Abstract

Diphthamide, the target of diphtheria toxin, is a unique posttranslational modification on translation elongation factor 2 (EF2) in archaea and eukaryotes. The biosynthesis of diphthamide was proposed to involve three steps. The first step is the transfer of the 3-amino-3-carboxypropyl group from S-adenosyl-l-methionine (SAM) to the histidine residue of EF2, forming a C–C bond. Previous genetic studies showed this step requires four proteins in eukaryotes, Dph1–Dph4. However, the exact molecular functions for the four proteins are unknown. Previous study showed that Pyrococcus horikoshii Dph2 (PhDph2), a novel iron-sulfur cluster-containing enzyme, forms a homodimer and is sufficient for the first step of diphthamide biosynthesis in vitro. Here we demonstrate by in vitro reconstitution that yeast Dph1 and Dph2 form a complex (Dph1-Dph2) that is equivalent to the homodimer of PhDph2 and is sufficient to catalyze the first step in vitro in the presence of dithionite as the reductant. We further demonstrate that yeast Dph3 (also known as KTI11), a CSL-type zinc finger protein, can bind iron and in the reduced state can serve as an electron donor to reduce the Fe-S cluster in Dph1-Dph2. Our study thus firmly establishes the functions for three of the proteins involved in eukaryotic diphthamide biosynthesis. For most radical SAM enzymes in bacteria, flavodoxins and flavodoxin reductases are believed to serve as electron donors for the Fe-S clusters. The finding that Dph3 is an electron donor for the Fe-S clusters in Dph1-Dph2 is thus interesting and opens up new avenues of research on electron transfer to Fe-S proteins in eukaryotic cells.
Diphthamide is a unique posttranslationally modified histidine residue in archaeal and eukaryotic translation elongation factor 2 (EF2), a protein required for ribosomal protein synthesis.1−4 Diphthamide is the target of Diphtheria toxin (DT) and Pseudomonas exotoxin A, which ADP-ribosylate diphthamide and inhibit ribosomal protein synthesis.5 Diphthamide modification is conserved in all eukaryotes and archaea. Several reports suggested that diphthamide helps to maintain the translation fidelity in yeast6 and mouse7 by preventing −1 frameshift. Previous studies have led to a three-step biosynthetic pathway for diphthamide (Scheme 1).8−11 The first step is the transfer of a 3-amino-3-carboxypropyl (ACP) group from S-adenosyl methionine (SAM) to the C2 carbon of the imidazole ring of the histidine residue in EF2 (Scheme 1). This step requires four proteins in eukaryotes, Dph1–Dph4. This is then followed by a trimethylation step (catalyzed by Dph5) and an amidation step (catalyzed by Dph612,13 and Dph714) to give diphthamide.
Scheme 1. Diphthamide Biosynthesis Pathway.
The first step of diphthamide biosynthesis is particularly interesting as the enzymatic reaction mechanism and the molecular function of each of the four proteins are still not well understood. We previously showed that Pyrococcus horikoshii Dph2 (PhDph2) is sufficient for the first step of diphthamide biosynthesis in vitro.15 PhDph2 is homologous to eukaryotic Dph1 and Dph2 (Dph1 and Dph2 share about 20% sequence identity), but no Dph3 or Dph4 homologues can be found in archaea by BLAST search. PhDph2 is a novel [4Fe-4S] cluster-containing radical SAM enzyme. Different from traditional radical SAM enzymes, existing evidence suggests that it cleaves a Cγ,Met–S bond in SAM and generates a 3-amino-3-carboxypropyl (ACP) radical.15 PhDph2 is a homodimer, and each monomer contains a [4Fe-4S] cluster. Previous report suggests that only one [4Fe-4S] cluster in this homodimer is needed for the activity in vitro.16
The studies on PhDph2 have begun to shed light on the molecular functions of eukaryotic Dph1–4. Specifically, we proposed that Dph1 and Dph2 form a heterodimer (Dph1-Dph2), which is equivalent to the PhDph2 homodimer and is the catalyst required for the C–C bond formation reaction. Dph3 and Dph4 may serve to assemble the Fe-S cluster on Dph1-Dph2 or to keep the Fe-S cluster in Dph1-Dph2 in a reduced state. Here we report the in vitro reconstitution of the first step of yeast diphthamide biosynthesis using purified recombinant Dph1 and Dph2. This reconstituted system provides the first direct experimental evidence supporting that the Dph1-Dph2 complex is the enzyme for the first step of diphthamide biosynthesis. This reconstituted system also allows us to directly test whether Dph3 or Dph4 can serve as an electron donor to reduce the Fe-S cluster in Dph1-Dph2. Our evidence suggests that Dph3, but not Dph4, is the electron carrier for Dph1-Dph2.
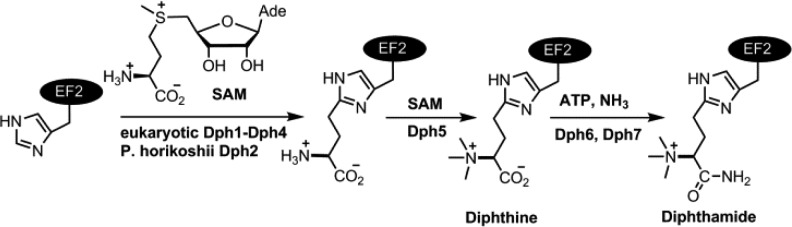
Attempts to overexpress yeast Dph1 and Dph2 separately in Escherichia coli were unsuccessful, yielding insoluble proteins. However, we readily obtained soluble Dph1 and Dph2 when we co-expressed them in E. coli, suggesting that they form a complex. To increase the Fe-S cluster loading, Dph1-Dph2 was coexpressed with the ISC (iron sulfur cluster) proteins, which are a set of bacterial proteins that assemble Fe-S clusters and transfer them to client proteins.17 Similar to PhDph2, anaerobically purified yeast Dph1-Dph2 complex displayed a dark-brown color and showed a broad absorption band at 410 nm, typical for a [4Fe-4S]2+ cluster. The 410 nm absorption decreased upon reduction with dithionite (Figure 1A). Quantification based on the 410 nm absorption suggested the presence of 0.6 [4Fe-4S]2+ clusters per Dph1-Dph2 complex. The EPR spectrum of the reduced Dph1-Dph2 complex showed g values of 2.04 and 1.93, consistent with a [4Fe-4S]+ cluster (Figure 1B). Size exclusion chromatography-multiangle light scattering (SEC-MALS) experiment of the Dph1-Dph2 complex showed that the molecular weight is ∼110 kDa (Figure S1), suggesting that Dph1 and Dph2 form a heterodimer.
Figure 1.
Spectroscopic characterization of the [4Fe-4S] cluster in Dph1-Dph2 heterodimer. (A) UV–vis absorption spectra of anaerobically isolated (purple) and dithionite-reduced (red) Dph1-Dph2. (B) X-band EPR spectra of dithionite reduced Dph1-Dph2 at 12 K.
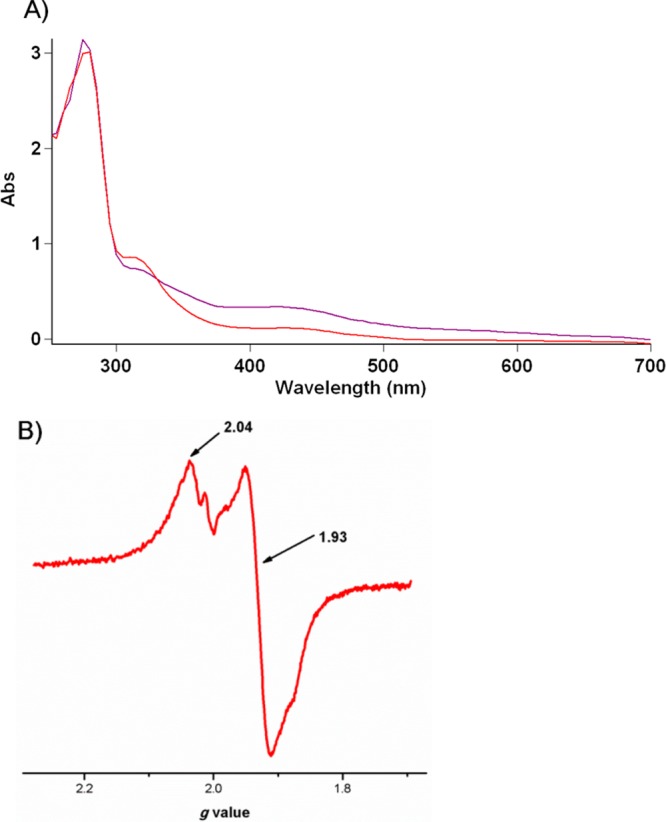
We next tested whether Dph1-Dph2 heterodimer is sufficient to catalyze the first step of diphthamide biosynthesis in vitro with dithionite as the reductant. We first tested whether Dph1-Dph2 could cleave SAM to generate 5′-deoxy-5′-methylthioadenosine (MTA) using high-performance liquid chromatography (HPLC). In the reaction containing Dph1-Dph2, SAM, dithionite, with or without EF2, SAM was cleaved and MTA was produced (Figure 2A). In control reactions without Dph1-Dph2 or dithionite, only background level of MTA was detected. This result showed that the Dph1-Dph2 complex was active and could cleave SAM, similar to PhDph2.15 To reconstitute the full reaction of the first step, we used carboxy-14C-labeled SAM (14C-SAM) to examine whether EF2 can be labeled by 14C. Indeed, EF2 was labeled in the reaction containing Dph1-Dph2, 14C-SAM, and dithionite (Figure 2B lane 2). As a negative control, EF2 could not be labeled in the reaction without dithionite (Figure 2B lane 3). These results suggested that Dph1-Dph2 complex is sufficient for the first step of diphthamide biosynthesis in vitro with dithionite as the reductant.
Figure 2.
Reconstitution of Dph1-Dph2 activity with dithionite as the reductant. (A)HPLC analysis showing that reduced Dph1-Dph2 was sufficient in the SAM cleavage reaction, generating MTA with or without EF2. SAM was not cleaved without Dph1-Dph2 or dithionite. (B) Activity assay using carboxy 14C-SAM. Left panel, Coomassie blue-stained gel; right panel, the autoradiography. Lane 1: protein standard; Lane 2: reaction containing Dph1-Dph2, SAM, and dithionite; Lane 3: negative control containing Dph1-Dph2 and SAM but without dithionite.
With this fully active Dph1-Dph2, we next tested whether Dph3 or Dph4 could serve as an electron carrier to reduce Dph1-Dph2. Eukaryotic Dph3 and Dph4 belong to the protein family PF05207, which share a conserved region encoding a CSL-type zinc finger. The CSL zinc finger was thought to use four conserved Cys residues to coordinate Zn2+,18 and the last Cys residue is followed by a Ser and a Leu residue (hence the name CSL zinc finger). Dph3 is a small protein (82 aa) that essentially just contains the CSL zinc finger, while Dph4 contains a DnaJ domain in addition to the zinc finger domain.
In the CSL zinc finger family, Dph3 is relativily well studied. It was first identified as the Kluyveromyces lactis killer toxin zymocin insensitive 11 (Kti11) gene from Saccharomyces cerevisiae.19 Later the Kti11 gene was found to be identical to Dph3. Further studies showed Dph3/Kti11 is involved in multiple biological processes and interacts with different proteins,20 such as Elongator subunits (Elp1–Elp3, Elp5),20,21 EF2, Dph1, and Dph2.21 Multiple data suggested that Dph1–Dph3 may form a multimeric complex.11,20,22 The solution structure of yeast Dph3, purified from E. coli grown on minimal medium M9 supplemented with ZnSO4, was solved by NMR.18 A single Zn2+ ion is bound by four conserved cysteine residues of Dph3. Yakunin and co-workers demonstrated that yeast Dph3 binds both zinc and iron when prepared from E. coli cells grown on rich medium.23 Mutagenasis and deletion studies showed that the Cys25, Cys27, Cys47, and Cys50 are responsible for metal coordination.
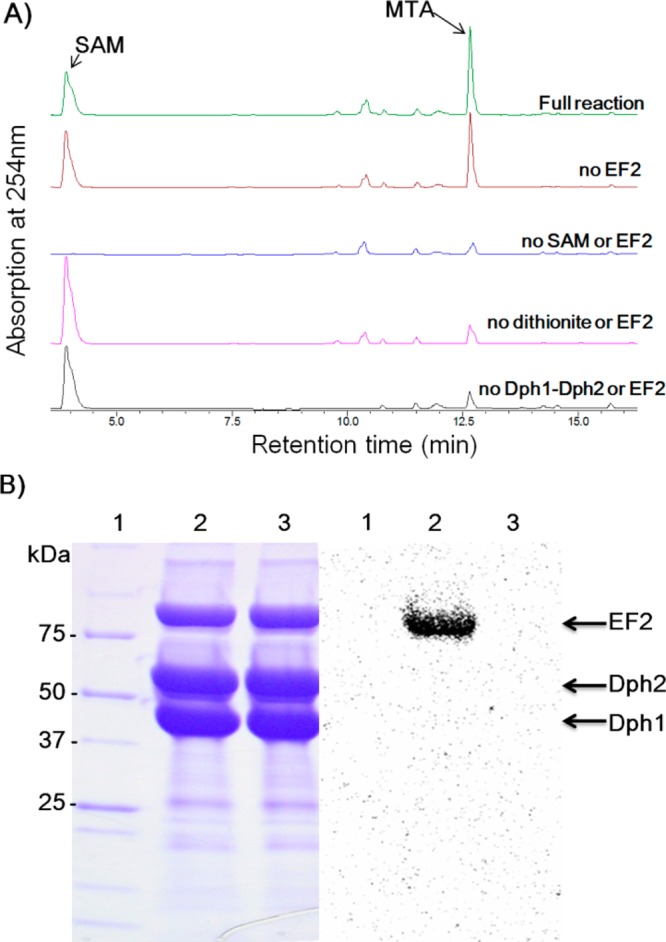
We expressed and purified yeast Dph3 in E. coli. Dph3 purified from E. coli in LB medium was purple, suggesting that it binds iron, consistent with previous report.23 The UV–vis spectrum showed two major peaks at 350 and 490 nm, with a shoulder at 570 nm (Figure 3A). When reduced by dithionite, the three peaks disappeared. After exposing the sample to air for 10 min, the three peaks came back. These results showed that Dph3 is redox active. In contrast, purified Dph4 (Figure S2) was colorless, even when we supplemented the growth medium with 200 μM FeCl3. These observations made us believe that Dph3, but not Dph4, is the electron carrier for Dph1-Dph2. Therefore, in later studies, we focused on testing Dph3.
Figure 3.
Dph3 binds iron and is redox active. (A) UV–vis absorption spectrum of Dph3; Dph3 reduced by dithionite and reoxidized by air. (B) Dph3 can be reduced by NorW and NADH (monitored at 488 nm).
It was reported that Dph3 can be reduced by E. coli rubredoxin reductase NorW in the presence of NADH and is redox active to cytochrome C from horse heart.23 We expressed and purified NorW (Figure S2), a bright yellow flavoprotein. Consistent with the previous report, NorW could reduce Dph3 (Figure 3B, monitored at 488 nm). The reduced Dph3 was slowly oxidized in air but was reduced when additional NADH was added. Therefore, NADH and NorW are a suitable reduction system that can be used to generate reduced Dph3.
With the system to generate the reduced form of Dph3, we then tested whether Dph3 could serve as an electron donor to reduce Dph1-Dph2 and enable Dph1-Dph2 to catalyze the first step of diphthamide biosynthesis in the absence of dithionite. Using 14C-SAM, EF2 was successfully labeled in the reaction containing Dph1-Dph2, Dph3/NorW/NADH (Figure 4, lane 4). The result was similar with the positive control when dithionite was used as the reductant (lane 2). NorW and NADH were both required for the reduction of Dph3, and EF2 was not labeled when any one of them was omitted in the reaction (Figure 4, lanes 5 and 6). It was reported that Dph3 can also be isolated in a Zn2+-bound form when expressed in M9 minimal medium supplemented with Zn2+. We also obtained the colorless Zn2+-bound Dph3 and found that it cannot serve as the electron donor for Dph1-Dph2 (Figure 4, lane 7). Our results thus demonstrated that the iron-bound form of Dph3 can serve as an electron carrier for Dph1-Dph2. To investigate the specificity of Dph3, we similarly tested the electron carrier activity of Dph3 on PhDph2 using 14C-SAM (Figure S3). PhDph2 could be reduced and PhEF2 could be labeled with dithionite as the reductant, but not with Dph3/NorW/NADH as the reductant.
Figure 4.
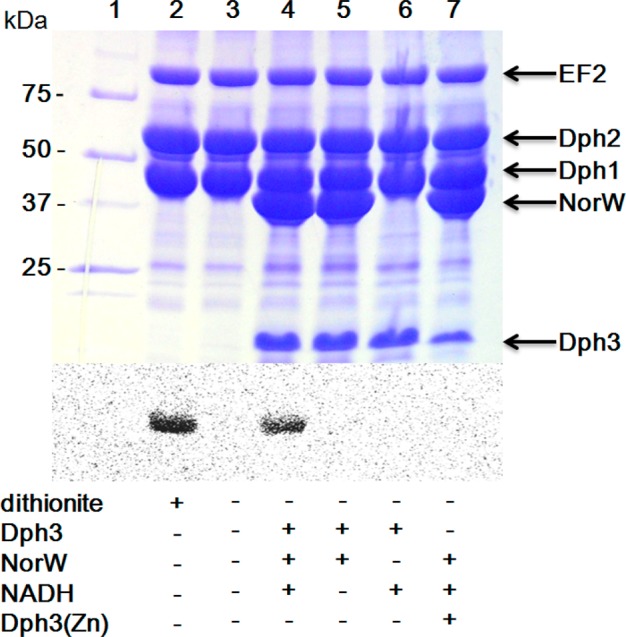
In vitro reconstitution of Dph1–Dph3 activity using carboxy-14C-SAM. The top panel shows the Coomassie-blue-stained gel; the bottom panel shows the autoradiography. All the reactions in lanes 2–7 contained EF2 and Dph1-Dph2. The presence of other reagents is indicated below each lane.
In summary, our data demonstrate that Dph1-Dph2 complex is the enzyme that catalyzes the first step of diphthamide biosynthesis in eukaryotic cells. Dph3 is an electron donor that reduces the [4Fe-4S] cluster in Dph1-Dph2. Although previous work suggested that Dph4 could behave similarly to Dph3,24 our studies suggest that Dph4 does not readily bind iron and thus is unlikely to be an electron donor for Dph1-Dph2. Thus the exact molecular function of Dph4 awaits further studies.
The discovery that Dph3 is an electron donor in diphthamide biosynthesis is the first time that the physiological role of a member from the CSL zinc finger family is convincingly demonstrated, paving the road for understanding the function of this protein family. In the Pfam database, the CSL type zinc finger family (PF05207) is listed with 560 sequences from 303 species, but no definite function has been assigned to CSL zinc fingers. In the 560 sequences, 307 sequences have only the CSL zinc finger domain like Dph3. Among the 303 species that contain CSL zinc finger proteins, there are 296 eukaryotes and 7 bacteria. As diphthamide modification only exits in archaea and eukaryotes, proteins in this family must have functions other than diphthamide biosynthesis. Knowing the electron-donor activity of Dph3 will help to understand the functions of other CSL zinc finger proteins.
Radical SAM enzymes require electron donors to keep the [4Fe-4S] clusters in the active and reduced state. In vitro, almost all radical SAM enzyme reactions were reconstituted using artificial electron donors, such as dithionite or illuminated deazaflavin. In E. coli, the flavodoxin/flavodoxin reductase/NADPH25 reducing system is shown to be the electron donor for radical SAM enzymes, which is also used in vitro for reduction of [4Fe-4S] in radical SAM enzymes such as lysine 2,3-aminomutase,26 DesII27 and LipA.28 However, very little is known about electron carriers for Fe-S cluster-containing enzymes in eukaryotes. The identification of Dph3 as an electron carrier thus will likely enable other studies in this direction. Interestingly, Dph3 is also required for tRNA modifications at the wobble position that involves another radical SAM enzyme, Elp3.29 Dph3 may also pass electrons to reduce Elp3. In our resonstitution experiment, an E. coli rubredoxin reductase NorW was used to reduce Dph3. The identity of the endogenous reductase for Dph3 is unknown. The discovery of Dph3′s activity will facilitate the discovery of the reductase and thus provide further understanding about the electron-transfer pathways for Fe-S enzymes in eukaryotes.
Acknowledgments
This work is supported by NIH (R01GM088276) and NIH/NIGMS P41-GM103521. H.L. and M.D. thank Prof. Steven Ealick for the use of the anaerobic chamber, Dr. Laura J. Byrnes for assistance with SEC-MALS experiment, and Jinzhao Ji for participation of protein purification.
Supporting Information Available
Experimental materials, methods, and figures. This material is available free of charge via the Internet at http://pubs.acs.org
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Robinson E. A.; Henriksen O.; Maxwell E. S. J. Biol. Chem. 1974, 249, 5088. [PubMed] [Google Scholar]
- Van Ness B. G.; Howard J. B.; Bodley J. W. J. Biol. Chem. 1980, 255, 10710. [PubMed] [Google Scholar]
- Van Ness B. G.; Howard J. B.; Bodley J. W. J. Biol. Chem. 1980, 255, 10717. [PubMed] [Google Scholar]
- Pappenheimer A. M. Jr.; Dunlop P. C.; Adolph K. W.; Bodley J. W. J. Bacteriol. 1983, 153, 1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier R. J. Toxicon 2001, 39, 1793. [DOI] [PubMed] [Google Scholar]
- Ortiz P. A.; Ulloque R.; Kihara G. K.; Zheng H.; Kinzy T. G. J. Biol. Chem. 2006, 281, 32639. [DOI] [PubMed] [Google Scholar]
- Liu S.; Bachran C.; Gupta P.; Miller-Randolph S.; Wang H.; Crown D.; Zhang Y.; Wein A. N.; Singh R.; Fattah R.; Leppla S. H. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring J. M.; Moehring T. J.; Danley D. E. Proc. Natl. Acad. Sci. U.S.A. 1980, 77, 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring T. J.; Danley D. E.; Moehring J. M. Mol. Cell. Biol. 1984, 4, 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Leppla S. H. Mol. Cell 2003, 12, 603. [DOI] [PubMed] [Google Scholar]
- Liu S.; Milne G. T.; Kuremsky J. G.; Fink G. R.; Leppla S. H. Mol. Cell. Biol. 2004, 24, 9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.; Lin Z.; Chen W.; Jiang H.; Zhang S.; Lin H. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 19983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uthman S.; Bar C.; Scheidt V.; Liu S.; Ten Have S.; Giorgini F.; Stark M. J.; Schaffrath R. PLoS Genet. 2013, 9, e1003334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.; Chen W.; Lee W.; Jiang H.; Zhang S.; Lin H. J. Am. Chem. Soc. 2012, 134, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Zhu X.; Torelli A. T.; Lee M.; Dzikovski B.; Koralewski R. M.; Wang E.; Freed J.; Krebs C.; Ealick S. E.; Lin H. Nature 2010, 465, 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; Dzikovski B.; Su X.; Torelli A. T.; Zhang Y.; Ealick S. E.; Freed J. H.; Lin H. Mol. Biosyst 2011, 7, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazzon J.; Dean D. R. Curr. Opin. Chem. Biol. 2003, 7, 166. [DOI] [PubMed] [Google Scholar]
- Sun J.; Zhang J.; Wu F.; Xu C.; Li S.; Zhao W.; Wu Z.; Wu J.; Zhou C.-Z.; Shi Y. Biochemistry 2005, 44, 8801. [DOI] [PubMed] [Google Scholar]
- Fichtner L.; Schaffrath R. Mol. Microbiol. 2002, 44, 865. [DOI] [PubMed] [Google Scholar]
- Bar C.; Zabel R.; Liu S.; Stark M. J.; Schaffrath R. Mol. Microbiol. 2008, 69, 1221. [DOI] [PubMed] [Google Scholar]
- Fichtner L.; Jablonowski D.; Schierhorn A.; Kitamoto H. K.; Stark M. J.; Schaffrath R. Mol. Microbiol. 2003, 49, 1297. [DOI] [PubMed] [Google Scholar]
- Abdel-Fattah W.; Scheidt V.; Uthman S.; Stark M. J.; Schaffrath R. Toxins 2013, 5, 958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot M.; Sanders S. A.; Singer A.; Zhang R.; Brown G.; Binkowski A.; Xu L.; Lukin J. A.; Murzin A. G.; Joachimiak A.; Arrowsmith C. H.; Edwards A. M.; Savchenko A. V.; Yakunin A. F. J. Mol. Biol. 2008, 375, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A.; Chitoor B.; Goswami A. V.; Pareek G.; Atreya H. S.; D’Silva P. J. Biol. Chem. 2012, 287, 13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi V.; Eliasson R.; Fontecave M.; Mulliez E.; Hoover D. M.; Matthews R. G.; Reichard P. Biochem. Biophys. Res. Commun. 1993, 197, 792. [DOI] [PubMed] [Google Scholar]
- Brazeau B. J.; Gort S. J.; Jessen H. J.; Andrew A. J.; Liao H. H. Appl. Environ. Microbiol. 2006, 72, 6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szu P. H.; Ruszczycky M. W.; Choi S. H.; Yan F.; Liu H. W. J. Am. Chem. Soc. 2009, 131, 14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchillo R. M.; Iwig D. F.; Jones A. D.; Nesbitt N. M.; Baleanu-Gogonea C.; Souder M. G.; Tu L.; Booker S. J. Biochemistry 2004, 43, 6378. [DOI] [PubMed] [Google Scholar]
- Huang B. O.; Johansson M. J. O.; Bystrom A. S. RNA 2005, 11, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


