Abstract
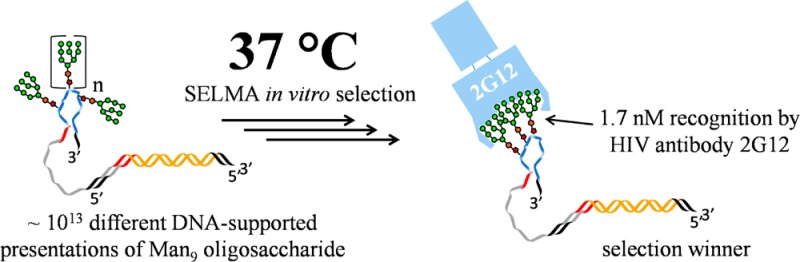
SELMA (SELection with Modified Aptamers) is a directed evolution method which can be used to develop DNA-supported clusters of carbohydrates in which the geometry of clustering is optimized for strong recognition by a lectin of interest. Herein, we report a modification of SELMA which results in glycoclusters which achieve dramatically stronger target recognition (100-fold) with dramatically fewer glycans (2–3-fold). Our first applications of SELMA yielded clusters of 5–10 oligomannose glycans which were recognized by broadly neutralizing HIV antibody 2G12 with moderate affinities (150–500 nM Kd’s). In the present manuscript, we report glycoclusters containing just 3–4 glycans, which are recognized by 2G12 with Kd’s as low as 1.7 nM. These glycoclusters are recognized by 2G12 as tightly as is the HIV envelope protein gp120, and they are the first constructs to achieve this tight recognition with the minimal number of Man9units (3–4) necessary to occupy the binding sites on 2G12. They are thus of great interest as immunogens which might elicit broadly neutralizing antibodies against HIV.
Monoclonal antibody 2G12, isolated from HIV+ patient serum in 1996,1 neutralizes a broad range of HIV isolates2 and is protective in animal models of HIV infection.3 2G12 binds to a cluster of high-mannose (Man5–9GlcNAc2) glycans on HIV envelope protein gp120,1,4 and synthetic glycoclusters which closely mimic this epitope are of interest as immunogens which may be able to elicit a 2G12-like antibody response through vaccination.
There have been many attempts to design clusters of oligomannose glycans which mimic the 2G12 epitope.5 Chemical synthesis has enabled construction of well-defined structures in which glycans are mounted on numerous backbones, including cyclic peptides,5d,5f PNA,5i,5m dendrimers,5h and Qβ phage particles.5j Additionally, yeast strains have been engineered to express primarily high mannose carbohydrates on their surface.5g Unfortunately, none of these immunogens has been used successfully to raise a 2G12-like antibody response in vivo. In the best cases, when mannose binding antibodies have been generated, their binding to gp120 or neutralization of HIV in vitro has still been weak or undetectable. Among several reasons for these failures is the likelihood that the clustering of oligomannose carbohydrates present in these immunogens did not sufficiently resemble the 2G12 epitope.5f,5g,5j,5l
We have attempted to develop immunogens with optimized clustering of carbohydrates for more faithful mimicry of the 2G12 epitope by using the antibody to recognize and select the best gp120 mimics from among a very diverse library. To achieve this, we have developed a new selection method, termed SELMA (SELection with Modified Aptamers, Figure 1), which uses diverse DNA backbones to cluster the glycans in various ways.6 The library is constructed using copper assisted alkyne/azide cycloaddition (CuAAAC) chemistry7 to attach glycans to a library of random DNA sequences containing alkynyl bases. In single-stranded form, each DNA sequence clusters the glycans in a unique geometry, and the clusters which are selected from the library by binding to the target lectin (2G12 in this case) are amplified by PCR to generate a new library for further selection. The process is then repeated for several cycles with increasingly stringent selection conditions. We have previously reported6 the use of this method to generate clusters of 7–10 oligomannose glycans which were moderately good mimics of the 2G12 epitope; our previous constructs were recognized by 2G12 with 150–500 nM Kd’s, whereas the HIV envelope protein, gp120, is recognized much more tightly, with a Kd of ∼6–9 nM.8 Moreover, only 3–4 glycans are likely involved in gp120–2G12 recognition. It is thus desirable that artificial constructs mimicking this epitope would achieve similar low nanomolar 2G12 recognition with just a few glycans.
Figure 1.
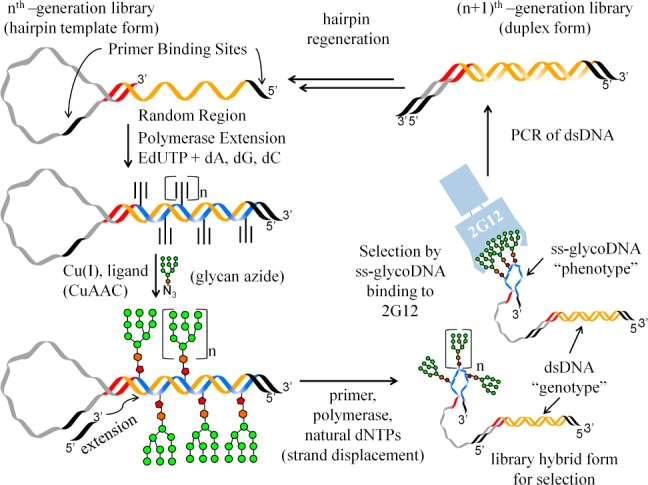
Overview of SELMA (SELection with Modified Aptamers). Selection begins with a library of DNA containing a template for the random sequence region (orange/blue section), flanked by primer binding sites and a hairpin structure terminating in a 3′ self-complementary patch of sequence. Polymerase extension with alkynyl base EdUTP substituted for TTP results in alkyne incorporation in the random region across from A’s in the template. A click reaction with glycan azide positions glycans across from A’s in the template, and then a primer is annealed inside the hairpin and extended with all natural dNTPs, displacing the glycosylated strand. The glycosylated ssDNA strand then folds in a sequence dependent manner, and the dsDNA region contains a copy of the same sequence as “natural” DNA, available for PCR. After selection by binding to immobilized target, the dsDNA of winning sequences is amplified, the hairpin section is rebuilt (not pictured), and the process is repeated.
In the current study, we examined the effect of running the selection step (binding to 2G12) at 37 °C rather than room temperature and found that the higher temperature led to dramatically improved results. Figure 2 shows the changing library multivalency profile throughout two selections carried out with the same starting library of ∼1013 sequences, one at room temperature and the other at 37 °C. In the room temperature selection (lanes 3–6), one could see a trend toward increased bands of high MW as the selection progressed (orange arrow), indicating increased multivalency. By contrast, the selection at 37 °C (lanes 7–10) resulted in very little increase in glycosylation.
Figure 2.
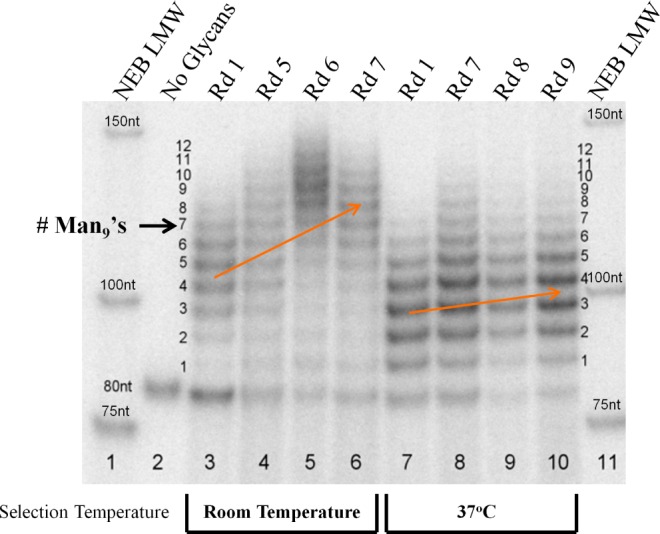
Trends in multivalency during selections at rt and 37 °C. Radiograph of 10% Denaturing PAGE of whole libraries (glycosylated with Man9) throughout selections at the two temperatures. Orange arrows depict the shift in average library multivalency throughout each selection.
After 7–9 rounds of selection, we obtained sequences of individual clones and found that the 37 °C library had mostly converged on a single highly related family (Table 1, entries 1–13). Confirming what was observed in the Figure 2 gel, these sequences contained only 3–5 glycosylation sites, contrasting with the average of ∼8 glycosylation sites resulting from the analogous room-temperature selection.9 Nitrocellulose filter binding assays10 showed that the sequences selected at 37 °C are very tightly recognized by 2G12, with Kd’s of 1.7–16 nM for all members of the major family, a vast improvement compared with the ∼300 nM Kd’s measured for the room temperature selection winners. This tight recognition is significant in that it is comparable to the strength of the natural interaction between 2G12 and the HIV envelope protein gp120.(8)Moreover, these glyco-DNAs are the first gp120 mimics to bind 2G12 tightly with a small number of Man9units, matching the number of glycan binding sites (3–4)4on 2G12. The only synthetic glycoclusters reported to exhibit similarly tight binding to 2G12 are Wong’s oligomannose dendrimers,5h but these required nine copies of Man9 (or 27 copies of Man4) to achieve Kd’s below 200 nM. By contrast, our clone 2 achieves a Kd of 1.7 nM with only 3 glycans and is, thus, in a qualitative sense likely a better gp120 mimic than the dendrimers.
Table 1. Sequences, Multivalency, and 2G12 Binding of Clones from 37 °C Selection.
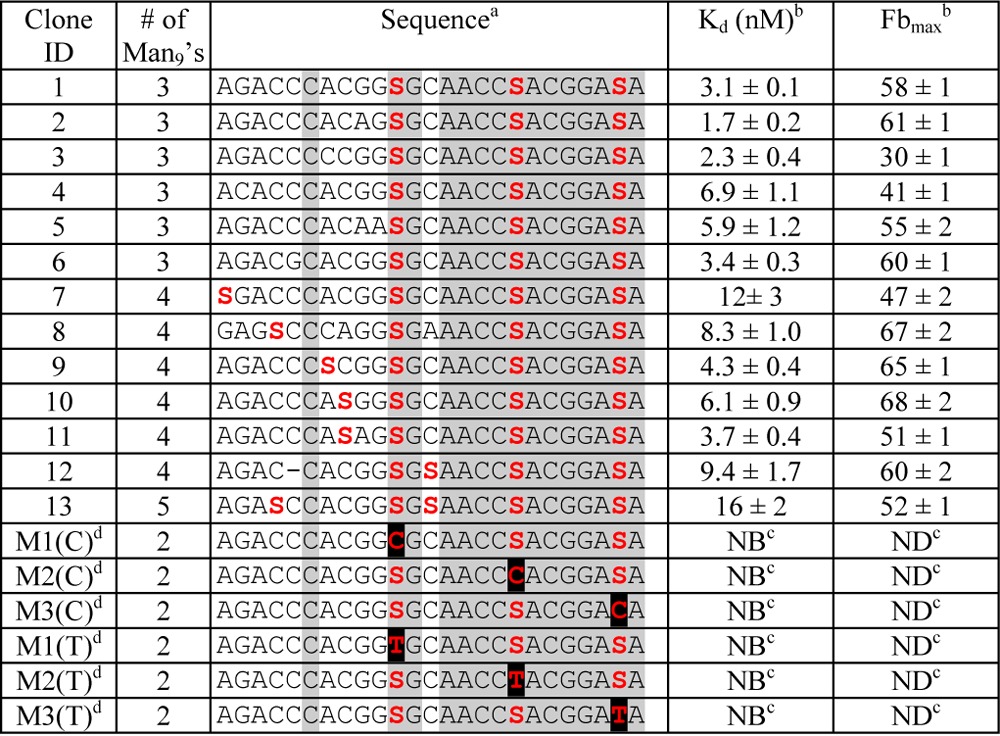
S = Man9-click-glycosylated EdU; gray shading indicates consensus sequence.
Kd and Fbmax determined by Nitrocellulose/PVDF filter binding assay; Fbmax expressed as percentages.
NB = no binding detected with up to 500 nM 2G12; ND = not determined.
Black highlights are positions of Man9 deletion.
We then ran several additional experiments to further delineate the binding interaction of selected clones with 2G12. A competition study (Figure 3) showed that the interaction of clone 1 with 150 nM 2G12 was greatly decreased in the presence of 200 nM gp120JR-FL, suggesting that binding to 2G12 occurs in its gp120 binding site. We also looked at the glycan dependence of 2G12 recognition. No binding was observed for clone 1 when glycans were not attached (SI Figure 4), indicating that the nucleic acid component alone was not responsible for the binding. Deletion of any of the three glycans by replacing an alkynyl base with cytosine (EdU→C) resulted in complete loss of binding (Table 1, M1–3(C)). To rule out the possibility that this loss of binding might be due to a change in pairing of the underlying base, we also prepared EdU→T mutants (Table 1, M1–3(T)). Again, none of the three mutants showed any binding to 2G12, further supporting the importance of all three glycans for 2G12 recognition.
Figure 3.
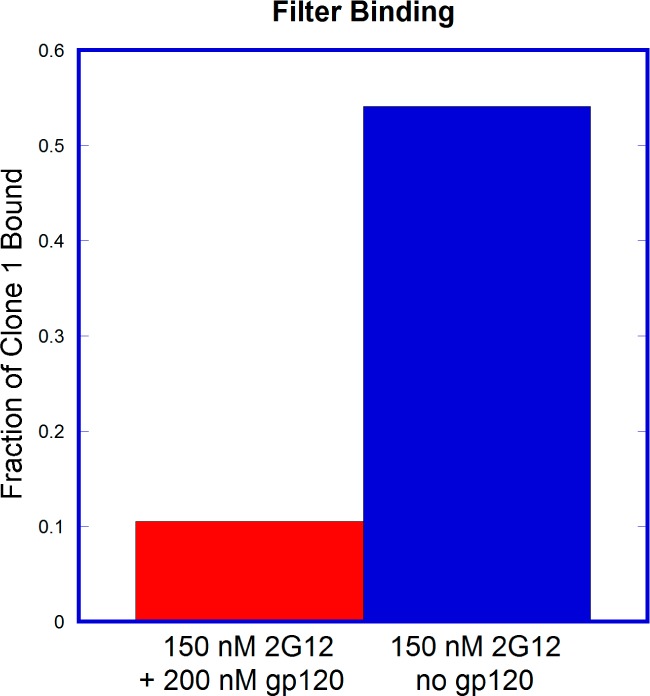
gp120 and clone 1 compete for binding to 2G12.
To determine whether our glycoclusters were good mimics of gp120 in terms of the kinetics of their interaction with 2G12, we also examined the binding of clone 1 to 2G12 in real time via biolayer interferometry (BLI).11 Clone 1, modified with a 5′-(A)5 spacer and biotin tag, was immobilized on a streptavidin sensor, and 2G12 was associated to the surface at several concentrations, followed by dissociation in blank buffer (Figure 4). The resulting response curves were fit globally to a 1:1 binding model and afforded rate constants of kon = 2.5 × 104 M–1 s–1 and koff = 3.1 × 10–4 s–1, which are both similar to values reported for the gp120–2G12 interaction (kon = 7 × 104 M–1 s–1, koff = 4 × 10–4 s–1). Our measured kon/koff rates correspond to a Kd of 12 nM, which is in reasonable agreement with the results of the nitrocellulose filter binding assay and also close to the Kd’s of 6–9 nM reported from studies of the 2G12–gp120 interaction.8 Interestingly, a mutant antibody 2G12 I19R12 showed no binding to clone 1 (SI Figure 8). This mutant differs from wt2G12 in that it lacks the domain exchanged structure, so that carbohydrate-binding sites are separated by a much greater distance. These data further support our hypothesis that the strong 2G12–clone 1 interaction is due to good matching of the glycan spacing with binding sites in 2G12.
Figure 4.
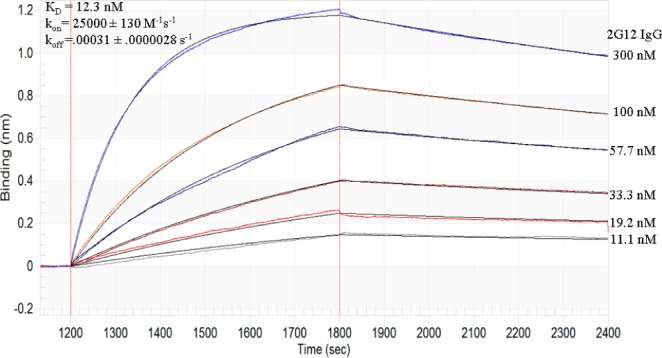
Biolayer interferometry (BLItz) kinetic binding sensorgrams for association/dissociation of 2G12 to clone 1 glycoDNA. Biotin-labeled clone 1 was immobilized on streptavidin biosensors, and sensorgrams were fit globally to a 1:1 binding model (see SI Figure 6).
In summary, we have shown that SELMA-based glycocluster selection with the temperature increased to 37 °C affords low-valent Man9 clusters whose affinity for 2G12 matches that of gp120 both thermodynamically and kinetically. From a standpoint of understanding multivalency,13 it is very interesting that that 37 °C selection winners are not only of higher affinity (1.7–16 nM vs 150–500 nM) but also contain fewer glycans than room temperature selection winners (3–5 vs 7–10). We speculate that flexibly linked, highly multivalent, moderate affinity binders must be so common in the starting library that, at low temperature, they overwhelm the very rare, high-affinity, low-valent, but rigidly linked binders. As the affinity of the high-valent binders is primarily due to statistical rebinding, they may pay a much greater entropic penalty for binding at higher temperatures, compared with low-valent, rigid binders. Consistent with this, we found that the 2G12 binding of some room-temperature selection winners became undetectable in the nitrocellulose assay at 37 °C (data not shown), whereas most binding was retained for clone 1 (SI Figure 6). Further investigation of this temperature effect, as well as structural and immunological investigation of the selection winners, will be reported in due course.
Acknowledgments
I.J.K. gratefully acknowledges Brandeis University and the NIH (R01 AI090745). We are grateful to Polymun Scientific for a gift of mAb 2G12 and to Prof. Dennis Burton for a gift of 2G12 I19R mutant.
Supporting Information Available
Full sequence data for selection winners, preparation of mutant clones, binding data, and control experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Trkola A.; Purtscher M.; Muster T.; Ballaun C.; Buchacher A.; Sullivan N.; Srinivasan K.; Sodroski J.; Moore J. P.; Katinger H. J. Virol. 1996, 70, 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley J. M.; Wrin T.; Korber B.; Zwick M. B.; Wang M.; Chappey C.; Stiegler G.; Kunert R.; Zolla-Pazner S.; Katinger H.; Petropoulos C. J.; Burton D. R. J. Virol. 2004, 78, 13232–13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Mascola J. R.; Stiegler G.; VanCott T. C.; Katinger H.; Carpenter C. B.; Hanson C. E.; Beary H.; Hayes D.; Frankel S. S.; Birx D. L.; Lewis M. G. Nat. Med. 2000, 6, 207–210. [DOI] [PubMed] [Google Scholar]; b Hessell A. J.; Rakasz E. G.; Poignard P.; Hangartner L.; Landucci G.; Forthal D. N.; Koff W. C.; Watkins D. I.; Burton D. R. PLoS Pathog 2009, 5, e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Sanders R. W.; Venturi M.; Schiffner L.; Kalyanaraman R.; Katinger H.; Lloyd K. O.; Kwong P. D.; Moore J. P. J. Virol. 2002, 76, 7293–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Scanlan C. N.; Pantophlet R.; Wormald M. R.; Ollmann Saphire E.; Stanfield R.; Wilson I. A.; Katinger H.; Dwek R. A.; Rudd P. M.; Burton D. R. J. Virol. 2002, 76, 7306–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Calarese D. A.; Scanlan C. N.; Zwick M. B.; Deechongkit S.; Mimura Y.; Kunert R.; Zhu P.; Wormald M. R.; Stanfield R. L.; Roux K. H.; Kelly J. W.; Rudd P. M.; Dwek R. A.; Katinger H.; Burton D. R.; Wilson I. A. Science 2003, 300, 2065–2071. [DOI] [PubMed] [Google Scholar]
- a Li H. G.; Wang L. X. Org. Biomol. Chem. 2004, 2, 483–488. [DOI] [PubMed] [Google Scholar]; b Wang L. X.; Ni J.; Singh S.; Li H. Chem. Biol. 2004, 11, 127–134. [DOI] [PubMed] [Google Scholar]; c Ni J. H.; Song H. J.; Wang Y. D.; Stamatos N. M.; Wang L. X. Bioconjugate Chem. 2006, 17, 493–500. [DOI] [PubMed] [Google Scholar]; d Krauss I. J.; Joyce J. G.; Finnefrock A. C.; Song H. C.; Dudkin V. Y.; Geng X.; Warren J. D.; Chastain M.; Shiver J. W.; Danishefsky S. J. J. Am. Chem. Soc. 2007, 129, 11042–11044. [DOI] [PubMed] [Google Scholar]; e Wang J.; Li H.; Zou G.; Wang L. X. Org. Biomol. Chem. 2007, 5, 1529–1540. [DOI] [PubMed] [Google Scholar]; f Joyce J. G.; Krauss I. J.; Song H. C.; Opalka D. W.; Grimm K. M.; Nahas D. D.; Esser M. T.; Hrin R.; Feng M. Z.; Dudkin V. Y.; Chastain M.; Shiver J. W.; Danishefsky S. J. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 15684–15689. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Luallen R. J.; Lin J. Q.; Fu H.; Cai K. K.; Agrawal C.; Mboudjeka I.; Lee F. H.; Montefiori D.; Smith D. F.; Doms R. W.; Geng Y. J. Virol. 2008, 82, 6447–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Wang S. K.; Liang P. H.; Astronomo R. D.; Hsu T. L.; Hsieh S. L.; Burton D. R.; Wong C. H. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 3690–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]; i Gorska K.; Huang K.-T.; Chaloin O.; Winssinger N. Angew. Chem., Int. Ed. 2009, 48, 7695–7700. [DOI] [PubMed] [Google Scholar]; j Astronomo R. D.; Kaltgrad E.; Udit A. K.; Wang S. K.; Doores K. J.; Huang C. Y.; Pantophlet R.; Paulson J. C.; Wong C. H.; Finn M. G.; Burton D. R. Chem. Biol. 2010, 17, 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]; k Doores K. J.; Fulton Z.; Hong V.; Patel M. K.; Scanlan C. N.; Wormald M. R.; Finn M. G.; Burton D. R.; Wilson I. A.; Davis B. G. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 17107–17112. [DOI] [PMC free article] [PubMed] [Google Scholar]; l Agrawal-Gamse C.; Luallen R. J.; Liu B.; Fu H.; Lee F.-H.; Geng Y.; Doms R. W. J. Virol. 2011, 85, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]; m Ciobanu M.; Huang K. T.; Daguer J. P.; Barluenga S.; Chaloin O.; Schaeffer E.; Mueller C. G.; Mitchell D. A.; Winssinger N. Chem. Commun. 2011, 47, 9321–9323. [DOI] [PubMed] [Google Scholar]; n Marradi M.; Di Gianvincenzo P.; Enriquez-Navas P. M.; Martinez-Avila O. M.; Chiodo F.; Yuste E.; Angulo J.; Penades S. J. Mol. Biol. 2011, 410, 798–810. [DOI] [PubMed] [Google Scholar]; o Clark B. E.; Auyeung K.; Fregolino E.; Parrilli M.; Lanzetta R.; De Castro C.; Pantophlet R. Chem. Biol. 2012, 19, 254–263. [DOI] [PubMed] [Google Scholar]
- a MacPherson I. S.; Temme J. S.; Habeshian S.; Felczak K.; Pankiewicz K.; Hedstrom L.; Krauss I. J. Angew. Chem., Int. Ed. 2011, 50, 11238–11242. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Temme J. S.; Drzyzga M. G.; MacPherson I. S.; Krauss I. J. Chem.—Eur. J 2013, 19, 17291–17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kolb H. C.; Finn M. G.; Sharpless K. B. Angew. Chem., Int. Ed. 2001, 40, 2004–2021. [DOI] [PubMed] [Google Scholar]; b Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. Angew. Chem., Int. Ed. 2002, 41, 2596–2599. [DOI] [PubMed] [Google Scholar]; c Gierlich J.; Burley G. A.; Gramlich P. M.; Hammond D. M.; Carell T. Org. Lett. 2006, 8, 3639–3642. [DOI] [PubMed] [Google Scholar]; d Gierlich J.; Gutsmiedl K.; Gramlich P. M.; Schmidt A.; Burley G. A.; Carell T. Chem.—Eur. J. 2007, 13, 9486–9494. [DOI] [PubMed] [Google Scholar]
- Hoorelbeke B.; van Montfort T.; Xue J.; LiWang P. J.; Tanaka H.; Igarashi Y.; Van Dammef E. J. M.; Sanders R. W.; Balzarini J. FEBS Lett. 2013, 587, 860–866. [DOI] [PubMed] [Google Scholar]
- For complete results of the room temperature selection, see ref (6b).
- Rio D. C.Cold Spring Harbor Protocols2012, 2012, pdb.prot071449. [DOI] [PubMed]
- Abdiche Y.; Malashock D.; Pinkerton A.; Pons J. Anal. Biochem. 2008, 377, 209–217. [DOI] [PubMed] [Google Scholar]
- a Huber M.; Le K. M.; Doores K. J.; Fulton Z.; Stanfield R. L.; Wilson I. A.; Burton D. R. J. Virol. 2010, 84, 10700–10707. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Doores K. J.; Huber M.; Le K. M.; Wang S. K.; Doyle-Cooper C.; Cooper A.; Pantophlet R.; Wong C. H.; Nemazee D.; Burton D. R. J. Virol. 2013, 87, 2234–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selected reviews about multivalency:; a Mammen M.; Choi S.-K.; Whitesides G. M. Angew. Chem., Int. Ed. 1998, 37, 2754–2794. [DOI] [PubMed] [Google Scholar]; b Kiessling L. L.; Gestwicki J. E.; Strong L. E. Angew. Chem., Int. Ed. 2006, 45, 2348–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Fasting C.; Schalley C. A.; Weber M.; Seitz O.; Hecht S.; Koksch B.; Dernedde J.; Graf C.; Knapp E. W.; Haag R. Angew. Chem., Int. Ed. 2012, 51, 10472–10498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


