate proteins listed in Online Supplement Table S1 and on Western blot analyses (Fig. 2A), we focused on identifying whether ANT and/or VDAC was the 35 kDa tyr-N protein. IP assays of mitochondrial protein extracts of IR groups were carried out with anti-3-NT antibody followed by Western blotting with antibodies against VDAC and ANT. Immunoreactive signals were detected when the VDAC antibody was used to probe anti-3-NT immunoprecipitates (Fig. 2C, Lane 4). This corresponded to a similar size 35 kDa band in the total mitochondrial protein lysate probed with anti-VDAC antibody (Fig. 2C, Lane 3, input) or probed with anti-3-NT antibody (Fig. 2A). In contrast, when ANT antibody (Fig. 2C, Lane 2) was used no immunoreactive signal was detected from the anti-3-NT immunoprecipitates. These data strongly implicated VDAC as the 35 kDa protein exhibiting enhanced tyr-N after IR injury. A 15 kDa mitochondrial protein also displayed enhanced tyr-N in IR groups on Western blot and we also found 3 proteins with molecular weights about 15 kDa (see Online Supplement Table S1); identity of these tyr-N proteins is as yet incomplete; our intent here was to focus only on VDAC.
Next we utilized an approach reported by Tran et al. [38] to compare the relative ratio of nitrated VDAC to total VDAC between the TC and IR groups. Based on Fig. 2A results, at the 35 kDa protein, the IR10 group displayed a similar tyr-N level as the IR60 group, but higher than the IR30 group. Therefore, we chose the IR10 group to compare with the TC in this and all other experiments. Mitochondria were isolated as described in Method (see Method 2.3) from TC and IR10 hearts. VDAC was first immunoprecipitated with anti-VDAC antibody (Sigma) from mitochondrial protein lysate of TC and IR10 groups individually. The immunoprecipitates were subjected to Western blotting with anti-3-NT antibody, and then the membranes were stripped and reblotted with anti-VDAC antibody (Cell Signaling). The resulting Western blots showed that similar amounts of VDAC protein were immunoprecipitated from the TC and IR10 groups (Fig. 2D, bottom panel), whereas immunoprecipitated tyr-N VDAC increased in the IR10 group compared to the TC group (Fig. 2D, upper panel). Comparison of the relative ratio of the tyr-N VDAC bands to the total VDAC bands clearly confirmed that IR increased VDAC nitration significantly by approximately 4 fold (Fig. 2E). This result (Fig. 2D, E) was similar to the result of Western blotting with anti-3-NT shown in Fig. 2A and B, which displayed a 35 kDa protein (identified as VDAC) with a higher tyr-N level in the IR than in the TC group.
To further confirm tyr-N of VADC, mitochondrial proteins from TC and IR10 groups were subjected to 2-DE individually, followed by Western blots with anti-3-NT antibody. After blotting with anti-3-NT antibody, the membranes were stripped and then reblotted with anti-VDAC antibody. The results showed that VDAC spots (solid tilted arrows) displayed strong immunoreactivity to anti-3-NT antibody in the IR10 group (Fig. S2B, D, Online Supplement), whereas in the TC group, the VDAC spot (dotted tilted arrows) showed only a slight immunoreactive signal with anti-3-NT antibody (Fig. S2A, C, Online Supplement). Why two VDAC spots showed immunoreactivity to anti-3-NT antibody (solid tilted arrows, Fig. S2, Online Supplement) in the IR10 group is unclear at this time.
3.4. ONOO− induces tyr-N of rVDAC in vitro
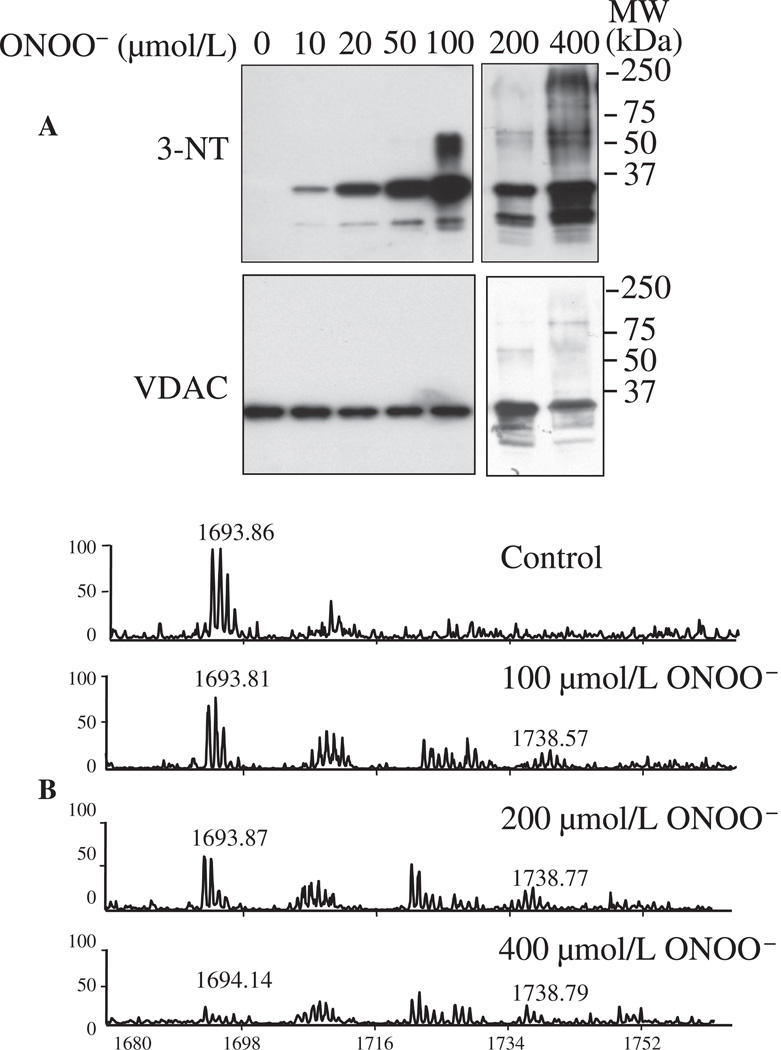
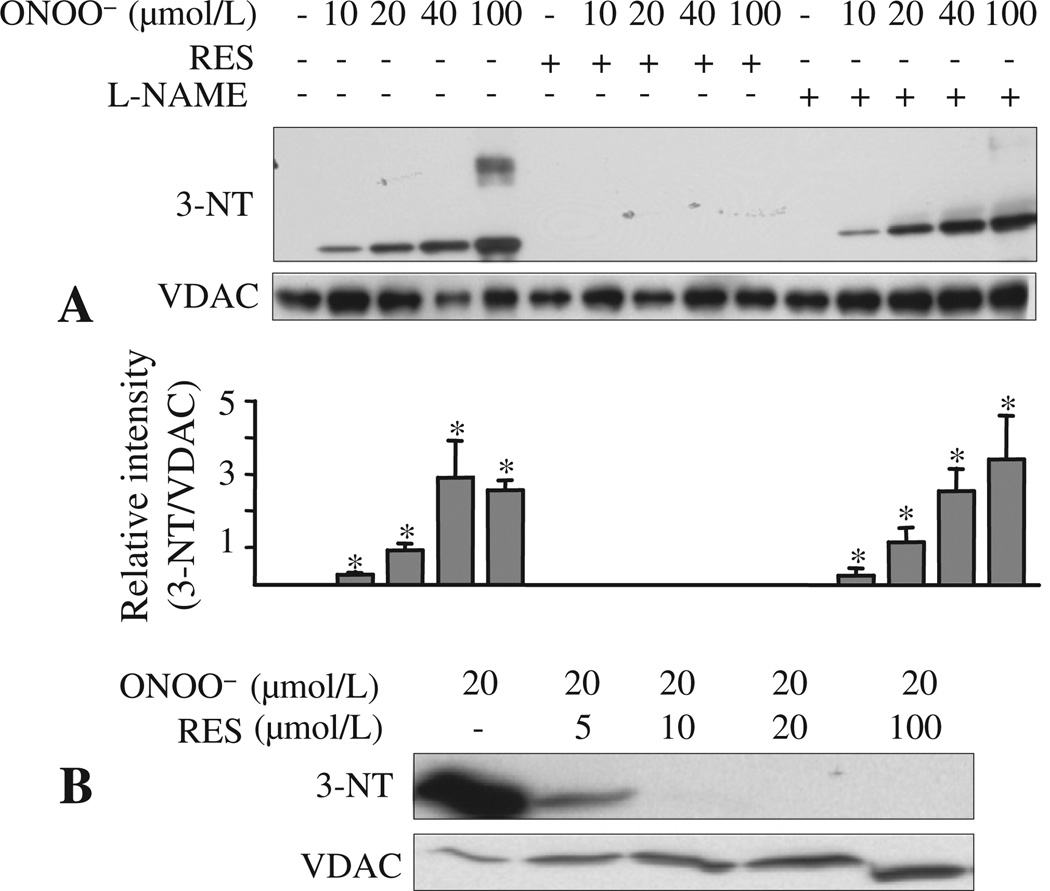
The aim of these experiments was to identify specific tyr-N residues and further confirm tyr-N of VDAC. Purified rVDAC was exposed to ONOO− (0–400 µmol/L) and the ONOO−-treated rVDAC protein was subjected to Western blotting with anti-3-NT antibody. Our approach showed that rVDAC can be nitrated by as low as 10 µmol/L of ONOO− (Fig. 3A); moreover, signal intensity of tyr-N of rVDAC increased as [ONOO−] increased. Some tyr-N of rVDAC protein exhibited slow mobility on the SDS-PAGE gel. In addition to the 35 kDa bands (rVDAC monomer), additional bands with molecular weights of approximately 65 kDa, 100 kDa, and higher appeared. This indicated formation of higher molecular mass species (Fig. 3A) and suggested that tyr-N induced a structural alteration of rVDAC that led to protein oligomerization.
Fig. 3.
Tyr-N of recombinant (r) VDAC induced by ONOO− in vitro. A: rVDAC was incubated with 0–400 µmol/L ONOO− and subjected to Western blot with anti-3-NT antibody. rVDAC loading was monitored with anti-VDAC antibody. B: Mass spectrometric characterization of the peptide fragment 62YRWTEYGLTFTEK74 determined by MALDI-TOF MS. Peaks labeled at m/z 1693 correspond to the peptide containing an unmodified tyr residue and peaks labeled at m/z 1738 correspond to the same peptide as m/z 1693, but with a tyr-N residue, i.e. one that displays a +45 mass unit shift. MW: molecular weight.
In addition, we examined the characteristics of rVDAC incubated with ONOO− by 2-DE. In this experiment, we chose one concentration of ONOO−, i.e. 200 µmol/L, to treat rVDAC. After treatment rVDAC appeared as 4 adjacent spots (Fig. S3B and D, Online Supplement); three of these spots revealed equal signal intensities to anti-3-NT antibody and to anti-VDAC antibody, whereas most of the control (non-nitrated) rVDAC were accumulated at two adjacent spots and showed no immunoreactivity to anti-3-NT antibody (Fig. S3A and C, Online Supplement).
To identify possible tyr-N residues in the rVDAC, we excised ONOO−-untreated (0 µmol/L) and 100, 200, 400 µmol/L ONOO−-treated rVDAC bands on silver stained SDS-PAGE gels, and then subjected them to MALDI-TOF MS. The sensitivity of the MALDI-TOF MS assay is lower than that of the Western blots, because in the MS analysis only 1–2 µl of trypsin-digested sample was loaded on the sample plate, and the laser beam hits peptide fragments partially and randomly. Therefore, for this experiment 100, 200, 400 µmol/L of ONOO− was used to treat rVDAC. Nine of 11 total tyr residues (Y) of rVDAC were detected by MALDI-TOF MS, specifically, 22Y, 62Y, 67Y, 146Y, 153Y, 173Y, 195Y, 225Y and 247Y (Online Supplement Table S2). Among the identified Y-containing peptide fragments, one fragment, 62YRWTEYGLTFTEK74, displayed a +45 mass units' shift from m/z 1693 to 1738 in ONOO−-treated rVDAC (Fig. 3B), consistent with a single NO2 group added to either 62Y or 67Y. In ONOO−-untreated (control) rVDAC, the intensity of spectrum m/z 1693 was highest among all four groups, while the spectrumm/z 1738 was absent. In ONOO−-treated rVDAC, the intensity of spectra m/z 1693 decreased gradually with increasing [ONOO−], while spectra m/z 1738 appeared among the three ONOO−-treated rVDAC groups (Fig. 3B). The site(s) identified in ONOO−-treated rVDAC may or may not be the same as in native VDAC after IR injury.
3.5. Resveratrol or L-NAME treatments reduce mitochondrial protein tyr-N
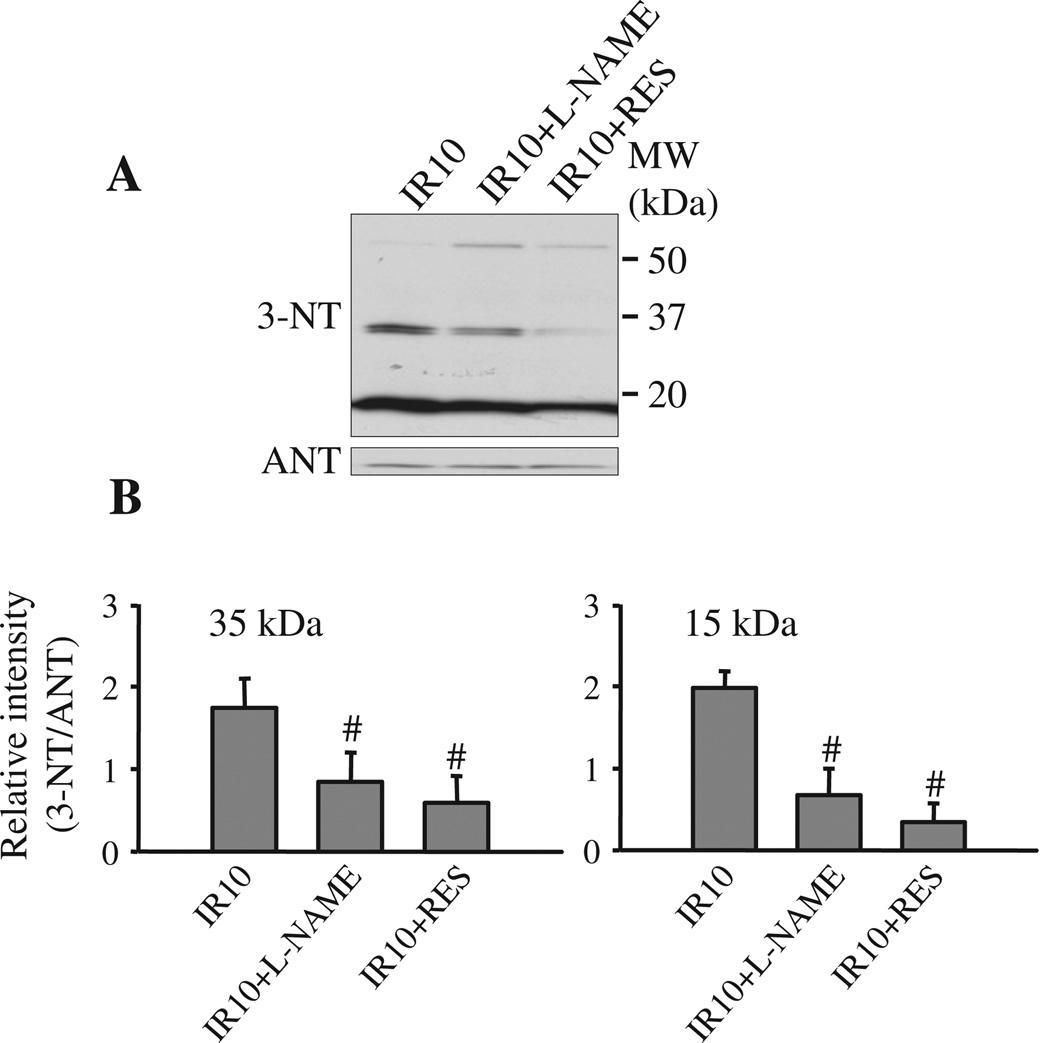
We next examined if the enhanced protein tyr-N during IR is attributable to the increases in ONOO− formation resulting from combined generation of NO• and . Hearts were treated with L-NAME or RES before 30 min ischemia, followed by IR, and tyr-N of mitochondrial proteins was again evaluated as described above (see Results 3.2) by Western blot with anti-3-NT antibody. Both RES and L-NAME reduced tyr-N of 35 kDa (identified as VDAC) and 15 kDa mitochondrial proteins compared to the untreated group (Fig. 4A, B). These data indicated that ONOO− produced during IR injury induced mitochondrial protein nitration, in particular tyr-N of VDAC.
Fig. 4.
Inhibitory effects of L-NAME and resveratrol (RES) on mitochondrial protein tyr-N. A: Isolated hearts were treated without or with L-NAME (100 µmol/L) or RES (10 µmol/L) for 10 min before the onset of ischemia. After reperfusion, mitochondria were isolated and tyr-N of mitochondrial protein was evaluated by Western blot with an anti-3-NT antibody. The amount of mitochondrial protein loaded was assessed by reprobing the membrane with anti-ANT (A, lower panel). B: Densities of anti-3-NT bands at about 35 kDa, 15 kDa and ANT bands were measured using the ImageJ system, and then density of each anti-3-NT bands was normalized to their corresponding ANT band. A is a representative blot and B is means ± SEM of all experiments performed in triplicates. #: p < 0.05, compared to IR10.
To correlate the preventive effects of L-NAME and RES on mitochondrial protein tyr-N with cardioprotection during IR injury, we monitored ONOO− and production online in isolated hearts. L-NAME attenuated DiTyr fluorescence (ONOO−) levels during ischemia (Fig. 5B). We showed previously that L-NAME reduced cold-induced ONOO− production, but that it did not alter emission [24]. Because RES interfered with the DiTyr fluorescence signal, we could only monitor the emission with DHE fluorescence. RES attenuated DHE fluorescence ( emission) induced during ischemia and completely blocked it during reperfusion (Fig. 5A). Reduction of DiTyr/DHE fluorescence by L-NAME and RES was associated with a significant improvement in cardiac function during reperfusion (Table 1). Collectively, reducing production during IR decreased protein tyr-N, and at the same time improved recovery of function on reperfusion.
Fig. 5.
Effects of L-NAME and resveratrol on and ONOO− production in isolated hearts. Levels of and ONOO− were measured online with DHE (A) and L-tyrosine (B), respectively, as described in section 2. DHE/DiTyr fluorescence intensities in arbitrary fluorescence units (afu) were normalized to zero at baseline. *: p < 0.05, compared to IR10 + RES/L-NAME. TC: time control; IR10: Ischemia 30 min, reperfusion 10 min; RES: resveratrol; L-NAME: NG-nitro-L-arginine methyl ester; MW: molecular weight.
3.6. Resveratrol reduces rVDAC tyr-N in vitro
To verify that L-NAME and RES attenuated VDAC tyr-N during IR, rVDAC was incubated with ONOO− (0–100 µmol/L), with or without RES or L-NAME; rVDAC tyr-N was evaluated again by Western blot using anti-3-NT antibody. Incubation with ONOO− resulted in tyr-N of rVDAC (Fig. 6A). RES, 100 µmol/L, abolished tyr-N of rVDAC, while L-NAME (100 µmol/L) had no effect. The inhibitory effect of RES on ONOO− induced rVDAC tyr-N was concentration-dependent, such that greater than 10 µmol/L RES inhibited rVDAC tyr-N induced by 20 µmol/L of ONOO− (Fig. 6B). This suggests that L-NAME inhibits only generation of NO• in the presence of NOS, whereas RES scavenges both and ONOO−.
Fig. 6.
Inhibition of recombinant (rVDAC) tyr-N by L-NAME and resveratrol (RES) in vitro. A: rVDAC was incubated with ONOO− (0–100 µmol/L), with or without L-NAME (100 µmol/L) or RES (100 µmol/L), and subjected to Western blot with an anti-3-NT antibody. rVDAC loading was monitored with VDAC antibody. B: Western blot analysis of rVDAC incubated with 20 µmol/L of ONOO− without or with 5–100 µmol/L RES with anti-3-NT antibody. Values are mean ± SEM of three experiments. *: p < 0.05 vs. non-treated VDAC.
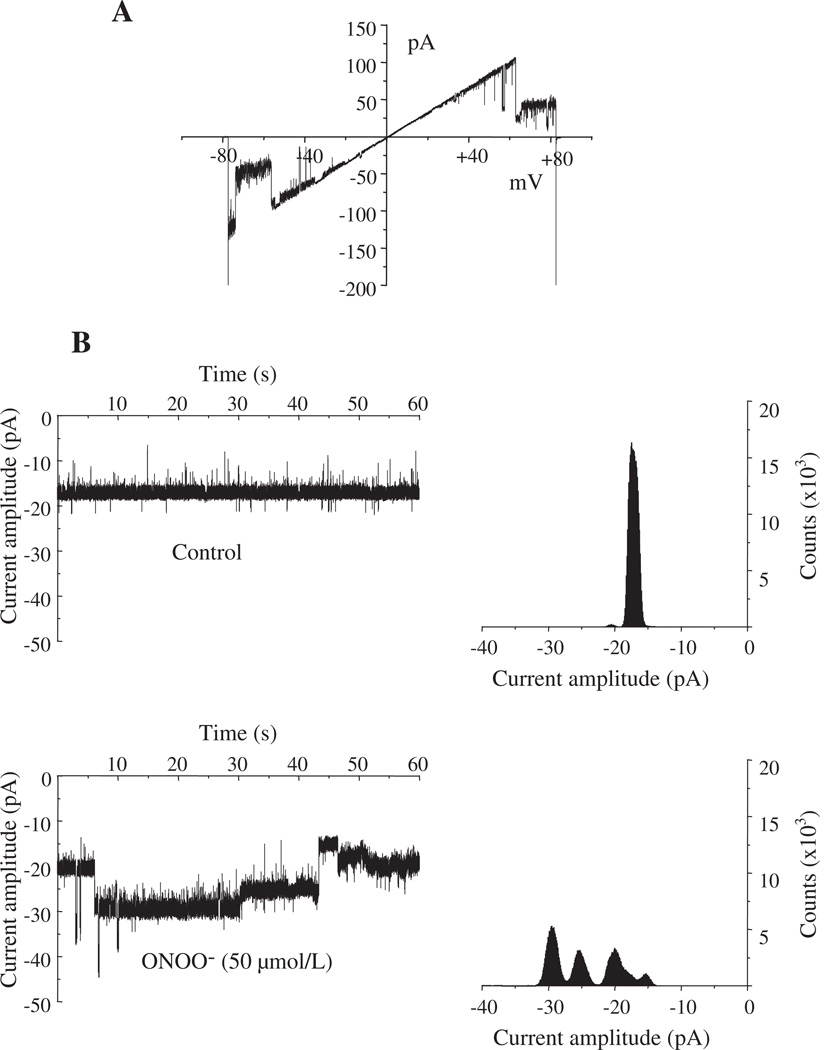
3.7. ONOO− enhances rVDAC conductance
We next investigated if ONOO− altered rVDAC activity by recording single channel conductance after reconstituting rVDAC into PLB. We assumed that ONOO− is the major species inducing tyr-N of rVDAC. rVDAC exhibited a characteristic voltage-dependent conductance (Fig. 7A), which is similar to native VDAC (Fig. S4, right panel, Online Supplement) [37]. The effect of ONOO− on rVDAC was monitored at a membrane potential of −10 mV. Under control conditions, primary rVDAC chord conductance was approximately 1.8 nS as depicted by the unitary current amplitude of 18 pA (Fig. 7B). When 50 µmol/L ONOO− was added, channel activity was enhanced with multiple higher conductance states, as evidenced by current amplitudes of −25 pA and −30 pA (Fig. 7B). These experiments demonstrate that 50 µmol/L ONOO− directly enhanced rVDAC conductance.
Fig. 7.
Effects of ONOO− on recombinant (r) VDAC channel activity. A: Characteristic voltage-dependence of rVDAC in response to a ramp voltage protocol from −80 to +80 mV under control conditions. B: Effects of adding ONOO− on rVDAC activity. Current traces show in control and after adding 50 µmol/L ONOO− obtained at a membrane potential of −10 mV. In control, primary conductance was 1.8 nS. Application of ONOO− triggered enhanced opening of VDAC as evidenced by the multiple conductance states. Corresponding all-points amplitude histograms are also shown.
4. Discussion
The functional and biological consequences of tyr-N depend on which proteins are nitrated as well as on the extent and sites of nitration in a given protein [39]. Identifying tyr-N of mitochondrial proteins should help to define the potential pathogenic role of this dPTM in disease states and to provide guidance on how and where to target therapies against these dPTMs. Although there are prior reports [8,17] that IR injury induces tyr-N of mitochondrial proteins, only a few proteins so far have been identified, and there has been no direct evidence for VDAC nitration or for specifically nitrated tyr residues. Here we used proteomic analyses and IP assays to search for specific proteins that undergo enhanced tyr-N during IR in isolated perfused hearts. We found that VDAC, a key and abundant outer mitochondrial membrane (OMM) protein was targeted for tyr-N during IR. Consistent with our isolated heart results, the in vitro experiments confirmed that rVDAC can be nitrated by ONOO− at either the 62Y or 67Y site. In addition, our results indicate that rVDAC tyr-N may trigger oligomerization of rVDAC and that ONOO− enhances rVDAC channel conductance. Thus, nitration of VDAC likely induces alterations in both its structure and function.
To further explore mechanisms of tyr-N induction, RES or L-NAME was perfused into isolated hearts before ischemia as well as applied to rVDAC protein. In isolated hearts, both RES and L-NAME decreased VDAC tyr-N, inhibited formation of ONOO−, and improved cardiac function after IR. In vitro, RES, but not L-NAME, directly prevented tyr-N of rVDAC induced by ONOO−. Lastly, in vitro tyr-N of rVDAC confirmed our ex vivo results that RES indeed can scavenge ONOO− to reduce tyr-N. Overall, our study suggests that VDAC tyr-N occurs during IR injury and that scavenging and attenuating NO• generation reduces tyr-N of VDAC during IR.
4.1. Sites of VDAC tyr-N and functional change
Previous studies suggested that VDAC tyr-N can occur in mouse hearts during IR injury [17], in old-aged rat hearts [31], and in rat hippocampal tissue after acute smoke inhalation [40]. In those studies, VDAC was suggested as a nitrated protein based on MS analysis for anti-3-NT positive protein spots on 2-DE gels or anti-3-NT IP protein mixtures. However, there was no direct evidence of VDAC tyr-N or of attenuated VDAC tyr-N by inhibitors/scavengers of ONOO−. Moreover, the impact of tyr-N on rVDAC channel conductance was unknown. Based on our multifaceted approaches involving MS analysis, IP, and electrophysiology, we provide firm evidence that VDAC tyr-N occurs during cardiac IR injury. Moreover, tyr-N appears to oligomerize rVDAC and ONOO− increases the rVDAC channel conductance state. We also newly report that partial prevention of VDAC tyr-N is associated with improved cardiac function.
Protein tyr-N is a selective process, in so far as not all proteins can be nitrated. The potential for a protein to become nitrated is affected by the cellular and subcellular localization of a given protein [41]. Most nitrating agents are short-lived [42,43], so proteins to be nitrated must lie in close proximity to the site of ONOO− formation. Because NO• is a diffusible molecule and is diffusion limited and generated primarily in mitochondria, ONOO− is most likely formed in mitochondria. As the most abundant protein in the OMM, VDAC is likely highly exposed to ONOO−. Thus it is probable that VDAC is an easy target during IR injury for attack by ONOO− with subsequent tyr-N.
We examined for a direct functional impact of ONOO− on VDAC by treating rVDAC incorporated into PLB with ONOO− and found that 50 µmol/L ONOO− enhanced rVDAC channel activity, as shown as multiple higher conductance states. This may allow increased transport of metabolites across a “leakier” OMM. It is possible that the higher conductance states permit release of cytochrome c across the OMM via VDAC, independent of mitochondrial permeability transition pore (mPTP). How ONOO− enhances VDAC channel conductance is unclear.
Despite our finding that ONOO− induced higher rVDAC channel conductance states, it is not established if higher or lower VDAC channel conductance states contribute to impaired mitochondrial function during cardiac IR injury. We reported recently a concentration-dependent, biphasic inhibitory effect of exogenous NO• on cardiac VDAC [37]. Peak inhibition occurred at a physiologically relevant level of NO•, and this correlated with a delay in mitochondrial permeability transition pore (mPTP) opening. Consequently, a NO•-induced delay in mPTP opening, in tandem with an induced low conducting state of VDAC, could potentially contribute to myocardial protection. This would then support our hypothesis that the ONOO−-induced high conductance state of VDAC is detrimental to cardiac function, whereas the putatively protective NO•-induced low conductance state is beneficial.
It is likely that ONOO−-induced VDAC tyr-N leads to structural alterations because tyr-N renders the neutral tyr residue a charged residue; this could alter the tertiary structure of VDAC. Indeed, we found that nitration of rVDAC caused it to oligomerize into dimers or trimers (Figs. 3, 6). Keinan [44] reported that oligomerization of VDAC1 was associated with cytochrome c release and apoptosis induced by various stimuli in many cell types. Although we did not show directly that VDAC tyr-N is associated with cell death, our recent data indicate that IR-induced release of cytochrome c [45] occurs at the same time as tyr-N of mitochondrial proteins (and in particular, VDAC based on our current study). Thus our study suggests that VDAC tyr-N may be associated with mitochondrial-mediated cell death.
A rVDAC residue that is a target of nitration was narrowed to either 62Y or 67Y. The specific tyr residues capable of tyr-N are determined by both the protein's structure and the local environment around the tyr (Y) residues [41]. Most tyr-N residues have been found in loop structures in the vicinity of turn-inducing amino acids such as proline or glycine [41,46]. The proximity of negatively charge residues, such as glutamic or aspartic acids, also benefits nitration of tyr [41,46]. In our tyr-N containing peptide sequence, 67Y is adjacent to the turn-inducing residue glycine and the negatively charge residue glutamic acid, which conforms well to common features of structural determinants of protein tyr-N. In addition, based on elucidation of the three-dimensional structure of VDAC, 67Y is located in the loop region between the β3 and β4 strands [47]. Consequently, 67Y is likely the nitrated residue in the rVDAC, but for now nitration of 62Y cannot be excluded.
4.2. Reduced mitochondrial protein tyr-N occurs with improved recovery after IR injury
There is now ample evidence that RNS/ROS-induced protein tyr-N is a critical factor underlying cardiac IR injury [19,48]. Inhibiting NOS activity reduced both the level of protein nitration and the extent of IR injury [5]. Our present and prior studies, in which we measured ONOO−, i.e. DiTyr fluorescence, in the left ventricle [24] and in coronary effluent [4], demonstrate that inhibiting NOS with L-NAME can attenuate the increase in ONOO− induced during IR injury (Fig. 5B). To support these prior heart studies, we report here that L-NAME also decreased IR-induced mitochondrial proteins tyr-N (Fig. 4A), while improving functional recovery during reperfusion (Table 1). The rapid fall in DiTyr fluorescence signal for ONOO− on reperfusion in isolated hearts is due to washout of free DiTyr in the effluent. Overall, reducing VDAC tyr-N by attenuating NO• generation, and thus ONOO−, by L-NAME may play a supportive role in ameliorating IR injury.
To further evaluate the effects of mitochondrial protein tyr-N in IR injury, we tested effects of an inhibitor/scavenger of RNS/ROS, i.e. RES, on reducing protein tyr-N and improving functional recovery. RES is purported to have anti-inflammatory, anti-aging, anti-neoplastic, and cardioprotective properties [49]. RES may react directly with ONOO− to act as a functional ONOO− scavenger in vitro and thereby to prevent or mitigate ONOO−-induced tyr-N [50]. Although RES reduced hypoxia-induced emission in cardiomyocytes has been reported [51], RES treatment during ischemia in intact hearts involved scavenging or inhibiting , with a subsequently decreased mitochondrial protein tyr-N, has not been reported.
We report here for the first time that RES treatment partially inhibited DHE fluorescence (emission of ) induced during ischemia, and abolished it during reperfusion in intact perfused hearts, as well as reduced mitochondrial protein tyr-N. Goh et al. [51] reported in isolated perfused rat hearts that RES treatment during IR reduced LDH release, improved post-ischemic recovery of LV function, and increased Akt and p38MAPK activity. In our study, we also found that RES, given just before ischemia and present in hearts during ischemia, improved cardiac function after IR, which is consistent with the report of Goh et al. [51]. In addition, our in vitro experiments with rVDAC showed that RES directly, and completely, inhibited tyr-N of rVDAC induced by ONOO− while L-NAME did not. Thus, RES is likely an inhibitor of emission, and/or a scavenger of ONOO−; it not only reduced tyr-N of mitochondrial proteins, specifically VDAC after IR injury, but also improved cardiac contractility and relaxation. Although RES has been shown to stimulate NOS to generate NO• when given to precondition hearts against IR injury [43], it is likely in our experiments that much of NO• produced was converted to ONOO− because of abundant , which then leads to nitration of VDAC.
We have implied that ONOO− is in large part responsible for the observed tyr-N and that the major effect of ONOO− is to induce tyr-N. Thus, limitations of our study are that ONOO− may induce PTMs of not only tyr, but also of other aromatic residues and that tyr may be additionally nitrated by non-ONOO− oxidants, such as other nitrites and peroxidases.
In summary, we have identified VDAC as one of the mitochondrial proteins that undergoes enhanced tyr-N during IR injury. L-NAME and RES likely attenuate VDAC tyr-N in the ex vivo model, but only RES abolished ONOO−-induced VDAC nitration in vitro, because there was likely no NO• present. Moreover, inhibition of tyr-N of VDAC by L-NAME, and especially by RES, not only demonstrates that VDAC tyr-N occurs, but also that attenuating ONOO− production reduces tyr-N of VDAC and concomitantly improves cardiac function after IR. Tyr-N in ONOO−-treated rVDAC is selective and site-specific, and was accompanied by rVDAC protein oligomerization. ONOO−-treated rVDAC also leads to an enhanced channel conductance state. The results obtained in this study indicate that VDAC, possibly with other unidentified mitochondrial tyr-N proteins, plays a critical role in mediating cardiac dysfunction during IR injury. Targeting against specific protein tyr-N sites may represent a unique way to protect mitochondrial proteins and organs against oxidative damage.
Supplementary Material
Acknowledgement
The authors kindly acknowledge Jim Heisner for conducting the isolated heart experiments, Deena Phadnis for technical assistance, and Drs. Martin Bienengraeber and Meetha Medhora for their helpful comments on reviewing this manuscript.
Sources of funding
This work was supported in part by grants from the National Institutes of Health (HL095122 to AKSC, HL089514 to DFS, PO1GM066730 to Z.J. Bosnjak PI), the American Heart Association (0855940G to DFS); and the Veterans Administration (Merit Review 8204-05P to DFS).
Abbreviations
superoxide
- NO•
nitric oxide
- ONOO−
peroxynitrite
- tyr
tyrosine
- tyr-N
tyrosine nitration
- IR
ischemia reperfusion
- IP
immunoprecipitation
- VDAC
voltage-dependent anion channel
- RES
resveratrol
- RNS
reactive nitrogen species
- L-NAME NG
nitro-L-arginine methyl ester
- PTM
post translational modifications
- ROS
reactive oxygen species
- ETC
electron transport chain
- 3-NT
3-nitrotyrosine
- DHE
dihydroethidium
- DiTyr
dityrosine
- LVP
left ventricular pressure
- PLB
planar lipid bilayers
Footnotes
Footnotes: Preliminary reports of this work: FASEB J LB589, 2010; FASEB J 639.14, 2011; FASEB J 678.19, 2012.
Disclosures
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbabio.2012.06.004.
References
- 1.Stowe DF, Camara AK. Mitochondrial reactive oxygen species production in excitable cells: modulators of mitochondrial and cell function. Antioxid. Redox Signal. 2009;11:1373–1414. doi: 10.1089/ars.2008.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camara AK, Lesnefsky EJ, Stowe DF. Potential therapeutic benefits of strategies directed to mitochondria. Antioxid. Redox Signal. 2010;13:279–347. doi: 10.1089/ars.2009.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burwell LS, Brookes PS. Mitochondria as a target for the cardioprotective effects of nitric oxide in ischemia–reperfusion injury. Antioxid. Redox Signal. 2008;10:579–599. doi: 10.1089/ars.2007.1845. [DOI] [PubMed] [Google Scholar]
- 4.Novalija E, Varadarajan SG, Camara AK, An J, Chen Q, Riess ML, Hogg N, Stowe DF. Anesthetic preconditioning: triggering role of reactive oxygen and nitrogen species in isolated hearts. Am. J. Physiol. Heart Circ. Physiol. 2002;283:H44–H52. doi: 10.1152/ajpheart.01056.2001. [DOI] [PubMed] [Google Scholar]
- 5.Zweier JL, Fertmann J, Wei G. Nitric oxide and peroxynitrite in postischemic myocardium. Antioxid. Redox Signal. 2001;3:11–22. doi: 10.1089/152308601750100443. [DOI] [PubMed] [Google Scholar]
- 6.Radi R, Cassina A, Hodara R, Quijano C, Castro L. Peroxynitrite reactions and formation in mitochondria. Free Radic. Biol. Med. 2002;33:1451–1464. doi: 10.1016/s0891-5849(02)01111-5. [DOI] [PubMed] [Google Scholar]
- 7.Kevin LG, Camara AK, Riess ML, Novalija E, Stowe DF. Ischemic preconditioning alters real-time measure of O2 radicals in intact hearts with ischemia and reperfusion. Am. J. Physiol. Heart Circ. Physiol. 2003;284:H566–H574. doi: 10.1152/ajpheart.00711.2002. [DOI] [PubMed] [Google Scholar]
- 8.Wang P, Zweier JL. Measurement of nitric oxide and peroxynitrite generation in the postischemic heart. Evidence for peroxynitrite-mediated reperfusion injury. J. Biol. Chem. 1996;271:29223–29230. doi: 10.1074/jbc.271.46.29223. [DOI] [PubMed] [Google Scholar]
- 9.Yasmin W, Strynadka KD, Schulz R. Generation of peroxynitrite contributes to ischemia–reperfusion injury in isolated rat hearts. Cardiovasc. Res. 1997;33:422–432. doi: 10.1016/s0008-6363(96)00254-4. [DOI] [PubMed] [Google Scholar]
- 10.Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am. J. Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 11.Agarwal R, MacMillan-Crow LA, Rafferty TM, Saba H, Roberts DW, Fifer EK, James LP, Hinson JA. Acetaminophen-induced hepatotoxicity in mice occurs with inhibition of activity and nitration of mitochondrial manganese superoxide dismutase. J. Pharmacol. Exp. Ther. 2011;337:110–116. doi: 10.1124/jpet.110.176321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacMillan-Crow LA, Crow JP, Thompson JA. Peroxynitrite-mediated inactivation of manganese superoxide dismutase involves nitration and oxidation of critical tyrosine residues. Biochemistry. 1998;37:1613–1622. doi: 10.1021/bi971894b. [DOI] [PubMed] [Google Scholar]
- 13.Souza JM, Peluffo G, Radi R. Protein tyrosine nitration–functional alteration or just a biomarker? Free Radic. Biol. Med. 2008;45:357–366. doi: 10.1016/j.freeradbiomed.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Gow AJ, Duran D, Malcolm S, Ischiropoulos H. Effects of peroxynitrite-induced protein modifications on tyrosine phosphorylation and degradation. FEBS Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 15.Halliwell GJ, Free B. Radicals in Biology and Medicine. New York: Oxford University Press; 1999. [Google Scholar]
- 16.Aulak KS, Miyagi M, Yan L, West KA, Massillon D, Crabb JW, Stuehr DJ. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12056–12061. doi: 10.1073/pnas.221269198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu B, Tewari AK, Zhang L, Green-Church KB, Zweier JL, Chen YR, He G. Proteomic analysis of protein tyrosine nitration after ischemia reperfusion injury: mitochondria as the major target. Biochim. Biophys. Acta. 2009;1794:476–485. doi: 10.1016/j.bbapap.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turko IV, Murad F. Protein nitration in cardiovascular diseases. Pharmacol. Rev. 2002;54:619–634. doi: 10.1124/pr.54.4.619. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X, He G, Chen YR, Pandian RP, Kuppusamy P, Zweier JL. Endothelium-derived nitric oxide regulates postischemic myocardial oxygenation and oxygen consumption by modulation of mitochondrial electron transport. Circulation. 2005;111:2966–2972. doi: 10.1161/CIRCULATIONAHA.104.527226. [DOI] [PubMed] [Google Scholar]
- 21.Zhu X, Liu B, Zhou S, Chen YR, Deng Y, Zweier JL, He G. Ischemic preconditioning prevents in vivo hyperoxygenation in postischemic myocardium with preservation of mitochondrial oxygen consumption. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H1442–H1450. doi: 10.1152/ajpheart.00256.2007. [DOI] [PubMed] [Google Scholar]
- 22.Peluffo G, Radi R. Biochemistry of protein tyrosine nitration in cardiovascular pathology. Cardiovasc. Res. 2007;75:291–302. doi: 10.1016/j.cardiores.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Aldakkak M, Stowe DF, Heisner JS, Riess ML, Camara AK. Adding ROS quenchers to cold K+ cardioplegia reduces superoxide emission during 2-hour global cold cardiac ischemia. J. Cardiovasc. Pharmacol. Ther. 2012;17:93–101. doi: 10.1177/1074248410389815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camara AK, Riess ML, Kevin LG, Novalija E, Stowe DF. Hypothermia augments reactive oxygen species detected in the guinea pig isolated perfused heart. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H1289–H1299. doi: 10.1152/ajpheart.00811.2003. [DOI] [PubMed] [Google Scholar]
- 25.Das S, Tosaki A, Bagchi D, Maulik N, Das DK. Resveratrol-mediated activation of cAMP response element-binding protein through adenosine A3 receptor by Akt-dependent and -independent pathways. J. Pharmacol. Exp. Ther. 2005;314:762–769. doi: 10.1124/jpet.105.084285. [DOI] [PubMed] [Google Scholar]
- 26.Riess ML, Camara AK, Kevin LG, An J, Stowe DF. Reduced reactive O2 species formation and preserved mitochondrial NADH and [Ca2+] levels during short-term 17 degrees C ischemia in intact hearts. Cardiovasc. Res. 2004;61:580–590. doi: 10.1016/j.cardiores.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Beckman JS, Chen J, Ischiropoulos H, Crow JP. Oxidative chemistry of peroxynitrite. Methods Enzymol. 1994;233:229–240. doi: 10.1016/s0076-6879(94)33026-3. [DOI] [PubMed] [Google Scholar]
- 28.Haumann J, Dash RK, Stowe DF, Boelens AD, Beard DA, Camara AK. Mitochondrial free [Ca2+] increases during ATP/ADP antiport and ADP phosphorylation: exploration of mechanisms. Biophys. J. 2010;99:997–1006. doi: 10.1016/j.bpj.2010.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinen A, Aldakkak M, Stowe DF, Rhodes SS, Riess ML, Varadarajan SG, Camara AK. Reverse electron flow-induced ROS production is attenuated by activation of mitochondrial Ca2+-sensitive K+ channels. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H1400–H1407. doi: 10.1152/ajpheart.00198.2007. [DOI] [PubMed] [Google Scholar]
- 30.Aldakkak M, Stowe DF, Cheng Q, Kwok WM, Camara AK. Mitochondrial matrix K+ flux independent of large-conductance Ca2+-activated K+ channel opening. Am. J. Physiol. Cell Physiol. 2010;298:C530–C541. doi: 10.1152/ajpcell.00468.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanski J, Behring A, Pelling J, Schoneich C. Proteomic identification of 3-nitrotyrosine-containing rat cardiac proteins: effects of biological aging. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H371–H381. doi: 10.1152/ajpheart.01030.2003. [DOI] [PubMed] [Google Scholar]
- 32.Newman DK, Hoffman S, Kotamraju S, Zhao T, Wakim B, Kalyanaraman B, Newman PJ. Nitration of PECAM-1 ITIM tyrosines abrogates phosphorylation and SHP-2 binding. Biochem. Biophys. Res. Commun. 2002;296:1171–1179. doi: 10.1016/s0006-291x(02)02060-0. [DOI] [PubMed] [Google Scholar]
- 33.Halligan BD, Greene AS. Visualize: a free and open source multifunction tool for proteomics data analysis. Proteomics. 2011;11:1058–1063. doi: 10.1002/pmic.201000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelhardt H, Meins T, Poynor M, Adams V, Nussberger S, Welte W, Zeth K. High-level expression, refolding and probing the natural fold of the human voltage-dependent anion channel isoforms I and II. J. Membr. Biol. 2007;216:93–105. doi: 10.1007/s00232-007-9038-8. [DOI] [PubMed] [Google Scholar]
- 36.Murray J, Taylor SW, Zhang B, Ghosh SS, Capaldi RA. Oxidative damage to mitochondrial complex I due to peroxynitrite: identification of reactive tyrosines by mass spectrometry. J. Biol. Chem. 2003;278:37223–37230. doi: 10.1074/jbc.M305694200. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Q, Sedlic F, Pravdic D, Bosnjak ZJ, Kwok WM. Biphasic effect of nitric oxide on the cardiac voltage-dependent anion channel. FEBS Lett. 2011;585:328–334. doi: 10.1016/j.febslet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran MH, Yamada K, Nakajima A, Mizuno M, He J, Kamei H, Nabeshima T. Tyrosine nitration of a synaptic protein synaptophysin contributes to amyloid beta-peptide-induced cholinergic dysfunction. Mol. Psychiatry. 2003;8:407–412. doi: 10.1038/sj.mp.4001240. [DOI] [PubMed] [Google Scholar]
- 39.Koeck T, Stuehr DJ, Aulak KS. Mitochondria and regulated tyrosine nitration. Biochem. Soc. Trans. 2005;33:1399–1403. doi: 10.1042/BST0331399. [DOI] [PubMed] [Google Scholar]
- 40.Lee HM, Reed J, Greeley GH, Jr, Englander EW. Impaired mitochondrial respiration and protein nitration in the rat hippocampus after acute inhalation of combustion smoke. Toxicol. Appl. Pharmacol. 2009;235:208–215. doi: 10.1016/j.taap.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abello N, Kerstjens HA, Postma DS, Bischoff R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 2009;8:3222–3238. doi: 10.1021/pr900039c. [DOI] [PubMed] [Google Scholar]
- 42.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. U. S. A. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koppenol WH, Moreno JJ, Pryor WA, Ischiropoulos H, Beckman JS. Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem. Res. Toxicol. 1992;5:834–842. doi: 10.1021/tx00030a017. [DOI] [PubMed] [Google Scholar]
- 44.Keinan N, Tyomkin D, Shoshan-Barmatz V. Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol. Cell. Biol. 2010;30:5698–5709. doi: 10.1128/MCB.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aldakkak M, Camara AK, Heisner JS, Yang M, Stowe DF. Ranolazine reduces Ca2+ overload and oxidative stress and improves mitochondrial integrity to protect against ischemia reperfusion injury in isolated hearts. Pharmacol. Res. 2011;64:381–392. doi: 10.1016/j.phrs.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 47.Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi Y, Sawa Y, Ohtake S, Fukuyama N, Nakazawa H, Matsuda H. Peroxynitrite formation from human myocardium after ischemia–reperfusion during open heart operation. Ann. Thorac. Surg. 2001;72:571–576. doi: 10.1016/s0003-4975(01)02668-6. [DOI] [PubMed] [Google Scholar]
- 49.Camara AK, Bienengraeber M, Stowe DF. Mitochondrial approaches to protect against cardiac ischemia and reperfusion injury. Front. Physiol. 2011;2:13. doi: 10.3389/fphys.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holthoff JH, Woodling KA, Doerge DR, Burns ST, Hinson JA, Mayeux PR. Resveratrol, a dietary polyphenolic phytoalexin, is a functional scavenger of peroxynitrite. Biochem. Pharmacol. 2010;80:1260–1265. doi: 10.1016/j.bcp.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goh SS, Woodman OL, Pepe S, Cao AH, Qin C, Ritchie RH. The red wine antioxidant resveratrol prevents cardiomyocyte injury following ischemia–reperfusion via multiple sites and mechanisms. Antioxid. Redox Signal. 2007;9:101–113. doi: 10.1089/ars.2007.9.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.